Abstract
Accumulation of mitochondrial electron transport chain (ETC) defects is a recognized hallmark of the age-associated decline in cardiac bioenergetics; however, the molecular events involved are only poorly understood. In the present work, we hypothesized that age-related ETC deterioration stemmed partly from disassociation of large solid-state macromolecular assemblies termed “supercomplexes”. Mitochondrial proteins from young and old rat hearts were separated by Blue Native-PAGE, protein bands analyzed by LC-MALDI-MS/MS, and protein levels quantified by densitometry. Results showed that supercomplexes comprised of various stoichiometries of complexes I, III and IV were observed, and declined significantly (p < 0.05, n = 4) with age. Supercomplexes displaying the highest molecular masses were the most severely affected. Considering that certain diseases (e.g. Barth Syndrome) display similar supercomplex destabilization as our results for aging, the deterioration in ETC supercomplexes may be an important underlying factor for both impaired mitochondrial function and loss of cardiac bioenergetics with age.
Keywords: aging, rat heart mitochondria, electron transport supercomplexes, blue native-polyacrylamide gel electrophoresis
INTRODUCTION
Mitochondrial decay has been implicated as one of the principal underlying factors of aging [1-6]. Both significant and subtle alterations in mitochondrial ultrastructure have been observed which correlate with lower electron transport efficiency, increased superoxide appearance, and oxidative damage [7-9]. Though certainly multifactorial in nature, it is clear that electron transport chain defects are part of the age-associated loss of mitochondrial function [10-14]. Activities of complexes I, III, and IV reportedly decline with age [3, 12-15] and many acute mitochondrial-derived diseases show similar, if not more severe, electron transport chain deficits than seen in aging [16-21]. Thus a central research focus has been to understand the cellular and molecular processes that ultimately lead to electron transport chain defects in mitochondria from aging tissues.
Four large protein complexes comprise the electron transport chain (ETC), which along with the F1FO-ATP synthase (complex V), constitute the machinery for converting metabolic energy transiently stored as reduced coenzymes into ATP. Until recently, the prevailing view was that the components of the ETC were distinct entities where rapid, random collisions allowed electron transfer between complexes [22]. However, with the advent of blue native-PAGE (BN-PAGE) technology, there is a growing awareness that the individual components of the ETC may actually exist as large macromolecular assemblies, or so-called supercomplexes [23-25]. Several biochemical and biophysical lines of evidence support the existence of these supramolecular assemblies [25-30], including a proposed three-dimensional structure for a macromolecule composed of complexes I, III and IV with a stoichiometry of 1:2:1, respectively [31]. Although physical evidence for mitochondrial supercomplexes now exists by a variety of techniques outside of BN-PAGE, the ramifications of such large protein supercomplexes on ETC function are not completely understood. In this regard, there have been no studies critically examining age-associated changes to mitochondrial supercomplex levels or alterations to their structural composition. Moreover, there is a paucity of knowledge as to the role that supercomplexes play in the aging mitochondrial phenotype. Therefore, the goal of the present work was to characterize age-dependent changes in supercomplex levels and composition. The aging rat heart was chosen for this analysis as this organ exhibits a significant mitochondrial-driven impairment in bioenergetics [32, 33], and supercomplexes are readily observed in mitochondria from this tissue [34, 35].
Using BN-PAGE separation of membrane proteins and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, this report shows that rat cardiac mitochondria display robust levels of ETC supercomplexes, but most of these assemblies, particularly those of the highest molecular weight, decline significantly with age. Thus, age-associated alterations in certain subtypes of supercomplexes must be considered as part of the etiology of mitochondrial alterations in the aging rat heart.
MATERIALS AND METHODS
Materials
6-Aminohexanoic acid and digitonin were purchased from Acros-Organics (Morris Plains, NJ, USA), and n-dodecyl-β-D-maltoside from Calbiochem (San Diego, CA, USA). Sucrose, D-mannitol, MOPS, Tricine, Tris HCl, Bis-Tris, subtilisin A, acetonitrile, ethylene glycol tetraacetic acid (EGTA), ethylenediaminetetraacetic acid (EDTA), KCl and other salts were from Sigma-Aldrich (St. Louis, MO, USA). All chemicals were reagent grade. Sequencing-grade modified trypsin was purchased from Promega (Madison, WI, USA). Coomassie G-250 and Bio-Safe Coomassie-G250 were from BioRad (Hercules, CA, USA). Mitochondrial complexes were detected using monoclonal antibodies against the following subunits: 39 kDa of complex I, 70 kDa of complex II, core 2 of complex III, subunit I of complex IV, and subunit α of complex V (MitoSciences, Eugene, OR, USA).
Ethical treatment of vertebrate animals
Young (5 months) and old (24 months) male Fischer 344 rats were obtained from the National Institute on Aging animal colonies. The animals were housed in approved facilities at the Linus Pauling Institute at Oregon State University and maintained by the Department of Laboratory Animal Resources. All procedures and animal handling were in keeping with approved institutional animal care and use guidelines.
Isolation of mitochondria
Heart interfibrillar mitochondria from young and old rats were isolated according to the protocol described by Palmer et al. [36]. One noted change from this method is the use of subtilisin A instead of nagarse to release mitochondria from the myofibrils, as the latter compound is no longer commercially available. All steps of the isolation were performed on ice or at 4°C. Protein content was determined by the Lowry method using bovine serum albumin as the standard (total protein kit from Sigma-Aldrich, St. Louis, MO, USA).
Separation of electron transport supercomplexes by BN-PAGE
Separation of electron transport supercomplexes was performed using BN-PAGE according to the protocol described by Wittig and Schagger [37, 38]. Mitochondrial membranes were solubilized with digitonin in a buffer composed of 750 mM 6-aminohexanoic acid, 50 mM Bis-Tris and 0.5 mM EDTA, pH 7.0 at 4°C. A digitonin-to-protein ratio of 8:1 (w/w) was empirically determined to be optimal for solubilizing mitochondrial membranes but maintaining mitochondrial supercomplexes. Following solubilization, samples were centrifuged for 20 min at 12000 × g at 4°C. Coomassie G-250 was added to the resulting supernatants using a detergent-to-dye ratio of 8:1 (w/w). Proteins (100 μg aliquots per lane) were separated on a NativePAGE Novex 3-12% Bis-Tris gradient gel (Invitrogen, Carsbad, CA, USA). In order to prevent an artifactual difference in the density of protein bands due to the position of lanes in the gel, samples were loaded following an alternating sequence between mitochondria from young versus old animals. Following electrophoresis, protein bands were visualized using Bio-Safe Coomassie-G250. A gel imaging system (Alpha Innotech, San Leandro, CA, USA) was used to digitize gel images and the densitometric in-gel quantification of bands was performed using ImageJ (National Institutes of Health).
Separation of individual electron transport complexes by BN-PAGE
In order to control for differences in protein-loading as well as to establish a means to quantify the levels of individual ETC complexes, n-dodecyl-β-D-maltoside (DDM) was used to solubilize mitochondrial membranes. Solubilization with DDM allows for separation of individual complexes rather than retaining ETC supercomplexes [23, 37]. A DDM-to-protein ratio of 2:1 (w/w) was empirically determined to optimally separate ETC complexes in their individual forms. Mitochondrial membranes were solubilized in a buffer composed of 750 mM 6-aminohexanoic acid, 50 mM Bis-Tris and 0.5 mM EDTA, pH 7.0 at 4°C. With the exception of DDM for membrane solubilization, all other conditions for BN-PAGE were the same as described for separation of electron transport supercomplexes (see above).
Identification of mitochondrial supercomplexes by nano-LC matrix assisted laser desorption ionization tandem mass spectrometry (MALDI-MS/MS)
Identification of proteins comprising individual bands on BN-PAGE gels were made using LC-MALDI-MS/MS analysis according to an adapted protocol from the method described by Fandiño et al. [39]. Prior to MALDI-MS/MS analysis, peptides were separated by nano-LC (column: PepMap 100, C-18, 3 μm, 100 Å, 75 μm i.d. × 15 cm, Dionex Corporation, Sunnyvale, CA, USA. Flow rate: 260 nl/min) and were spotted on 144-well MALDI target plates. Analysis of peptides was performed using a 4700 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Carlsbad, CA, USA). MASCOT (Matrix Science, Boston, MA, USA) was used to search the MS/MS data against the Swiss-Prot database (taxonomy rodentia) from which protein subunits of the oxidative phosphorylation system (OXPHOS) complexes were identified. Protein hits were considered significant with a significance threshold of p < 0.05 for peptide fingerprint search (i.e. a peptide match has less than 5% probability of being a random event), using an ion cut off score equal to 20. Searching of a decoy database resulted in a false positive discovery rate of less than 5%.
Identification of mitochondrial complexes by Western blot
Following the electrophoretic separation of proteins (30 μg of total protein added per lane), gels were transferred onto PVDF membranes (Immobilon Millipore, Billerica, MA, USA). Immunodetection of each OXPHOS complex was independently performed. Mitochondrial complexes were detected using monoclonal antibodies against the following subunits: 39 kDa of complex I, 70 kDa of complex II, core 2 of complex III, subunit I of complex IV, and subunit α of complex V.
Statistical analysis
All results were analyzed using a two-sided two-sample t-test. Differences were considered statistically significant at p < 0.05. When performing the search of the MS/MS data using MASCOT, peptide matches were considered significant with a significance threshold of p < 0.05 for peptide fingerprint search (i.e. a peptide match has less than 5% probability of being a random event).
RESULTS
Levels of individual ETC complexes do not change in the aging heart
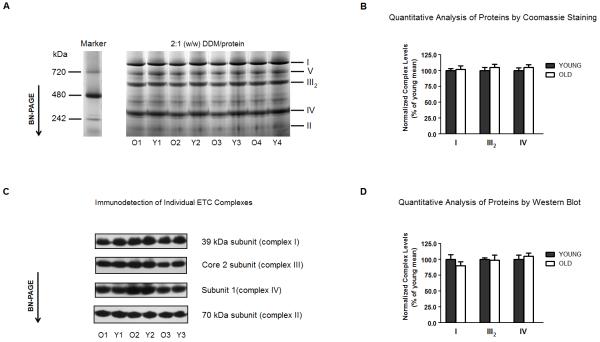
Figure 1A shows the separation pattern of proteins by BN-PAGE of DDM-solubilized rat heart interfibrillar mitochondria. From the protein separation achieved, individual OXPHOS complexes were identified in mitochondria isolated from both young and old rat hearts. To investigate potential age-related differences in the abundance of electron transport complexes, protein content of the Coomassie-stained bands was analyzed using standard densitometric methods (see Materials and Methods). Because complex II is not associated with supercomplexes in cardiac mitochondria [23, 30, 31, 34], it was chosen as a means to normalize against differences in protein loading. As shown in Figure 1B, no significant differences (p > 0.05) in the levels of complexes I, III1 and IV were evident in mitochondria isolated from old rats relative to young controls. Because of the potential for contaminating proteins to comigrate with ETC complexes, Western blot analysis was also used as to confirm the densitometric results (Fig. 1C). Using antibodies specific for each complex, no significant differences (p > 0.05) in the levels of any complex was evident with age (Fig. 1D). Thus, aging does not result in changes to levels of any individual electron transport complex.
Figure 1.
The levels of individual electron transport complexes of rat heart interfibrillary mitochondria do not change with age. (A) Heart interfibrillar mitochondria were isolated from young (Y: 5 months, n = 4) and old (O: 24 months, n = 4) Fischer 344 rats, and membranes were solubilized with a DDM-to-protein ratio of 2:1 (w/w). NativeMark (Invitrogen, Carsbad, CA, USA) was used as a molecular weight standard for proteins separated by BN-PAGE. (B) Levels of complexes I, III and IV were calculated using the density of Coomassie-stained proteins. In order to control for differences in protein-loading, the density of each complex was normalized to the density of complex II from the corresponding lane. (C) In separate experiments, mitochondrial proteins were separated by BN-PAGE as indicated in (A), and Western blot analysis was used for identification of individual electron transport complexes. Mitochondrial complexes were detected using monoclonal antibodies against the following subunits: 39 kDa of complex I, 70 kDa of complex II, core 2 of complex III and subunit I of complex IV, as described in Materials and Methods. (D) Levels of complexes I, III and IV were calculated using densitometric analysis after identification of proteins by Western blot. In order to control for differences in protein-loading, the density of each complex was normalized to the density of complex II from the corresponding lane. I, II, III2, IV and V denote individual OXPHOS complexes. All results are presented as the mean ± SEM, and plotted as a percentage of the mean from young controls.
Separation of rat cardiac mitochondrial supercomplexes by BN-PAGE and LC-MALDI-MS/MS analysis
Solubilization of mitochondrial membranes with digitonin versus DDM resulted in a more complex separation profile of proteins by BN-PAGE. Protein bands of very high mass were observed (S1 to S4, Fig. 2), which suggested the presence of supramolecular assemblies. Careful removal of these protein bands followed by LC-MALDI-MS/MS analysis showed that complexes I, III, and IV were constituents of these high-mass protein bands (Fig. 2). Thus, electron transport supercomplexes were identified in heart interfibrillar mitochondria. As multiple high molecular weight protein bands were observed using BN-PAGE, our data further suggest the presence of supercomplexes composed of complexes I, III and IV with differing stoichiometries. Another protein assembly comprising complexes I and III was also observed (designated “SC” in Fig. 2). There is a potential for complex V to also be associated with this particular supercomplex, but because of poor band separation by BN-PAGE, it is not currently possible to determine whether complex V is an integral part of the SC assembly or merely comigrates with it on the gel. Regardless, the SC protein complex displayed the lowest molecular mass with respect to the other identified supercomplexes. As noted previously, no association of complex II with other complexes was observed, indicating that it is not part of any respiratory supercomplex in cardiac mitochondria [23, 30, 31, 34] (Fig. 2).
Figure 2.
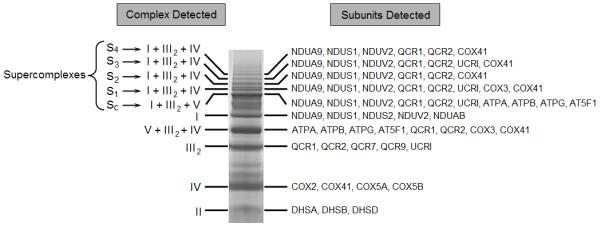
Identification of electron transport supercomplexes in rat heart interfibrillar mitochondria by LC-MALDI-MS/MS analysis. Mitochondrial proteins were separated on a 3-12% Bis-Tris gradient polyacrylamide gel by one-dimensional BN-PAGE. Individual mitochondrial OXPHOS complexes were detected using LC-MALDI-MS/MS analysis. The most representative protein subunits detected for each OXPHOS complex are displayed. I, II, III2, IV and V denote individual OXPHOS complexes. SC to S4 indicate different mitochondrial supercomplexes. All proteins were identified by MASCOT (Matrix Science, Boston, MA, USA) using the parameters described in Materials and Methods. Subunits are named according to the designation approved by the Protein Knowledgebase (UniProtKB) (http://www.uniprot.org/). Results are representative of three mitochondrial preparations.
In addition to supercomplex identification, LC-MALDI-MS/MS analysis also denoted protein bands consisting solely of complexes I, II, III or IV (Fig. 2). Separation of complex V as a single unit could not be optimally achieved using digitonin at the concentrations used. The identity of all proteins separated by BN-PAGE of digitonin-solubilized rat heart interfibrillar mitochondria was also confirmed using Western blot analysis of individual OXPHOS complexes (data not shown).
Mitochondrial supercomplexes are diminished in the aging heart
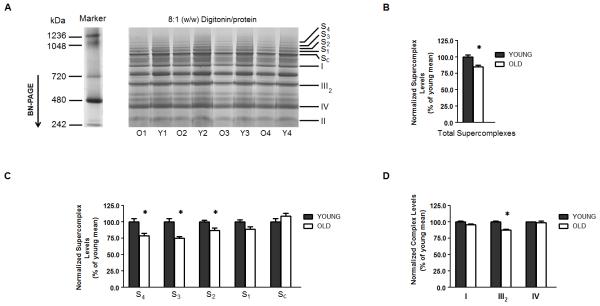
The same overall separation pattern of proteins was observed by BN-PAGE of digitonin-solubilized cardiac mitochondria isolated from both young and old rats (Fig. 3A). Nevertheless, with age, most of the electron transport supercomplexes declined in heart interfibrillar mitochondria (S1 to S4, Fig. 3A). In order to quantify the extent of such an age-related loss, supercomplexes were collectively examined. Summing the normalized densities of protein bands corresponding to S1, S2, S3 and S4 (Fig. 3A) revealed that interfibrillar mitochondrial supercomplexes collectively diminished by 15% relative to young controls (p < 0.05, n = 4) (Fig. 3B).
Figure 3.
Mitochondrial electron transport supercomplexes are diminished in the aging rat heart. (A) Heart interfibrillar mitochondria were isolated from young (Y: 5 months, n = 4) and old (O: 24 months, n = 4) Fischer 344 rats, and solubilized with a digitonin-to-protein ratio of 8:1 (w/w). NativeMark (Invitrogen, Carsbad, CA, USA) was used as a molecular weight standard for proteins separated by BN-PAGE. (B) From each lane, the total levels of supercomplexes were estimated by summing the densities of the four protein bands corresponding to supercomplexes (i.e. S1-S4). In order to control for differences in protein-loading, the density of each supercomplex was normalized to the density of complex II from the corresponding lane. (C) Levels of mitochondrial electron transport supercomplexes. (D) Levels of electron transport complexes in a free form. S1-SC indicate different mitochondrial electron transport supercomplexes. I, II, III2, IV and V denote individual OXPHOS complexes. All results are presented as the mean ± SEM, and plotted as a percentage of the mean from young controls. n = 4; *p < 0.05 compared to young controls.
Although practically all the identified high molecular weight assemblies were comprised of complexes I, III and IV (Fig. 2), not all these supercomplexes deteriorated to the same degree as a function of age. As shown in Figure 3C, the assemblies displaying the highest molecular masses underwent the largest age-related declines. Relative to young controls, supercomplexes S4, S3 and S2 declined by 21%, 25%, and 13%, respectively (p < 0.05, n = 4) (Fig. 3, A and D). There was a trend (p < 0.07, n = 4) for the protein band designated S1 to also decline with age, but no significant difference (p > 0.05) in the multicomplex assembly named SC was observed. On the other hand, no significant differences (p > 0.05) in the free forms of complexes I and IV were detected; however complex III was diminished by ~12% (p < 0.05, n = 4), relative to young controls (Fig. 3D). Thus, aging results in lower levels of mitochondrial supercomplexes without a concomitant loss in individual components of the ETC.
DISCUSSION
BN-PAGE was used to separate but maintain mitochondrial proteins in a native architecture [23, 34]. Using this technique, we observed both individual electron transport complexes as well as high molecular weight protein bands previously identified as supercomplexes [23, 34]. Additionally, both LC-MALDI-MS/MS analysis and immunodetection were employed to confirm that these high molecular weight protein bands were indeed macromolecular assemblies of individual ETC components. Thus, analytical techniques of sufficient resolving power were used to investigate age-associated changes to mitochondrial supercomplexes. Nevertheless, we recognize that even though BN-PAGE has been instrumental in showing that individual ETC complexes assemble into supercomplexes, the necessity of membrane solubilization with specific detergents may cause artifactual protein aggregates which in turn affect interpretation of data.
It is clear from our data that significant age-related losses in most identified supercomplexes occur, with those of the highest molecular mass the most severely diminished. As the profile of mitochondrial proteins separated by BN-PAGE was not affected with age (Fig. 3A), it is reasonable to assume that the loss of supercomplexes is not caused by any age-related deficit of constituent subunits of ETC complexes. However, because the present work constitutes the first characterization of age-dependent changes in supercomplexes, we are aware that BN-PAGE as a technique might produce variations in results when using mitochondrial preparations from young versus old rats. Therefore, the potential contribution of other factors such as a difference in the protein yield between heart mitochondria from young and old animals during solubilization of membranes for BN-PAGE analysis cannot be excluded. The mechanism(s) for the general deterioration in supercomplexes are currently unknown and were not explored in the present work. However, as complex IV is a constituent of all supercomplexes in rat heart mitochondria and also the last component to be incorporated into them [40, 41], it is enticing to speculate that age-related supercomplex decay is connected with alterations in complex IV. In this regard, Oswald et al. established an association with complex IV and destabilization of supercomplexes when they showed that knockdown of COX17 in HeLa cells resulted in supercomplex loss [42]. Moreover, D’Aurelio and colleagues also observed supercomplex destabilization in human mtDNA cybrids containing a mutation in the cytochrome c oxidase subunit 1 gene (MT-COX1) [43]. Finally, we previously showed that complex IV activity declined with age, which correlated with extensive oxidative modification of this complex [15]. These latter results, together with our present data, may therefore establish a rationale for oxidative protein modification of complex IV and decline in supercomplex assembly. We are presently exploring the connection between alterations in complex IV components and the age-related loss of supercomplexes.
While the age-associated loss of interfibrillary mitochondrial supercomplexes was significant, the degree of diminishment was relatively modest. Thus it will be important to ascertain how these changes are part of the aging mitochondrial phenotype. For perspective, supercomplex deterioration has been observed for both Barth Syndrome [44] and in a patient with classic Leigh Syndrome [40]. Despite severe mitochondrial ultrastructural and respiratory chain defects evident in both cases, only relatively subtle declines in supercomplex levels were noted (~20% for Barth Syndrome, [44]). Additionally, in an acute heart failure model in dogs, disassembly of mitochondrial supercomplexes only reached 50% relative to control animals [45]. Thus, the apparent subtle loss of supercomplex levels observed in our present study (i.e. ~15%) is not far below that seen in overt pathologies connected to mitochondrial dysfunction. Therefore, even seemingly small deficits in mitochondrial supercomplex levels may significantly impact ETC function. In this regard, our results may at least partly account for the decline in oxidative capacity evident in the aging rat heart [3, 13, 32, 33, 46].
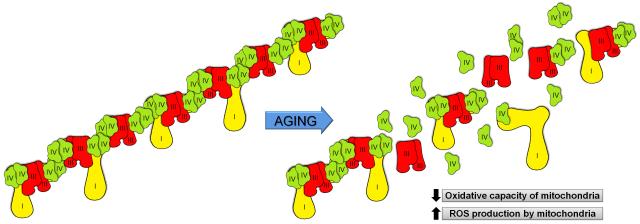
The data presented herein are also important for the so-called “respiratory string model” proposed by Schagger and colleagues [23-25]. This model postulates that the ETC exists as long super-assemblies that wrap the cristae and efficiently transfer electrons to O2. If this model is correct, then the supercomplexes noted by BN-PAGE are themselves fragments of even higher ordered ETC strings [25, 47]. Our results interpreted in relation to this model indicate that aging leads to respiratory string destabilization in rat heart mitochondria. A schematic representation of this scenario is depicted in Figure 4. An extension of this concept further suggests that if electron transport supercomplexes limit formation of reactive oxygen intermediates as previously suggested [23], age-associated destabilization of respiratory strings would promote production of oxidants and oxidative damage, which have been observed in isolated rat cardiac myocytes in general [32], and interfibrillar mitochondria in particular [15, 33]. Nevertheless, this interpretation requires further investigation as the current evidence in support of supercomplexes has been not sufficient to mitigate all doubts about the physiological existence of such a supramolecular organization of the ETC.
Figure 4.
Schematic representation of the age-related destabilization of the mitochondrial ETC “respiratory string”. “ROS” denotes reactive oxygen species.
Despite the implications for deficits to ETC function, the precise mechanism(s) involved in the age-associated loss of rat heart mitochondrial supercomplexes have yet to be elucidated. As individual ETC components do not decline with age, our data suggest that either supercomplex formation is impaired or the rate of decomposition is accelerated. Aside from changes to complex IV, evidence has accumulated suggesting that cardiolipin promotes supercomplex assembly [44, 48, 49]. Because cardiolipin has been observed to decline and/or its acyl side-chain composition changes with age [13, 50], it is tempting to think that age-related deficits in cardiolipin content may be responsible for a destabilization of supercomplexes. However, Hoppel and colleagues provided evidence that interfibrillary mitochondria of the aging rat heart have no age-associated decrements in cardiolipin, belying previous reports to the contrary [51]. Thus, additional work is warranted in order to define the role that cardiolipin plays in age-associated supercomplex deficits.
Another possibility contributing to loss of supercomplexes may result from an age-related alteration of the mitochondrial proteome. In this regard, prohibitins have become very interesting targets since such proteins have been observed to regulate the assembly of the ETC [52], and appear to play an important role during aging [53-55]. We are currently examining the potential for both cardiolipin and/or age-associated alterations to ETC assembly to constitute the mechanism(s) associated with supercomplex degradation with age.
ACKNOWLEDGMENTS
This research was funded by the National Institute on Aging, grant number 2R01AG017141-06A2. The mass spectrometry-based proteomics methods were developed in part with funds from the National Institutes of Health and the National Institute on Aging, grant number R01AG025372. We would also like to acknowledge the contribution of the Mass Spectrometry Facility of the Environmental Health Sciences Center (supported in part by the National Institutes of Health and the National Institute of Environmental Health Sciences, grant number P30ES00210) at Oregon State University. The authors would like to thank Judy A. Butler for careful reading of the manuscript.
Footnotes
Hereafter, complex III will denote the individual form of this mitochondrial complex that corresponds to a dimer (i.e. III2) when separated by BN-PAGE.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Kim JH, Woldgiorgis G, Elson CE, Shrago E. Mech. Ageing. Dev. 1988;46:263–277. doi: 10.1016/0047-6374(88)90129-7. [DOI] [PubMed] [Google Scholar]
- [2].Kim JH, Shrago E, Elson CE. Mech. Ageing. Dev. 1988;46:279–290. doi: 10.1016/0047-6374(88)90130-3. [DOI] [PubMed] [Google Scholar]
- [3].Fannin SW, Lesnefsky EJ, Slabe TJ, Hassan MO, Hoppel CL. Arch. Biochem. Biophys. 1999;372:399–407. doi: 10.1006/abbi.1999.1508. [DOI] [PubMed] [Google Scholar]
- [4].Shigenaga MK, Hagen TM, Ames BN. Proc. Natl. Acad. Sci. USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hagen TM, Yowe DL, Bartholomew JC, Wehr CM, Do KL, Park JY, Ames BN. Proc. Natl. Acad. Sci. USA. 1997;94:3064–3069. doi: 10.1073/pnas.94.7.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hagen TM, Wehr CM, Ames BN. Ann. N. Y. Acad. Sci. 1998;854:214–223. doi: 10.1111/j.1749-6632.1998.tb09904.x. [DOI] [PubMed] [Google Scholar]
- [7].Amicarelli F, Ragnelli AM, Aimola P, Bonfigli A, Colafarina S, Di Ilio C, Miranda M. Biochim. Biophys. Acta. 1999;1453:105–114. doi: 10.1016/s0925-4439(98)00088-x. [DOI] [PubMed] [Google Scholar]
- [8].de Cavanagh EM, Piotrkowski B, Basso N, Stella I, Inserra F, Ferder L, Fraga CG. FASEB J. 2003;17:1096–1098. doi: 10.1096/fj.02-0063fje. [DOI] [PubMed] [Google Scholar]
- [9].Yasuda K, Ishii T, Suda H, Akatsuka A, Hartman PS, Goto S, Miyazawa M, Ishii N. Mech. Ageing. Dev. 2006;127:763–770. doi: 10.1016/j.mad.2006.07.002. [DOI] [PubMed] [Google Scholar]
- [10].Muller-Hocker J. Am. J. Pathol. 1989;134:1167–1173. [PMC free article] [PubMed] [Google Scholar]
- [11].Choksi KB, Nuss JE, Boylston WH, Rabek JP, Papaconstantinou J. Free Radic. Biol. Med. 2007;43:1423–1438. doi: 10.1016/j.freeradbiomed.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lesnefsky EJ, Gudz TI, Moghaddas S, Migita CT, Ikeda-Saito M, Turkaly PJ, Hoppel CL. J. Mol. Cell. Cardiol. 2001;33:37–47. doi: 10.1006/jmcc.2000.1273. [DOI] [PubMed] [Google Scholar]
- [13].Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. FEBS Lett. 1997;406:136–138. doi: 10.1016/s0014-5793(97)00264-0. [DOI] [PubMed] [Google Scholar]
- [14].Petrosillo G, Matera M, Casanova G, Ruggiero FM, Paradies G. Neurochem. Int. 2008;53:126–131. doi: 10.1016/j.neuint.2008.07.001. [DOI] [PubMed] [Google Scholar]
- [15].Suh JH, Heath SH, Hagen TM. Free Radic. Biol. Med. 2003;35:1064–1072. doi: 10.1016/s0891-5849(03)00468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Barth PG, Scholte HR, Berden JA, Van der Klei-Van Moorsel JM, Luyt-Houwen IE, Van ’t Veer-Korthof ET, Van der Harten JJ, Sobotka-Plojhar MA. J. Neurol. Sci. 1983;62:327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- [17].Barth PG, Wanders RJ, Vreken P, Janssen EA, Lam J, Baas F. J. Inherit. Metab. Dis. 1999;22:555–567. doi: 10.1023/a:1005568609936. [DOI] [PubMed] [Google Scholar]
- [18].Bentlage HA, Wendel U, Schagger H, ter Laak HJ, Janssen AJ, Trijbels JM. Neurology. 1996;47:243–248. doi: 10.1212/wnl.47.1.243. [DOI] [PubMed] [Google Scholar]
- [19].Figarella-Branger D, Pellissier JF, Scheiner C, Wernert F, Desnuelle C. J. Neurol. Sci. 1992;108:105–113. doi: 10.1016/0022-510x(92)90195-q. [DOI] [PubMed] [Google Scholar]
- [20].Pitkanen S, Robinson BH. J. Clin. Invest. 1996;98:345–351. doi: 10.1172/JCI118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hackenbrock CR, Chazotte B, Gupte SS. J. Bioenerg. Biomembr. 1986;18:331–368. doi: 10.1007/BF00743010. [DOI] [PubMed] [Google Scholar]
- [23].Schagger H, Pfeiffer K. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wittig I, Carrozzo R, Santorelli FM, Schagger H. Biochim. Biophys. Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- [25].Wittig I, Schagger H. Biochim. Biophys. Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- [26].Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. Proc. Natl. Acad. Sci. USA. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dudkina NV, Sunderhaus S, Boekema EJ, Braun HP. J. Bioenerg. Biomembr. 2008;40:419–424. doi: 10.1007/s10863-008-9167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Marques I, Dencher NA, Videira A, Krause F. Eukaryot. Cell. 2007;6:2391–2405. doi: 10.1128/EC.00149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vonck J, Schafer E. Biochim. Biophys. Acta. 2009;1793:117–124. doi: 10.1016/j.bbamcr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- [30].Bianchi C, Genova ML, Parenti Castelli G, Lenaz G. J. Biol. Chem. 2004;279:36562–36569. doi: 10.1074/jbc.M405135200. [DOI] [PubMed] [Google Scholar]
- [31].Schafer E, Dencher NA, Vonck J, Parcej DN. Biochemistry. 2007;46:12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- [32].Hagen TM, Moreau R, Suh JH, Visioli F. Ann. N. Y. Acad. Sci. 2002;959:491–507. doi: 10.1111/j.1749-6632.2002.tb02119.x. [DOI] [PubMed] [Google Scholar]
- [33].Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. J. Mol. Cell. Cardiol. 2001;33:1065–1089. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- [34].Wittig I, Schagger H. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- [35].Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. J. Proteome Res. 2006;5:1117–1132. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- [36].Palmer JW, Tandler B, Hoppel CL. J. Biol. Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- [37].Wittig I, Braun HP, Schagger H. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- [38].Wittig I, Schagger H. Methods Cell Biol. 2007;80:723–741. doi: 10.1016/S0091-679X(06)80033-6. [DOI] [PubMed] [Google Scholar]
- [39].Fandiño AS, Rais I, Vollmer M, Elgass H, Schagger H, Karas M. J. Mass Spectrom. 2005;40:1223–1231. doi: 10.1002/jms.903. [DOI] [PubMed] [Google Scholar]
- [40].Schagger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U. J. Biol. Chem. 2004;279:36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- [41].Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- [42].Oswald C, Krause-Buchholz U, Rodel G. J. Mol. Biol. 2009;389:470–479. doi: 10.1016/j.jmb.2009.04.034. [DOI] [PubMed] [Google Scholar]
- [43].D’Aurelio M, Gajewski CD, Lenaz G, Manfredi G. Hum. Mol. Genet. 2006;15:2157–2169. doi: 10.1093/hmg/ddl141. [DOI] [PubMed] [Google Scholar]
- [44].McKenzie M, Lazarou M, Thorburn DR, Ryan MT. J. Mol. Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- [45].Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiovasc. Res. 2008;80:30–39. doi: 10.1093/cvr/cvn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chen JC, Warshaw JB, Sanadi DR. J. Cell. Physiol. 1972;80:141–148. doi: 10.1002/jcp.1040800115. [DOI] [PubMed] [Google Scholar]
- [47].Schagger H, Pfeiffer K. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- [48].Zhang M, Mileykovskaya E, Dowhan W. J. Biol. Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- [49].Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- [50].Lee HJ, Mayette J, Rapoport SI, Bazinet RP. Lipids Health Dis. 2006;5:2. doi: 10.1186/1476-511X-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Moghaddas S, Stoll MS, Minkler PE, Salomon RG, Hoppel CL, Lesnefsky EJ. J. Gerontol. A Biol. Sci. Med. Sci. 2002;57:B22–28. doi: 10.1093/gerona/57.1.b22. [DOI] [PubMed] [Google Scholar]
- [52].Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Coates PJ, Nenutil R, McGregor A, Picksley SM, Crouch DH, Hall PA, Wright EG. Exp. Cell Res. 2001;265:262–273. doi: 10.1006/excr.2001.5166. [DOI] [PubMed] [Google Scholar]
- [54].Piper PW, Jones GW, Bringloe D, Harris N, MacLean M, Mollapour M. Aging Cell. 2002;1:149–157. doi: 10.1046/j.1474-9728.2002.00018.x. [DOI] [PubMed] [Google Scholar]
- [55].Merkwirth C, Langer T. Biochim. Biophys. Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]