Abstract
To initiate fertilization, mouse sperm bind to Ser- (O-) linked oligosaccharides located at the sperm combining site of zona pellucida glycoprotein mZP3. Apparently, the oligosaccharides are present on one or more of five Ser residues clustered in the carboxyl-terminal region of the mZP3 polypeptide. Here, each of the Ser residues, as well as an intervening Asn residue, was converted to a small, nonhydroxy amino acid by site-directed mutagenesis. Mouse embryonal carcinoma (EC) cells were then stably transfected with the wild-type and mutated mZP3 genes. In each case, transfected cells synthesized and secreted recombinant EC-mZP3 into the culture medium. The glycoproteins were partially purified and assayed for their ability to inhibit binding of sperm to ovulated eggs in vitro. As compared with wild-type EC-mZP3, mutations of Ser-329, Ser-331, or Ser-333 had no effect on sperm receptor activity. Mutation of Asn-330, a potential N-linked glycosylation site, also had no effect on sperm receptor activity. On the other hand, mutation of either Ser-332 or Ser-334, or mutation of Ser-332, Ser-333, and Ser-334, resulted in complete inactivation of EC-mZP3 as a sperm receptor. These results suggest that Ser-332 and Ser-334, residues conserved in mouse, hamster, and human ZP3, are essential for sperm receptor activity.
The plasma membrane of mouse eggs is surrounded by a thick extracellular coat, the zona pellucida (ZP), that consists of three glycoproteins, called mZP1, mZP2, and mZP3 (1). During fertilization, free-swimming mouse sperm recognize and bind to specific Ser- (O-) linked oligosaccharides located close together in the carboxyl-terminal region of the mZP3 polypeptide (called the “sperm combining site”) (2, 3). Although not a great deal is known about the structure of mZP3 O-linked oligosaccharides that are recognized by sperm, it is clear that they are essential for sperm binding and induction of cellular exocytosis, the acrosome reaction (4).
Mouse embryonal carcinoma (EC) cells (F9), stably transfected with the mZP3 gene fused to a constitutive mouse promoter (pgk-1), synthesize and secrete an active form of EC-mZP3 into the culture medium (5, 6). On the other hand, EC cells transfected with mZP3 in which five Ser residues, Ser-329, -331, -332, -333, and -334, are converted to small, nonhydroxy amino acids by site-directed mutagenesis, synthesize and secrete an inactive form of EC-mZP3 (7). These and other results (8, 9) strongly suggest that one or more of five Ser residues encoded by mZP3 exon 7 carry oligosaccharides recognized by sperm.
Here we mutated Ser-329, -331, -332, -333, and -334, as well as Asn-330 individually by site-directed mutagenesis of mZP3, produced stably transfected EC cell lines carrying the mutated genes, and analyzed wild-type and mutated forms of recombinant EC-mZP3 secreted by EC cells. The results suggest that only two of the five Ser residues, Ser-332 and Ser-334, are essential for sperm receptor activity (i.e., binding of sperm to mZP3). Interestingly, both of these residues are conserved in mouse, hamster, and human ZP3 (10–13). A preliminary report has appeared describing some of these results (14).
MATERIALS AND METHODS
Construction of mZP3 Genes Containing Ser Mutations.
Plasmids containing single as well as multiple Ser or other amino acid residue mutations at the mZP3 combining-site (Fig. 1) were constructed essentially as described (7). Plasmid DNA carrying mutated mZP3 sequences were produced by annealing complementary oligonucleotides, digesting them with PstI and AatII, and then ligating the DNA to pZP3BHId. The complementary oligonucleotides were designed such that each set of oligonucleotides generated either single or triple amino acid residue changes located from Ser-329 to Ser-334 in exon 7 of mZP3 (11, 12). Sequencing of the plasmids confirmed that the predicted changes had occurred. The resulting plasmids are designated as the following: pZP3BHI-[Ser-329], -[Ser-331], -[Ser-332], -[Ser-333], -[Ser-334], and -[Asn-330], representing mutations at Ser-329, -331, -332, -333, -334, and Asn-330 of the mZP3 polypeptide, respectively. pZP3BHI-[Ser-332–334] contains a triple mutation at Ser-332, -333, and -334. Each of the above plasmids was digested with HindIII and XbaI. A 1.55-kb HindIII–XbaI fragment was identified on gels, isolated, and ligated in the presence of a 15-mer XbaI–NotI linker to an 11.5-kb HindIII–NotI fragment resulting from partial HindIII digestion of NotI-linearized pPGK/mZP3. The seven constructs produced are designated as pPGK/mZP3-[Ser-329], -[Ser-331], -[Ser-332], -[Ser-333], -[Ser-334], -[Ser-332–334], and -[Asn-330]. In addition, a construct in which all five Ser residues were mutated to nonhydroxy amino acids, pPGK/mZP3-[Ser-329–334], was used as described (7).
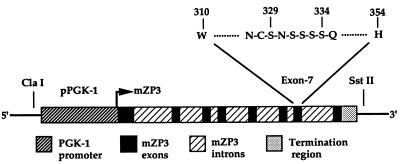
Figure 1.
Schematic diagram of the PGK/mZP3 recombinant gene used to generate stably transfected EC-mZP3 cell lines. pPGK represents the mouse phosphoglycerate kinase-1 promoter region. Restriction enzymes ClaI and SstII were used to generate a linearized DNA fragment for electroporation. Arrow indicates the transcriptional start site on the PGK-1 promoter. The sites of mutagenesis in exon 7 of the mZP3 gene are indicated as amino acids 329–334.
Production of Transfected EC Cell Lines.
EC cells were cultured and EC cell lines harboring pPGK/mZP3 recombinant genes were generated and screened essentially as described (5–7). Briefly, plasmids of pPGK/mZP3 and pPKJ-1 linearized by appropriate restriction enzyme digestions were combined at an 8:1 molar ratio for subsequent transfections of EC cells by electroporation (Gene Pulser; 400 V/cm, 500 μF; Bio-Rad). Individual G418-resistant colonies were selected at 0.5 mg/ml G418 and the cells were amplified to a suitable size for isolation of expressing cell lines. Cell lysates were prepared from 2 × 105 cells essentially as described (5–7). Protein from lysates was subjected to SDS/PAGE and recombinant EC-mZP3 was detected by Western immunoblotting by using a polyclonal goat anti-mZP3 IgG (Pocono Rabbit Farms, Canadensis, PA), followed by a rabbit anti-goat IgG-alkaline phosphatase conjugate (Bio-Rad) and nitroblue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indolyphosphate p-toluidine salt (BCIP) (GIBCO/BRL), essentially as described (7). Two to three cell lines that represented each mZP3 gene construct, which gave the highest levels of secreted EC-mZP3 in the culture medium, were chosen for production of recombinant glycoprotein. Cells expressing recombinant EC-mZP3 were cultured overnight in serum-free medium and cell supernatants were collected by passing medium through a 0.22 μm filter (Corning).
Purification of Recombinant EC-mZP3.
Purification of EC-mZP3 by HPLC on a size-exclusion column (Bio-Sil SEC-250, 600 × 25 mm; Bio-Rad) was carried out essentially as described (5–7). Samples were quantified by Western immunoblotting and laser densitometry (LKB UltroScan-XL). Briefly, known amounts of egg mZP3, determined spectrophotometrically, were immunoblotted following SDS/PAGE together with unknown amounts of EC-mZP3. The density of stained bands with known amounts of egg mZP3 was compared with that of EC-mZP3 by laser scanning of the blots. EC-mZP3 concentration was estimated from a standard curve.
In certain experiments, wild-type EC-mZP3 (EC-mZP3-[wt]) and EC-mZP3-[Ser-332] were purified from culture medium by immunoaffinity chromatography by using a goat anti-mouse IgG (anti-mZP3) column, prepared according to the supplier’s instructions (ImmunoPure IgG Orientation kit; Pierce). Culture medium (≈18 ml) was applied to a 1-ml column equilibrated with PBS (pH 7.5). The flow rate was maintained at ≈5 ml/hr. After extensive washing with PBS, the protein was eluted from the column either with elution buffer provided by the supplier (Pierce) or with 0.2 M glycine (pH 3.2) containing 0.5 M NaCl and 6% glycerol. The eluted fractions were neutralized with 1 M Tris⋅HCl (pH 8.0). Fractions containing the protein peak were pooled, concentrated, and the buffer exchanged with 0.2 M phosphate (pH 7.2). The concentrated protein was then subjected to HPLC on a size-exclusion column and protein concentration determined spectrophotometrically. HPLC- and immunoaffinity-purified EC-mZP3 was dialyzed against 8 M urea and then extensively against distilled water before assaying sperm receptor activity.
Assaying Sperm Receptor Activity.
Assays for sperm receptor activity were carried out in vitro by using gametes and embryos obtained from randomly bred, Swiss albino mice (CD-1; Charles River Breeding Laboratories), essentially as described (15–17). In most cases, mouse sperm (capacitated in M199-M at 37°C for 1 hr) in 20–30 μl of culture medium were incubated in the presence of EC-mZP3 (≈10–15 ng/μl, final concentration), egg mZP3 (≈10–15 ng/μl, final concentration), or M199-M alone for 15 min at 37°C in a humidified atmosphere of 5% CO2/95% air. In some cases, capacitated sperm were incubated in the presence of higher concentrations of egg mZP3 and EC-mZP3 (≈20–50 ng/μl, final concentration). Then, ovulated eggs and two-cell embryos were added to the cultures and the incubation was continued for an additional 30–40 min. At the end of the incubation, eggs and two-cell embryos were washed, fixed, and the number of sperm bound per egg in the largest plane of focus was determined by light microscopy, essentially as described (15–17).
RESULTS
Experimental Rationale.
Previously, Wassarman and colleagues provided experimental evidence for the involvement of one or more of five Ser residues of mZP3, Ser-329, -331, -332, -333, and -334, in sperm binding (7). This portion of polypeptide was chosen for site-directed mutagenesis because previous experiments revealed that proteolysis of this region resulted in inactivation of mZP3 in vitro (8, 9) and that antibodies directed specifically against this region prevented binding of sperm to eggs both in vivo (18) and in vitro (ref. 8, and S. Mortillo and P.M.W., unpublished results). Apparently, this is the only region of mZP3 polypeptide that carries O-linked oligosaccharides essential for sperm binding and induction of the acrosome reaction. To characterize this region of mZP3 polypeptide in more detail, we constructed mZP3 genes carrying single and multiple mutations in this portion of the polypeptide (amino acids 329–334). A description of the eight mutations that were analyzed in stably transfected EC cells is presented in Table 1. The five Ser residues, Ser-329, -331, -332, -333, and -334, were converted as individual mutations to Gly, Ala, or Val residues. In addition, Ser-332, -333, and -334 were converted in a triple mutation to Gly or Ala residues. Finally, Asn-330, a potential N-linked glycosylation site (i.e., part of the consensus sequence Asn-X-Ser/Thr, where X is any amino acid other than Pro; refs. 19 and 20), was converted to an Ala residue. EC-mZP3 carrying each of these seven mutations, together with a mutant in which all five Ser residues were converted to Gly, Ala, or Val (7), were tested for sperm receptor activity in vitro and compared with that of wild-type EC-mZP3 (EC-mZP3-[wt]).
Table 1.
Summary of site-directed mutagenesis of EC-mZP3
| EC-mZP3 designation | Position of EC-mZP3 mutation* |
|---|---|
| 329-330-331-332-333-334 | |
| EC-mZP3-[wt] | -Ser-Asn-Ser-Ser-Ser-Ser- |
| EC-mZP3-[Ser-329] | -Ala-Asn-Ser-Ser-Ser-Ser- |
| EC-mZP3-[Asn-330] | -Ser-Ala-Ser-Ser-Ser-Ser- |
| EC-mZP3-[Ser-331] | -Ser-Asn-Val-Ser-Ser-Ser- |
| EC-mZP3-[Ser-332] | -Ser-Asn-Ser-Gly-Ser-Ser- |
| EC-mZP3-[Ser-333] | -Ser-Asn-Ser-Ser-Ala-Ser- |
| EC-mZP3-[Ser-334] | -Ser-Asn-Ser-Ser-Ser-Ala- |
| EC-mZP3-[Ser-332-334] | -Ser-Asn-Ser-Gly-Ala-Ala- |
| EC-mZP3-[Ser-329-334] | -Ala-Asn-Val-Gly-Ala-Ala- |
Mutated amino acids are underlined.
Characterization of Transfected EC Cell Lines.
To select cell lines that synthesized and secreted recombinant EC-mZP3, EC cell lysates and culture medium collected from candidate colonies were screened by Western immunoblotting by using a goat anti-mZP3 IgG (see Materials and Methods). From the EC cell colonies for each mutation that tested positive for EC-mZP3, the two to three lines that produced the highest levels of secreted EC-mZP3 were chosen for further analysis. Recombinant glycoproteins were partially purified from the culture media by HPLC on a size-exclusion column and were then quantitated and tested for sperm receptor activity.
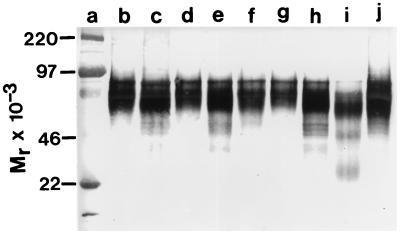
As seen in Fig. 2, the stably transfected EC cells secreted relatively large amounts of wild-type and mutated EC-mZP3 into the culture medium. The average apparent Mr (range of molecular weights) of EC-mZP3 for each of the recombinants were as follows: EC-mZP3-[wt], 79 kDa (67–91); -[Ser-329], 73 kDa (62–84); -[Ser-331], 79 kDa (67–91); -[Ser-332], 75 kDa (62–88); -[Ser-333], 79 kDa (67–91); -[Ser-334], 77 kDa (66–88); -[Ser-332–334], 74 kDa (61–88); -[Ser-329–334], 67 kDa (59–76); -[Asn-330], 78 kDa (66–91). Because mZP3 is synthesized as an ≈44 kDa Mr polypeptide (10, 12), it is apparent that the wild-type and mutant forms of EC-mZP3 are glycosylated. The Mr of EC-mZP3-[Ser-329] is significantly lower than that of EC-mZP3-[wt] and is probably attributable to elimination of the consensus N-linked glycosylation sequence, Asn327Cys328Ser329 by conversion of Ser-329 to Ala. A similar case can be made for mutant EC-mZP3-[Ser-329–334], in which Ser-329 was converted to Ala and disrupted the consensus sequence. Overall, the average apparent Mr remains about the same as EC-mZP3-[wt] (79 kDa) for four of the mutations (77–79 kDa) and is decreased significantly for the other four mutations (67–75 kDa).
Figure 2.
Western immunoblot analysis of wild-type and mutated forms of EC-mZP3. Stably transfected cell lines were cultured overnight in serum-free medium, and concentrated culture medium was subjected to HPLC purification on a size-exclusion column. Fractions containing EC-mZP3 were identified by Western immunoblotting by using a goat polyclonal antiserum directed against mZP3 (anti-mZP3). These fractions were pooled, dialyzed, and quantitated by Western immunoassays and by spectrophotometry. Shown are HPLC-purified EC-mZP3 samples (≈200–300 ng) probed with anti-mZP3 and stained as described. Lanes: a, size standards; b, EC-mZP3-[wt]; c, EC-mZP3-[Ser-329]; d, EC-mZP3-[Ser-331]; e, EC-mZP3-[Ser-332]; f, EC-mZP3-[Ser-333]; g, EC-mZP3-[Ser-334]; h, EC-mZP3-[Ser-332–334]; i, EC-mZP3-[Ser-329–334]; and j, EC-mZP3-[Asn-330].
Analysis of Recombinant EC-mZP3 Sperm Receptor Activity.
In vitro competition assays were carried out to assess the ability of different forms of recombinant EC-mZP3 to interfere with binding of sperm to ovulated eggs. Capacitated sperm were treated with HPLC-purified or immunoaffinity-purified wild-type and mutated EC-mZP3, ovulated eggs and two-cell embryos were added, and the extent of sperm binding was determined by light microscopy (refs. 15–17; see Materials and Methods).
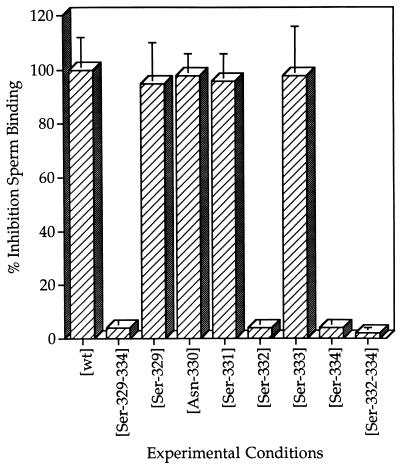
Whereas EC-mZP3-[wt] at a concentration of ≈10–15 ng/μl inhibited binding of sperm to eggs, EC-mZP3-[Ser-329–334] at the same concentration had virtually no effect on sperm binding (Fig. 3). This result is consistent with previous findings (7) and with the proposal that sperm recognize and bind to O-linked oligosaccharides located in the carboxyl-terminal region of mZP3 polypeptide. On the other hand, the single amino acid mutations EC-mZP3-[Ser-329], -[Ser-331], -[Ser-333], and -[Asn-330], also at a concentration of ≈10–15 ng/μl, were just as effective as EC-mZP3-[wt] at inhibiting binding of sperm to eggs in vitro (Fig. 3). The findings suggest that these four amino acid residues are not essential for mZP3 sperm receptor activity.
Figure 3.
Sperm receptor activity expressed as percent inhibition of sperm binding. Assays for sperm binding activity were carried out in vitro, essentially as described, by using gametes and embryos obtained from randomly bred, Swiss albino mice (CD-1; Charles River Breeding Laboratories). Mouse sperm (capacitated in M199-M at 37°C for 1 hr) in 20–30 μl of medium were incubated for 15 min in the presence of EC-mZP3 (≈10–15 ng/μl, final concentration), egg mZP3 (≈10–15 ng/μl, final concentration), or M199-M alone. Then, ovulated eggs and two-cell embryos were added to the cultures and the incubation was continued for an additional 30–40 min. At the end of the incubation, eggs and two-cell embryos were washed, fixed, and the number of bound sperm per egg was determined by light microscopy (dark field). Shown is the average percent inhibition of sperm binding for eight mutated forms of EC-mZP3. These values were calculated by setting the average value for the number of sperm bound per egg in the presence of EC-mZP3-[wt] at 100% inhibition. The values represent the average of at least three separate experiments with each form of mutated EC-mZP3. The SD of the mean for all test samples ranged from ±2 to ±9%.
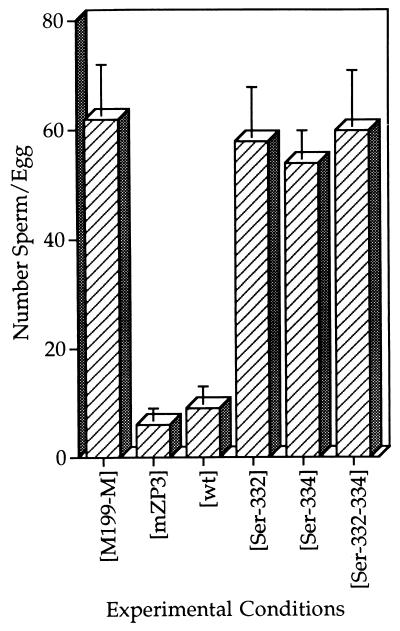
Only two of the single amino acid mutations had a significant effect on EC-mZP3 sperm receptor activity. EC-mZP3-[Ser-332] and Ser-[Ser-334], at concentrations of ≈10–30 ng/μl, failed to inhibit binding of sperm to eggs as compared with EC-mZP3-[wt] (Figs. 3 and 4; identical results were obtained at ≈50 ng/μl). Because EC-mZP3-[Ser-332] exhibited no sperm receptor activity as compared with EC-mZP3-[wt], the mutant glycoprotein was purified by immunoaffinity chromatography followed by HPLC on a size-exclusion column, and was then tested in the competition assay. As expected, immunoaffinity-purified EC-mZP3-[Ser-332] (≈10–20 ng/μl) also exhibited insignificant levels of sperm receptor activity (0–6%) as compared with EC-mZP3-[wt] (90–100%). This also was seen with the triple mutation EC-mZP3-[Ser-332–334] at the same concentrations. Such a mutant was unable to inhibit sperm binding (Figs. 3 and 4). Collectively, results with these three mutants strongly suggest that Ser-332 and Ser-334 are essential for mZP3 sperm receptor activity.
Figure 4.
Sperm receptor activity expressed as number of sperm bound per egg. These assays were carried out by using test samples at a final concentration of ≈20–30 ng/μl, essentially as described in the legend to Fig. 3. The values represent the average of three separate experiments with each form of mutated EC-mZP3, EC-mZP3-[wt], egg mZP3, and M199-M alone. The SD of the mean for all test samples ranged from ±2 to ±10 sperm per egg.
DISCUSSION
Fertilization in mice begins with species-specific binding of sperm to ovulated eggs. Sperm recognize and bind to mZP3, one of three glycoproteins that constitute the egg ZP (2, 4, 21). In mice, mZP3 is an ≈83 kDa Mr glycoprotein that serves as a primary receptor for acrosome-intact sperm and as an acrosome reaction-inducer following the binding of sperm (1, 2, 15, 22). Sperm recognize and bind to a specific class of mZP3 O-linked oligosaccharides (9, 17, 23–26) located at the sperm combining site. Although there is evidence that these oligosaccharides have either an essential galactose (17, 25, 27) or N-acetylglucosamine (26) residue at their nonreducing terminus, results obtained with null mutant mice are not completely consistent with this evidence (28–30).
Previously, Wassarman and colleagues determined the location of the sperm combining site of mZP3 by proteolytic digestion (8, 9) and by exon swapping and site-directed mutagenesis (7). The site is located relatively close to the carboxyl-terminus of the polypeptide and is encoded by exon 7 of mZP3. A glycopeptide (≈55 kDa) derived from the carboxyl-terminus of mZP3 is about as effective an inhibitor of sperm-egg interaction and inducer of the acrosome reaction as native mZP3 (≈100 nM) (8, 9). Similarly, synthetic oligosaccharides related to O-linked oligosaccharides of mZP3 also inhibit binding of sperm to eggs, albeit at much higher concentrations (≈10 μM), but do not induce sperm to undergo the acrosome reaction in vitro (17). Thus, mZP3 polypeptide influences binding of sperm, possibly by affecting the conformation of essential O-linked oligosaccharides, and is necessary for induction of the acrosome reaction. In the latter context, small mZP3 glycopeptides and mZP3 O-linked oligosaccharides bind to the head of acrosome-intact sperm, but do not induce the acrosome reaction (1, 2).
Here we identified amino acid residues at the sperm combining-site of mZP3 that are essential for sperm receptor activity (i.e., binding of sperm). We found that conversion of either of two Ser residues, Ser-332 or Ser-324, to nonhydroxy amino acids, Gly or Ala, respectively, resulted in the loss of the ability of EC-mZP3 to inhibit binding of sperm to ovulated eggs in vitro (Figs. 3 and 4). On the other hand, conversion of several neighboring amino acids, Ser-329, Asn-330, Ser-331, and Ser-333 to Ala or Val residues, had no effect on sperm receptor activity of EC-mZP3 (Fig. 3). Thus, although there are five clustered Ser residues in this region, a situation typical of heavily O-glycosylated regions of many glycoproteins (31, 32), mutation of only two of the five affected sperm receptor activity. It is tempting to suggest that Ser-332 and Ser-334 carry oligosaccharides essential for binding of sperm to mZP3. Of course, it is possible that mutation of either Ser-332 or Ser-334 could affect glycosylation of the other Ser residue; such a situation could account for the observed effects on EC-mZP3 sperm receptor activity. Because sperm are induced to undergo the acrosome reaction only after binding to mZP3, as expected, mutation of either Ser-332 or Ser-334 also resulted in the loss of the ability of EC-mZP3 to act as an acrosome reaction-inducer in vitro.
It is of interest that Ser-332 and Ser-334, the two residues thought to be essential for mZP3 sperm receptor activity, are conserved residues in mouse, hamster, and human ZP3 (10–13). They are conserved residues in a region of polypeptide that has undergone considerable changes during the course of evolution (3, 7). It has been suggested that these changes could direct the addition of alternative oligosaccharide structures to nascent ZP3 and, in this manner, affect species specificity of sperm–egg interaction (3, 7). There is sufficient experimental evidence to suggest that amino acids flanking a Ser or Thr residue influence addition of sugars to the site (33–36). In this context, it is well documented that mouse sperm bind to hamster eggs and hamster sperm bind to mouse eggs, but that human sperm do not bind to either mouse or hamster eggs in vitro (21). If Ser-332 and Ser-334 are glycosylated in hamster and human ZP3, it is likely that the structures of the oligosaccharides differ such that hamster, but not human, ZP3 is recognized by mouse sperm in vitro.
In summary, by using site-directed mutagenesis of mZP3 we have identified two Ser residues, Ser-332 and Ser-334, at the sperm combining site that are essential for sperm receptor activity. Taken together with other evidence, it is likely that these Ser residues carry O-linked oligosaccharides that are recognized by free-swimming, acrosome-intact sperm during fertilization of ovulated eggs in vivo. Interestingly, these two Ser residues are conserved in mouse, hamster, and human ZP3, consistent with their proposed essential role in fertilization.
Acknowledgments
We are grateful to members of our laboratory for helpful advice and constructive criticism throughout the course of this research. We thank the anonymous reviewers for their positive comments and helpful suggestions.
ABBREVIATIONS
- ZP
zona pellucida
- EC
embryonal carcinoma
References
- 1.Wassarman P M. Annu Rev Biochem. 1988;57:415–442. doi: 10.1146/annurev.bi.57.070188.002215. [DOI] [PubMed] [Google Scholar]
- 2.Wassarman P M. Development (Cambridge, UK) 1990;108:1–17. doi: 10.1242/dev.108.Supplement.1. [DOI] [PubMed] [Google Scholar]
- 3.Wassarman P M, Litscher E S. Curr Top Dev Biol. 1995;30:1–19. doi: 10.1016/s0070-2153(08)60562-1. [DOI] [PubMed] [Google Scholar]
- 4.Wassarman P M. Curr Opin Cell Biol. 1995;7:658–664. doi: 10.1016/0955-0674(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 5.Kinloch R A, Mortillo S, Stewart C L, Wassarman P M. J Cell Biol. 1991;115:655–664. doi: 10.1083/jcb.115.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litscher E S, Wassarman P M. Zygote. 1996;4:229–236. doi: 10.1017/s0967199400003142. [DOI] [PubMed] [Google Scholar]
- 7.Kinloch R A, Sakai Y, Wassarman P M. Proc Natl Acad Sci USA. 1995;92:263–267. doi: 10.1073/pnas.92.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosiere T K, Wassarman P M. Dev Biol. 1992;154:309–317. doi: 10.1016/0012-1606(92)90070-w. [DOI] [PubMed] [Google Scholar]
- 9.Litscher E S, Wassarman P M. Biochemistry. 1996;35:3980–3985. doi: 10.1021/bi952722m. [DOI] [PubMed] [Google Scholar]
- 10.Kinloch R A, Roller R J, Fimiani C M, Wassarman D A, Wassarman P M. Proc Natl Acad Sci USA. 1988;85:6409–6413. doi: 10.1073/pnas.85.17.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinloch R A, Ruiz-Seiler B, Wassarman P M. Dev Biol. 1990;142:414–421. doi: 10.1016/0012-1606(90)90363-n. [DOI] [PubMed] [Google Scholar]
- 12.Ringuette M J, Chamberlin M E, Baur A W, Sobieski D A, Dean J. Dev Biol. 1988;127:287–295. doi: 10.1016/0012-1606(88)90315-6. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlin M E, Dean J. Proc Natl Acad Sci USA. 1990;87:6014–6018. doi: 10.1073/pnas.87.16.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Litscher E S, Wassarman P M. Mol Biol Cell. 1995;6:320. doi: 10.1091/mbc.6.5.577. (abstr.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bleil J D, Wassarman P M. Cell. 1980;20:873–882. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 16.Moller C C, Bleil J D, Kinloch R A, Wassarman P M. Dev Biol. 1990;137:276–286. doi: 10.1016/0012-1606(90)90254-g. [DOI] [PubMed] [Google Scholar]
- 17.Litscher E S, Juntunen K, Seppo A, Penttilä L, Niemelä R, Renkonen O, Wassarman P M. Biochemistry. 1995;34:4662–4669. doi: 10.1021/bi00014a020. [DOI] [PubMed] [Google Scholar]
- 18.Millar S E, Chamow S M, Baur A, Oliver C, Robey F, Dean J. Science. 1989;246:935–938. doi: 10.1126/science.2479101. [DOI] [PubMed] [Google Scholar]
- 19.Bause E. Biochem J. 1983;209:331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–654. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 21.Yanagimachi R. In: The Physiology of Reproduction. 2nd Ed. Knobil E, Neill J D, editors. Vol. 1. New York: Raven; 1994. pp. 189–317. [Google Scholar]
- 22.Bleil J D, Wassarman P M. Dev Biol. 1983;95:317–324. doi: 10.1016/0012-1606(83)90032-5. [DOI] [PubMed] [Google Scholar]
- 23.Florman H M, Bechtol K, Wassarman P M. Dev Biol. 1984;106:243–255. doi: 10.1016/0012-1606(84)90079-4. [DOI] [PubMed] [Google Scholar]
- 24.Florman H M, Wassarman P M. Cell. 1985;41:313–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bleil J D, Wassarman P M. Proc Natl Acad Sci USA. 1988;85:6778–6782. doi: 10.1073/pnas.85.18.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller D J, Macek M B, Shur B D. Nature (London) 1992;357:589–593. doi: 10.1038/357589a0. [DOI] [PubMed] [Google Scholar]
- 27.Johnston D S, Wright W W, Shaper J H, Hokke C H, Van den Eijnden D H, Joziasse D H. J Biol Chem. 1998;273:1888–1895. doi: 10.1074/jbc.273.4.1888. [DOI] [PubMed] [Google Scholar]
- 28.Thall A D, Maly P, Lowe J B. J Biol Chem. 1995;270:21437–21440. doi: 10.1074/jbc.270.37.21437. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q, Hasty P, Shur B D. Dev Biol. 1997;181:257–267. doi: 10.1006/dbio.1996.8444. [DOI] [PubMed] [Google Scholar]
- 30.Asano M, Furukawa F, Kido M, Matsumoto S, Umesaki Y, Kochibe N, Uwakura Y. EMBO J. 1997;16:1850–1857. doi: 10.1093/emboj/16.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadler J E. In: Biology of Carbohydrates. Ginsburg V, Robbins P W, editors. Vol. 2. New York: Wiley; 1984. pp. 199–288. [Google Scholar]
- 32.Wilson I B H, Gavel Y, von Heijne G. Biochem J. 1991;275:529–534. doi: 10.1042/bj2750529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elhammer A P, Poorman R A, Brown E, Maggiora L L, Hoogerheide J G, Kézdy F J. J Biol Chem. 1993;268:10029–10038. [PubMed] [Google Scholar]
- 34.Wang Y, Agarwal N, Eckhardt A E, Stevens R D, Hill R L. J Biol Chem. 1993;268:22979–22983. [PubMed] [Google Scholar]
- 35.Gooley A A, Williams K L. Glycobiology. 1994;4:413–417. doi: 10.1093/glycob/4.4.413. [DOI] [PubMed] [Google Scholar]
- 36.Nehrke K, Hagen F K, Tabak L A. J Biol Chem. 1996;271:7061–7065. doi: 10.1074/jbc.271.12.7061. [DOI] [PubMed] [Google Scholar]