Abstract
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world. The clinical heterogeneity of HCC, and the lack of good diagnostic markers and treatment strategies, has rendered the disease a major challenge. Patients with HCC have a highly variable clinical course, indicating that HCC comprises several biologically distinctive subgroups reflecting a molecular heterogeneity of the tumors. Transforming growth factor β (TGF-β) is known to exhibit tumor stage dependent suppressive (that is, growth inhibition) and oncogenic (that is, invasiveness) properties. Here, we asked if a TGF-β specific gene expression signature could refine the classification and prognostic predictions for HCC patients. Applying a comparative functional genomics approach we demonstrated that a temporal TGF-β gene expression signature established in mouse primary hepatocytes successfully discriminated distinct subgroups of HCC. The TGF-β positive cluster included two novel homogeneous groups of HCC associated with early and late TGF-β signatures. Kaplan-Meier plots and log-rank statistics indicated that the patients with a late TGF-β signature showed significantly (P < 0.005) shortened mean survival time (16.2 ± 5.3 months) compared to the patients with an early (60.7 ± 16.1 months) TGF-β signature. Also, tumors expressing late TGF-β-responsive genes displayed invasive phenotype and increased tumor recurrence. We also showed that the late TGF-β signature accurately predicted liver metastasis and discriminated HCC cell lines by degree of invasiveness. Finally, we established that the TGF-β gene expression signature possessed a predictive value for tumors other than HCC.
Conclusion
These data demonstrate the clinical significance of the genes embedded in TGF-β expression signature for the molecular classification of HCC.
Hepatocellular carcinoma (HCC) is the fifth most common cancer world-wide, accounting for at least 600,000 deaths annually.1 Although most frequent in southeast Asia and sub-Sahara Africa, the incidence and mortality rate of HCC have doubled in the United States and Europe in the past 4 decades and are expected to double again over the next 10 to 20 years.2 As the incidence of HCC has increased, the age-distribution of HCC has shifted toward relatively younger ages.2 These observations make it clear that liver cancer is becoming a major heath problem in the United States and Europe.
Although recent advances in genomics analysis of HCC provide an increasingly comprehensive portrayal of hepatocarcinogenesis,3 the molecular pathogenesis of HCC remains poorly understood. Patients have a highly variable clinical course, indicating that HCC comprises several biologically distinctive subgroups reflecting the molecular heterogeneity of the tumors.4,5 Previously, we have demonstrated that gene expression signatures estab-lished in well-controlled experimental conditions can successfully identify new subgroups of tumors and thus improve the molecular classification of human HCC.6–9 For example, a gene signature specific for hepatocyte growth factor/c-Met signaling pathway allowed us to define a clinically relevant subset of human HCC characterized by a poor prognosis and an aggressive phenotype.10 Therefore, this approach provides a practical experimental framework for a systematic analysis of the contributions of growth factor signaling pathways or oncogenic signaling pathways to the tumor phenotype, as well as identifying homogeneous subgroups of heterogeneous cancers such as HCC.
Transforming growth factor beta (TGF-β) is a pleiotropic cytokine that controls many fundamental aspects of cell biology, including proliferation, migration, adhesion, differentiation, and modification of the cellular microenvironment.11,12 TGF-β exerts its effects through the TGFBR1 and TGFBR2 serine-threonine kinases receptors and the small mothers against decapentaplegic (SMAD) family of transcriptional regulators. TGF-β binding to TGFBR2 induces phosphorylation and the activation of TGFBR1. The activated TGFBR1 then phosphorylates SMAD2 and SMAD3, which in turn bind to SMAD4 and move into the nucleus to form complexes that regulate transcription.12,13 Genetic alterations in the TGF-β signaling pathway have been reported in cancer, but the implication of TGF-β signaling in carcinogenesis is complex, because TGF-β may exhibit both tumor suppressive and oncogenic properties.14–17 TGF-β acts as a tumor suppressor at the early stages of tumor development by inhibiting proliferation and inducing apoptosis, but TGF-β also possesses oncogenic potential, which contributes to tumor progression later in carcinogenesis. Notably, as a potent stimulator of epithelial-mesenchymal transitions (EMT), TGF-β promotes tumor cell invasiveness and metastasis.18 Based on these considerations, we hypothesized that a TGF-β gene expression signature established under well-controlled experimental conditions in vitro may contain gene sets characteristic of both tumor suppressive and oncogenic properties and thus be relevant for the molecular classification of tumors. Here we report that the genes embedded in the TGF-β expression signature could identify novel homogeneous and clinically distinct subgroups of human HCC.
Materials and Methods
Hepatocyte Isolation and Culture
Animal experiments were in accordance with the National Institutes of Health guidelines for animal care and approved by the National Cancer Institute Animal Care and Use Committee. We isolated hepatocytes from 8-week-old Tgfbr2+/+/AlbCre+/− (wild-type [WT]) and Tgfbr2fl/fl/AlbCre+/− (knockout [KO]) mice19 by two-step collagenase perfusion of the mouse livers followed by isodensity purification in Percoll gradient.20 At this stage, liver histology was normal in KO animals. We seeded the cells at 2 × 106 in 10-cm dishes in the plating medium supplemented with 10% fetal bovine serum. After 4 hours, we replaced the plating medium with serum-free medium. The following day, we treated the hepatocytes with 1 ng/mL of recombinant TGF-β (R&D Systems) versus vehicle alone for 0.5, 1, 2, 4, 12, and 24 hours. Triplicate cell cultures were established from three individual mice.
DNA Microarrays
We used a genome-wide set of longmer mouse oligonucleotides obtained from Illumina and printed at the Advanced Technology Center (National Cancer Institute) in this study. This set is based on well-annotated sequence information derived largely from the Mouse Exonic Evidence-Based Oligonucleotide consortium (http://mmc.ucsf.edu/Meebo.html). Target preparation and hybridization on microarrays were as described.21 Briefly, 15 µg of each target RNA was labeled with Cy-3 or Cy-5 and then hybridized with the same batch of reference RNA. We carried out at least two hybridizations for each sample using a dye-swap strategy to eliminate dye-labeling bias. We identified differentially expressed genes by a univariate two-sample t test with a random variance model. We computed permutation P values for significant genes (P < 0.01) based on 10,000 random permutations as described.21
Pathway Analysis
We used the Ingenuity Pathway Analysis tool to examine functional associations between genes and generated the gene networks with high significance. We estimated the significance of each network by the scoring system provided by Ingenuity. The scores are determined by the number of differentially expressed genes within each of the networks and the strength of the associations among network members.
Invasion Assay
We measured the invasive and migrating activity of HCC cell lines using the BD BioCoat Matrigel Invasion Chambers as recommended by the manufacturer (BD Biosciences). We plated 2.5 × 104 cells in 500 µL of serum-free medium in triplicate either on an 8-µm pore size positron emission tomography membrane insert coated with a layer of matrigel basement membrane matrix or on an 8-µm pore size positron emission tomography control membrane insert. The lower compartment contained Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum as chemoattractant. After incubation for 22 hours, we removed the cells remaining on the upper surface of membrane with a cotton swab, and we fixed cells that had passed through to the lower surface of membrane in 100% methanol and stained with 1% toluidine. We scored invasive and migrating cells by counting at least six fields per membrane under a light microscope at ×100 magnification.
Results
Establishment of the TGF-β Gene Expression Signature
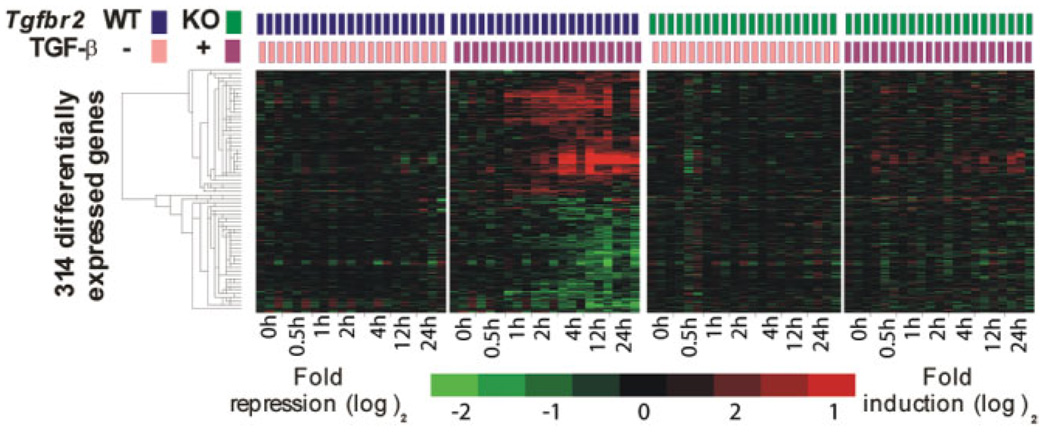
To profile human HCC with regard to the TGF-β signaling pathway, we first established a TGF-β-specific gene expression signature in mouse primary hepatocytes. To increase the specificity of the signature, we performed gene expression profiling on primary hepatocytes isolated from Tgfbr2+/+/AlbCre+/− (WT) and Tgfbr2fl/fl/AlbCre+/− (TGF-β type II receptor conditional KO) mice; the latter was used as a validation of TGF-β dependency. Hepatocytes from KO mice lack exon 4 of the Tgfbr2 gene essential for receptor activity and therefore are refractory to TGF-β stimulation.20 We assessed specific disruption of the Tgfbr2 gene as described,20 and we confirmed deficient TGF-β signaling in KO hepatocytes by resistance to the TGF-β induced growth inhibition (Supplementary Fig. 1). We isolated hepatocytes from WT and KO livers, and we exposed paired primary cultures to 1 ng/mL of recombinant TGF-β versus vehicle alone for 0.5, 1, 2, 4, 12, and 24 hours. As described,21 we quantified mRNA abundance using genome-wide mouse microarrays to achieve an exhaustive covering of TGF-β responsive genes. At each time point, only genes for which expression was significantly altered by TGF-β in WT but not in KO hepatocytes (P < 0.01, random-variance t test) were included as part of the TGF-β signature. In total, the TGF-β signature contained 314 genes (Fig. 1; Supplementary Table 1). Genes that were persistently differentially expressed between the two genotypes but not sensitive to TGF-β treatment (Supplementary Fig. 2) were omitted from the signature.
Fig. 1.
Hierarchical cluster analysis of 314 genes included in the mouse TGF-β signature. Data are presented in a matrix format in which rows and columns represent genes and samples, respectively. Hepatocytes were isolated from Tgfbr2+/+/AlbCre+/− (WT, two left panels) and Tgfbr2fl/fl/AlbCre+/− (KO, two right panels) mice. Time-course gene expression profiling was performed on primary cultures challenged with TGF-β (+) versus vehicle alone (−) up to 24 hours. Genes included in the TGF-β signature were significantly up-regulated or down-regulated by TGF-β in WT hepatocytes (second panel) but no fluctuation was observed in KO hepatocytes (two right panels).
We used the Ingenuity Pathway Analysis tool to examine the functional associations between the genes included in the signature. Validating our gene selection, this analysis identified TGF-β as the central regulator of genes included in the top-rank signaling network (Supplementary Fig. 3). Functional classification of target genes revealed a temporal organization of TGF-β transcriptional response in mouse hepatocytes. Early induction of Smad corepressors Skil and Tgif and the inhibitory Smad7 suggested that one of the of the early responses to TGF-β in primary hepatocytes was to turn on a negative feedback loop to desensitize TGF-β action. The early response was also characterized by transcriptional activation of inducers of cell cycle arrest and apoptosis (e.g., Bag3, Btg1, Clic4, Gadd45-a, Gadd45-b, Gadd45-g, Igfbp3, and Tnfrsf1b), in agreement with the antiproliferative and proapoptotic properties of TGF-β.16 At the later time points, modulation of genes involved in cytoskeleton organization (e.g., Net1, Rhob, Svil, and Vim), cell adhesion (e.g., Alcam, Itga6, Pcdh1, and Snai1), and matrix remodeling (e.g., Col18a1 and Ctgf) became more prominent, supporting the important role for TGF-β in the control of the cellular microenvironment and EMT.4,18,22 Interestingly, among the genes regulated after 4 hours of TGF-β treatment were also those associated with maintenance of lipid and redox homeostasis, suggesting a new role for TGF-β. Thus, genes encoding the transcription factors Srebp and Nfe2l2 (also known as Nrf2), which respectively control the expression of sterol-regulated and antioxidant genes,23,24 were strongly repressed. Accordingly, genes involved in cholesterol biosynthesis (e.g., Dhcr24, Dhcr7, Hmgcs1, and Sqle) and glutathione metabolism (e.g., Gss, Gsta1, Gstm6, and Gsto1) were simultaneously repressed. Taken together, these data implicate TGF-β as an important regulator of several pathways involved in cellular homeostasis and alterations known to contribute to cancer progression.
More importantly, the TGF-β signature included gene sets characteristic for both tumor suppressive and oncogenic properties of TGF-β , and therefore could be relevant for the molecular classification of tumors. Comparison with published gene expression data sets revealed that at least 30% of genes included in our signature were previously described as TGF-β or SMAD target genes in rodent and human (Supplementary Table 1). This overlap suggested that the gene subset in the TGF-β core signature was partly conserved between species, further indicating its relevance for profiling of human HCC and possibly other cancers.
Comparative Functional Genomic Analysis Identifies Subgroups of Human HCC Positive and Negative for the Mouse TGF-β Gene Expression Signature
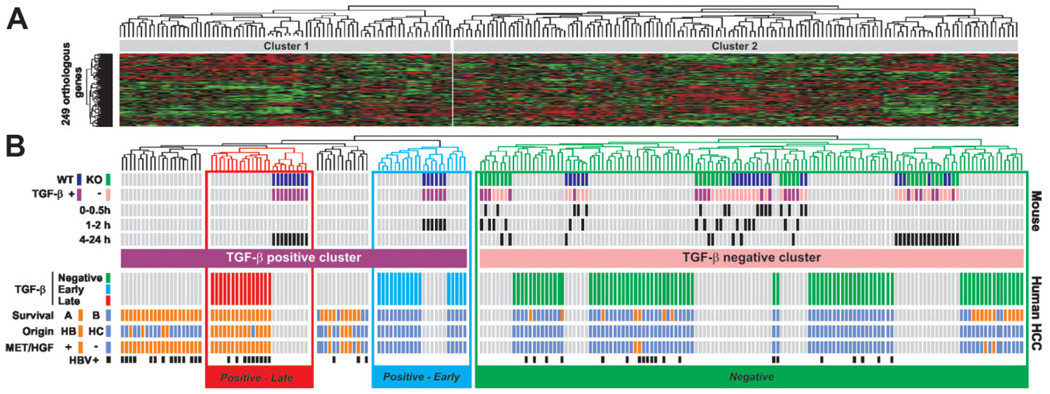
Using our previous comparative functional genomics paradigm,6,8,10 we integrated the gene expression data from mouse hepatocytes with 139 cases of human HCC.25 Curated mammalian gene orthology from The Jackson Laboratory produced 249 human orthologs out of the 314 mouse genes included in the TGF-β signature. For a direct cross-species comparison, we independently standardized gene expression values from human and mouse data sets by adjusting the mean and standard deviation to 0 and 1, respectively, as described. Based on the expression of the 249 orthologous genes (Supplementary Table 2), hierarchical clustering of the integrated data set established the degree of similarity between human and mouse samples, and identified two major clusters (Fig. 2A). Except for the 0.5-hour data point, cluster 1 included all mouse samples corresponding to WT hepatocytes challenged with TGF-β and therefore was referred to as TGF-β-positive (Fig. 2B, upper part). Cluster 2 included all samples derived from KO mice (treated and untreated) as well as all untreated WT samples, and was accordingly defined as TGF-β-negative (Fig. 2B, upper part). Human liver tumors were divided into positive and negative TGF-β clusters, indicating that the TGF-β gene expression signature effectively discriminates distinct subgroups of HCC (Fig. 2B, lower part). Nevertheless, the analysis of the distribution of several clinical variables revealed only trend differences between HCC harboring positive versus negative TGF-β signatures (data not shown), suggesting that HCCs defined by the complete TGF-β signature exhibit a high variability and may likely include additional subtypes.
Fig. 2.
Discrete temporal mouse TGF-β gene expression signatures define distinct subtypes of human HCC. (A) Dendrogram and heat-map overview of mouse data set integrated with 139 cases of human HCC. Clustering was based on the expression of 249 orthologous genes and the data are presented in a matrix format in which rows and columns represent genes and samples, respectively. (B) Detailed view of mouse and human samples cluster analysis. Upper part: Clustering of mouse samples corresponding to WTand KO hepatocytes challenged (+) or not (−) with TGF-β for a given time identified at the beginning of each row. Clustering analysis identified a TGF-β-positive cluster corresponding to WT-treated hepatocytes and a TGF-β-negative cluster corresponding to untreated and treated KO hepatocytes as well as untreated WT hepatocytes. Lower part: Clustering of human HCC samples. Integration of mouse and human samples sorted human HCC into TGF-β-positive and TGF-β-negative clusters. Within the TGF-β-positive cluster, the analysis of dendrogram branches revealed two homogeneous subtypes of HCC associated with WT hepatocytes treated for 1 to 2 hours and 4 to 24 hours and annotated Positive-Early and Positive-Late, respectively. Distribution of HCC between previously described subgroups with respect to survival (A, bad prognosis versus B, good prognosis), cell origin (HB, hepatoblast versus HC, hepatocyte), c-Met/hepatocyte growth factor (HGF) signature (− versus +) or HBV infection status is indicated at the beginning of each row.
TGF-β Signature Includes Two Independent Signatures
Given that our gene expression signature included genes characteristic of the tumor suppressive and oncogenic properties of TGF-β, we hypothesized that the TGF-β-positive cluster may include additional homogeneous subtypes of HCC. Interestingly, within the TGF-β-positive cluster, the mouse samples were not randomly distributed (Fig. 2B, upper part). Two distinct homogeneous clusters were revealed, corresponding to WT hepatocytes challenged with TGF-β either for short (1–2 hours) or long (4–24 hours) periods of time, suggesting that the TGF-β signature may in fact include two independent signatures. Based on their tight correlation with the temporally defined TGF-β mouse core clusters, two homogeneous subgroups of human HCC were consequently refined and referred to as Positive-Early and Positive-Late (Fig. 2B, lower part). To test the predictive value of early and late TGF-β signatures in the identification of specific HCC subtypes, six different prediction algorithms (SVM, CCP, 1NN, 3NN, NC, and LDA) were applied. HCC with early or late TGF-β signatures were first randomly divided into training and testing sets. When the trained classifier genes were applied to the testing set, all six algorithms could predict HCC with late or early TGF-β signature, with a prediction rate ranging from 93% to 100% (Supplementary Table 3). As an independent validation data set, we also analyzed the expression profiles of 104 HCC and seven liver metastases26 uploaded from the Stanford Microarray Database (http://genome-www5.stanford.edu). Cluster analysis based on the expression of 176 orthologous genes present in the mouse and human data sets similarly identified (1) two subsets of HCC, positive and negative for TGF-β signature; and (2) additional subtypes of HCC within the positive cluster based on early and late TGF-β signatures (Supplementary Fig. 4), indicating that early and late TGF-β signatures may effectively discriminate relevant subtypes of human HCC within the TGF-β-positive cluster.
Late TGF-β Signature Predicts Liver Metastasis In Vivo and Is Associated with an Invasive Phenotype In Vitro
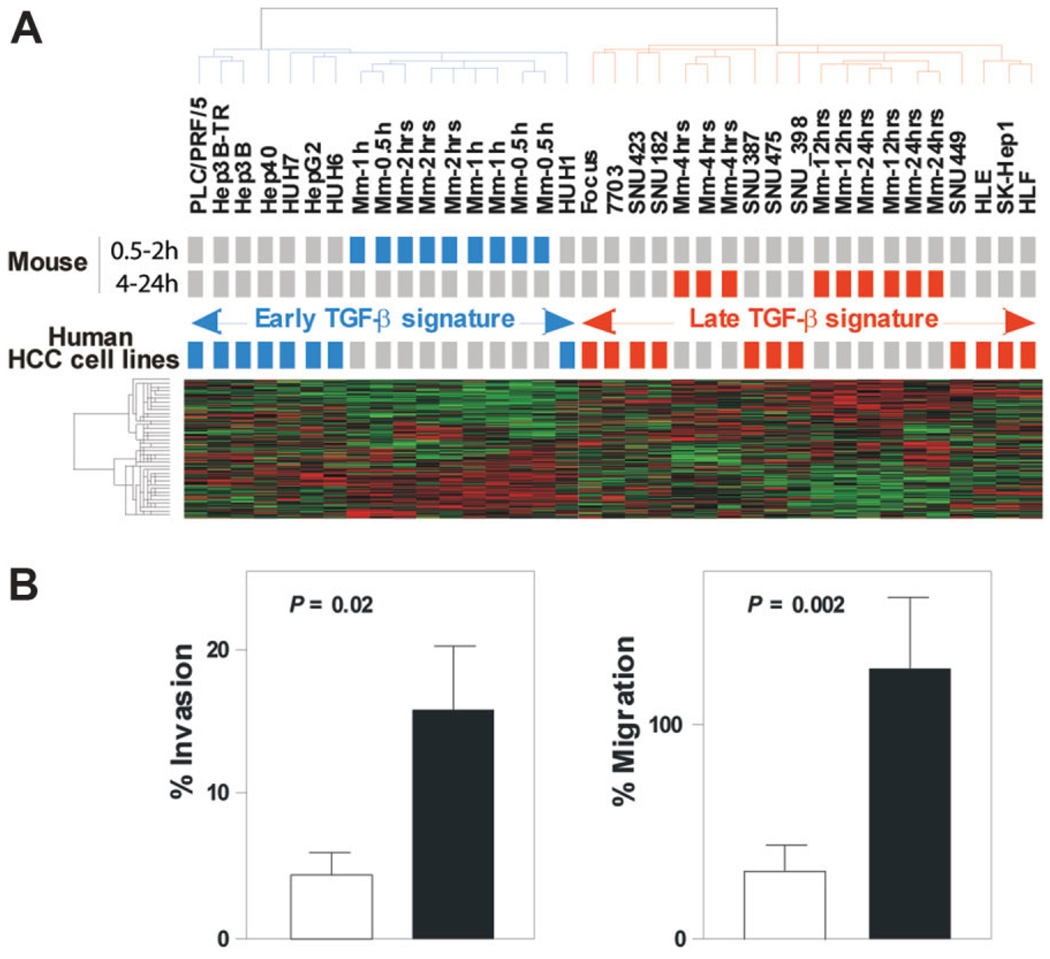
Of particular interest, all seven liver metastasis included in the Stanford microarray data set exhibited the late TGF-β signature (Supplementary Fig. 4). When the prediction models were applied to this independent data set with the HCC training set used previously, all six algorithms associated the metastatic liver lesions with the late TGF-β signature, with a prediction rate of 100% (Supplementary Table 3), suggesting that late TGF-β signature genes may be directly linked to the invasive potential of TGF-β. To test this hypothesis, early and late TGF-β signatures were integrated with the gene expression profiles of 19 human HCC-derived cell lines that are known to exhibit variable tumorigenic and invasive phenotypes.27 Remarkably, early and late TGF-β signatures successfully discriminated two subtypes of HCC cell lines (Fig. 3A) with a prediction rate ranging from 89% to 100% (Supplementary Table 4). A functional invasion assay confirmed that cell lines associated with the late TGF-β signature exhibited a more invasive phenotype than those associated with the early TGF-β signature (Fig. 3B). Moreover, gene expression analysis revealed that HCC cell lines with the late TGF-β signatures expressed genes characteristic of TGF-β-induced metastasis and EMT (e.g., matrix metalloproteinase [MMP] 1, Snail1, vimentin [VIM]) (Supplementary Fig. 5). Together, these results strongly indicate that early and late TGF-β gene signatures may represent a reliable indicator for prognosis in human liver cancer.
Fig. 3.
Early and late mouse TGF-β gene expression signatures discriminate two subtypes of human HCC-derived cell lines with distinct invasiveness behaviors. (A) Dendrogram and heat-map overview of early and late mouse TGF-β signatures integrated with the gene expression profiles of 19 human HCC-derived cell lines. Clustering was based on the expression of 249 orthologous genes and the data are presented in a matrix format in which rows and columns represent genes and samples, respectively. Cell lines that cluster with mouse samples corresponding to WT hepato-cytes challenged with TGF-β for 0.5 to 2 hours (early signature) or for 4 to 24 hours (late signature) are colored in blue and red, respectively. (B) Early and late TGF-β signatures predict invasiveness of HCC-derived cell lines. Invasive and migrating activity of HCC cell lines was measured by using the BD BioCoat Matrigel Invasion Chambers as described in Materials and Methods. Invasion and migration of HCC cell lines defined by the late TGF-β signature (black column) were higher than those defined by the early TGF-β signature (white column). Columns, mean; bars, standard error of the mean (SEM), n = 3.
Early and Late TGF-β Signatures Predict Different Clinical Outcomes
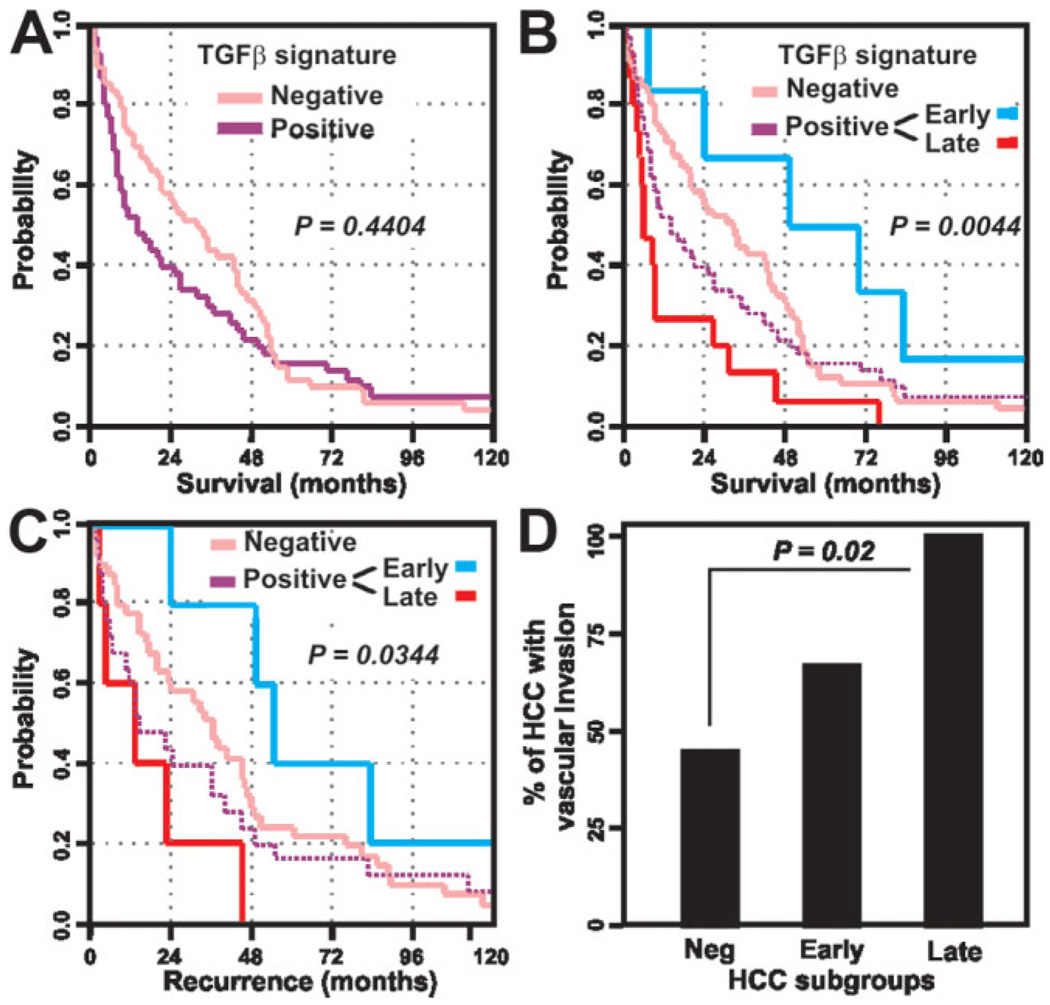
To evaluate the clinical significance of early and late TGF-β signatures in the molecular classification of HCC, we then compared the distribution of several clinical and pathological variables between HCC harboring early, late, or negative TGF-β signatures. The three subtypes of HCC were similar with respect to the individuals’ gender, age, presence of cirrhosis in surrounding tissues, tumor size, Edmondson grade, and plasma level of α-fetoprotein (Supplementary Table 5). Early and late TGF-β signatures were found to be significantly associated with patient survival and recurrence. Although no significant differences were observed between HCC included in the TGF-β-positive and TGF-β-negative clusters (Fig. 4A), Kaplan-Meier plots and logrank statistics revealed that within the TGF-β-positive cluster, patients with late TGF-β signature showed a significantly (P < 0.005) shortened mean survival time (16.2 ± 5.3 months) compared to the patients with early (60.7 ± 16.1 months) or negative (37.3 ± 4.3 months) TGF-β signatures (Fig. 4B; Supplementary Table 5). As an independent parameter of prognosis evaluation, we also compared the recurrence rate in the three subgroups of HCC. In accordance with poor prognosis demonstrated by survival analysis, patients with late TGF-β signature suffered recurrence significantly (P < 0.05) earlier (18.1 ± 2.7 months) than patients with an early (68.2 ± 17.6 months) or negative (43.2 ± 16.6 months) TGF-β signature (Fig. 4C; Supplementary Table 5). Evaluation of the vascular invasion rate in the subgroups of HCC defined by the TGF-β signatures indicated that the late TGF-β signature is also associated with an invasive phenotype (Fig. 4D; Supplementary Table 5), recapitulating the results obtained in HCC cell lines. Interestingly, a significant association between hepatitis B virus (HBV) infection and the late TGF-β signature was also observed (Supplementary Table 5). Thus, the late TGF-β signature is revealed as an indicator of bad prognosis characterized by short patient survival and an aggressive tumor phenotype. These observations confirmed the presence of clinically relevant subtypes of human HCC within the TGF-β-positive cluster, which were defined by early and late TGF-β signatures.
Fig. 4.
Clinical relevance of the HCC subtypes defined by the TGF-β signatures. (A) Kaplan-Meier plots and log-rank statistics analysis of overall survival do not reveal significant differences between patients defined by positive and negative TGF-β signatures. (B) Within the TGF-β-positive group (dashed purple line) the subtypes of HCC defined by early (blue line) and late (red line) TGF-β signatures show marked differences in the overall survival of patients. (C) Similar results are obtained by the analysis of patient recurrence. (D) Vascular invasion rate in HCC defined by negative, early, and late TGF-β signatures.
Early and Late TGF-β Signatures Predict Clinical Outcome in Lung Cancer
Finally, early and late TGF-β signatures were integrated with gene expression profiles obtained from human lung adenocarcinomas.28 Based on the expression of 174 orthologous genes, hierarchical clustering analysis successfully identified two distinct subtypes of lung tumors that harbored early and late mouse TGF-β signatures (Supplementary Fig. 6A). Remarkably, when applied to patients with lung adenocarcinomas, Kaplan-Meier plots and log-rank statistics recapitulated the results obtained for patients with HCC. For both liver and lung cancer, patients with late TGF-β signature showed a significantly shortened mean survival time compared to the patients with early TGF-β signature (Supplementary Fig. 6B). Thus, early and late TGF-β signatures identified in this study possess a predictive power for tumors other than HCC.
Discussion
Patients with liver cancer have a highly variable clinical course, indicating that HCC may comprise several biologically distinctive subtypes reflecting a molecular heterogeneity of tumors.4,5 In this study, we explored the potential of TGF-β gene expression signature to refine the diagnosis and prognostic predictions of patients with HCC. Based on the fact that TGF-β possesses both tumor-suppressive and tumor-promoting properties,4,15,17 we hypothesized that the application of a gene expression signature specific for the TGF-β signaling pathway to human HCC could identify more homogeneous and clinically relevant subgroups of patients with liver cancer.
To generate a robust TGF-β gene expression signature, we first profiled transcriptional responses to TGF-β in well-controlled experimental conditions. Specific TGF-β-responsive genes were identified through the use of primary hepatocytes that were isolated either from WT livers or livers in which the TGF-β type II receptor has been conditionally silenced specifically in hepatocytes to abolish their response to TGF-β. The direct cross-comparison of the effect of TGF-β on WT versus KO primary hepatocytes allowed the stringent detection of TGF-β target genes in a well-controlled experimental environment that closely mimicked the in vivo events following TGF-β stimulation. This approach led to the discovery of functionally diverse subsets of TGF-β-responsive genes, reflecting both the suppressive and oncogenic properties of TGF-β. Then, by using an integrative functional genomics strategy,8–10 we demonstrated that the TGF-β gene expression signature established in mouse hepatocytes successfully discriminated TGF-β-positive from TGF-β-negative human HCC. More importantly, within the group of HCC characterized by the presence of the TGF-β signature, we identified two distinct subsets of tumors that preferentially expressed either early or late TGF-β-responsive genes. We established further validation of the discriminatory power of the temporal pattern of activation of TGF-β-responsive genes in an independent cohort of 104 HCC samples. In addition, seven liver metastases (from colon cancer) exhibited the late TGF-β signature. Also, the late TGF-β signature accurately identified HCC cell lines according to the degree of invasiveness. Comparison of clinical variables revealed that the subtypes of HCC defined by early and late TGF-β signatures showed significant differences in survival and recurrence. Notably, patients with the late TGF-β signature had a considerably shortened mean survival time and increased tumor recurrence compared to the patients with the early TGF-β signature.
The mechanisms by which the late TGF-β signature genes support the more aggressive tumor phenotype are not fully elucidated. Functional analysis of genes differentially expressed between human HCC harboring early versus late TGF-β signature revealed distinct expression programs (Supplementary Fig. 7–Supplementary Fig. 9). Although the induction of a cytostatic program by TGF-β is a well-described process in the control of tumor progression,16 the fluctuations in the expression levels of cyclin-dependent kinase inhibitors such as p21, p15, or p16 did not reach statistical significance based on our stringent criteria. However, one of the most prominent and significant characteristics of HCC displaying the late TGF-β signature was overexpression of positive cell-cycle regulators (e.g., cyclins and cyclin-dependent kinases) (Supplementary Fig. 7). Furthermore, the comparison with previously published gene expression profiles established a direct association of late TGF-β signature with genes that mediate metastasis.29 Overexpression of genes involved in angiogenesis (e.g., hypoxia-inducible factor 1, alpha subunit [HIF1A]), vascular endothelial growth factor [VEGF]) and EMT (e.g., MMP1, VIM) was consistent with the more aggressive phenotype of HCC harboring the late TGF-β signature (Supplementary Fig. 7). In addition, down-regulation of genes characteristic for liver-specific functions (e.g., glucose and lipid metabolism, or detoxication) (Supplementary Fig. 8), and genes involved in antioxidant response (e.g., catalase and superoxide dismutase 1) (Supplementary Fig. 9) suggests that HCC defined by the late TGF-β signature are less differentiated and more sensitive to oxidative damage.
Of particular interest, we found a significant association between HBV infection and the late TGF-β signature (Supplementary Table 5). Although still debated, it has been suggested that if HBV DNA integration into the host genome occurs nonrandomly, then HBV may exert cis-effects on the activation of TGF-β-responsive genes.30 On the other hand, the products of HBV genes may exert trans-effects directly or indirectly on the cellular genes involved in tumor development, including those associated with the TGF-β signaling pathway. Supporting this hypothesis, the HBV-encoded HBx oncoprotein has been shown not only to enhance TGF-β signaling by potentiating the nuclear translocation of Smads31 but also to directly trans-activate TGF-β-promoter as well as TGF-β-responsive genes.31,32 HBx has been also found to induce MMPs33,34 and repress E-cadherin expression by inducing methylation-mediated promoter inactivation,35 two hallmarks of TGF-β-induced EMT and tumor cell invasion. Together, the data support the idea that HBx may provide a selective TGF-β -driven advantage for acquisition of metastatic properties in a more aggressive subtype of HCC associated with the late TGF-β signature.
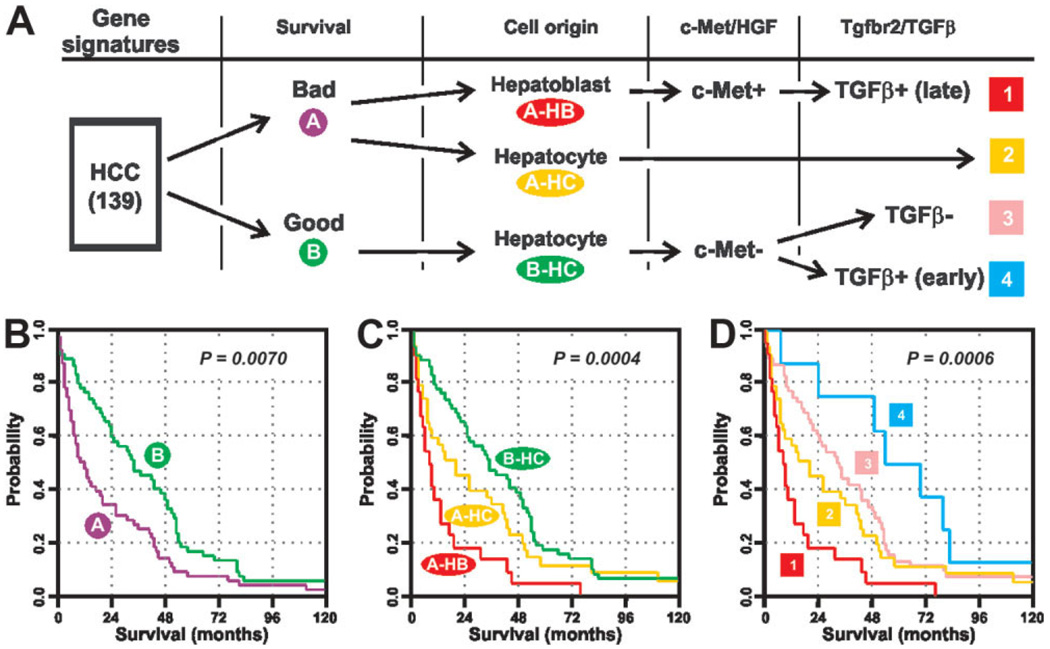
The results of this study confirm the power of the comparative and integrative functional genomic strategy to uncover subclasses of HCC and the molecular basis underlying the biological differences between the subclasses. Integration of several gene expression signatures allowed refinement of the molecular stratification of liver tumors into several distinct homogeneous subclasses that showed significant differences in biological properties and clinical outcome (Fig. 5). Thus, the use of gene expression signature derived from hepatic progenitor cells8 led to the identification of two new clinically relevant subtypes of tumors harboring either hepatoblast (A-HB) or hepatocyte (A-HC) signature within a previously identified HCC group A characterized by poor survival (Fig. 5A–C).25 The A-HB subtype was also distinguished by a positive hepatocyte growth factor/c-Met signature and the worst patient outcome. Here we have shown that this HCC subtype also harbored a late TGF-β signature (Supplementary Table 5) associated with the induction of genes involved in cell-cycle, metastasis, and angiogenesis, along with the repression of gene characteristics of well-differentiated hepatocytes (Supplementary Fig. 7–Supplementary Fig. 9). The A-HB c-Met+ and the late TGF-β signatures are not, due to the considerable overlap, independent. However, introduction of the new TGF-β signature identified in this study further categorized the HCC group B hepatocyte (B-HC) subtype into two novel homogeneous HCC subtypes characterized by TGF-β-positive (early) and TGF-β-negative signature (Fig. 5D). In addition, the observation that the late TGF-β signature overlaps with the HB signature strongly links a subset of the TGF-β signaling pathway with tumors possibly derived from stem/progenitor cells in the liver exhibiting the worst patient outcome.
Fig. 5.
Comparative and integrative functional genomic strategy improves the molecular prognostication of liver cancer. (A) Stratification of a cohort of 139 cases of human HCC based on the successive integration of gene expression signatures covering relevant genes with respect to survival (A, bad prognosis versus B, good prognosis), cell origin (HB, hepatoblast versus HC, hepatocyte), c-Met/hepatocyte growth factor (HGF), and Tgfbr2/TGF-β signaling pathways. (B-D) Kaplan-Meier plots and log-rank statistics demonstrate the difference in the overall survival between individuals with distinct subtypes of HCC defined by the successive integration of gene expression signatures.
In conclusion, we have discovered gene sets embedded in the TGF-β signaling pathway that can be used to identify clinically relevant subgroups of HCC patients which differ greatly in term of survival. Similar data were obtained for patients with lung adenocarcinomas indicating a general applicability of the TGF-β gene expression signature in the molecular prognostication of cancer. In the future, integration of multiple gene expression signatures specific for pathways or processes which are known to contribute to cancer development will greatly advance the molecular classification of tumors and provide a new basis for development of predictive personalized care based on targeted therapy.
Supplementary Material
Acknowledgment
Supported in part by the Intramural Research Program of the Center for Cancer Research.
We thank Dr. S. Karlsson for kindly providing us with the Tgfbr2fl/fl mouse strain and Dr. E.A. Conner for her comments and critical review of the manuscript. C.C. dedicates this work to the memory of Dr. Jean-Philippe Salier, an outstanding researcher, a great mentor, and a beloved friend.
Abbreviations
- B-HC
HCC group B hepatocyte subtype
- c-Met
mesenchymal epithelial transition factor, receptor for hepatocyte growth factor
- EMT
epithelial-mesenchymal transition
- HB
hepatoblast
- HBV
hepatitis B virus
- HBx
HBV-encoded oncoprotein
- HCC
hepatocellular carcinoma
- HGF
hepatocyte growth factor
- KO
knockout
- MMP
matrix metalloproteinase
- SMAD
small mothers against decapentaplegic
- TGF-β
transforming growth factor beta
- TGFBR
serine-threonine kinases receptor
- WT
wild-type
Footnotes
Potential conflict of interest: Nothing to report.
Supplementary material for this article can be found on the Hepatology Web site (http://interscience.wiley.com/jpages/0270-9139/suppmat/index.html).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.El Serag HB. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology. 2004;127(5 Suppl 1):S27–S34. doi: 10.1053/j.gastro.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Boix L, Sala M, Llovet JM. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 7.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127(Suppl 1):S51–S55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology. 2006;43(2 Suppl 1):S145–S150. doi: 10.1002/hep.21063. [DOI] [PubMed] [Google Scholar]
- 10.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 13.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 14.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 15.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 17.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 18.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesen-chymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 19.Leveen P, Larsson J, Ehinger M, Cilio CM, Sundler M, Sjostrand LJ, et al. Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood. 2002;100:560–568. doi: 10.1182/blood.v100.2.560. [DOI] [PubMed] [Google Scholar]
- 20.Oe S, Lemmer ER, Conner EA, Factor VM, Leveen P, Larsson J, et al. Intact signaling by transforming growth factor beta is not required for termination of liver regeneration in mice. HEPATOLOGY. 2004;40:1098–1105. doi: 10.1002/hep.20426. [DOI] [PubMed] [Google Scholar]
- 21.Coulouarn C, Gomez-Quiroz LE, Lee JS, Kaposi-Novak P, Conner EA, Goldina TA, et al. Oncogene-specific gene expression signatures at preneo-plastic stage in mice define distinct mechanisms of hepatocarcinogenesis. HEPATOLOGY. 2006;44:1003–1011. doi: 10.1002/hep.21293. [DOI] [PubMed] [Google Scholar]
- 22.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 24.Giudice A, Montella M. Activation of the Nrf2-ARE signaling pathway: a promising strategy in cancer prevention. Bioessays. 2006;28:169–181. doi: 10.1002/bies.20359. [DOI] [PubMed] [Google Scholar]
- 25.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. HEPATOLOGY. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JS, Thorgeirsson SS. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. HEPATOLOGY. 2002;35:1134–1143. doi: 10.1053/jhep.2002.33165. [DOI] [PubMed] [Google Scholar]
- 28.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pa-cyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami Y, Saigo K, Takashima H, Minami M, Okanoue T, Brechot C, et al. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162–1168. doi: 10.1136/gut.2004.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DK, Park SH, Yi Y, Choi SG, Lee C, Parks WT, et al. The hepatitis B virus encoded oncoprotein pX amplifies TGF-beta family signaling through direct interaction with Smad4: potential mechanism of hepatitis B virus-induced liver fibrosis. Genes Dev. 2001;15:455–466. doi: 10.1101/gad.856201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo YD, Ueda H, Park K, Flanders KC, Lee YI, Jay G, et al. Regulation of transforming growth factor-beta 1 expression by the hepatitis B virus (HBV) X transactivator. Role in HBV pathogenesis. J Clin Invest. 1996;97:388–395. doi: 10.1172/JCI118427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lara-Pezzi E, Gomez-Gaviro MV, Galvez BG, Mira E, Iniguez MA, Fresno M, et al. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygen-ase-2 expression. J Clin Invest. 2002;110:1831–1838. doi: 10.1172/JCI200215887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou DP, Tao YM, Tang FQ, Yang LY. The hepatitis B virus X protein promotes hepatocellular carcinoma metastasis by upregulation of matrix metalloproteinases. Int J Cancer. 2007;120:1208–1214. doi: 10.1002/ijc.22452. [DOI] [PubMed] [Google Scholar]
- 35.Lee JO, Kwun HJ, Jung JK, Choi KH, Min DS, Jang KL. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyl-transferase 1. Oncogene. 2005;24:6617–6625. doi: 10.1038/sj.onc.1208827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.