Abstract
MicroRNAs (miRNA) are approximately 22-nucleotide non-coding RNAs that negatively regulate protein-coding gene expression in a sequence-specific manner via translational inhibition or mRNA degradation. Our recent studies showed that miRNAs exhibit genomic alterations at a high frequency and their expression is remarkably deregulated in ovarian cancer, strongly suggesting that miRNAs are involved in the initiation and progression of this disease. In the present study, we performed miRNA microarray to identify the miRNAs associated with chemotherapy response in ovarian cancer and found that let-7i expression was significantly reduced in chemotherapy-resistant patients (n = 69, P = 0.003). This result was further validated by stem-loop real-time reverse transcription-PCR (n = 62, P = 0.015). Both loss-of-function (by synthetic let-7i inhibitor) and gain-of-function (by retroviral overexpression of let-7i) studies showed that reduced let-7i expression significantly increased the resistance of ovarian and breast cancer cells to the chemotherapy drug, cis-platinum. Finally, using miRNA microarray, we found that decreased let-7i expression was significantly associated with the shorter progression-free survival of patients with late-stage ovarian cancer (n = 72, P = 0.042). This finding was further validated in the same sample set by stem-loop real-time reverse transcription-PCR (n = 62, P = 0.001) and in an independent sample set by in situ hybridization (n = 53, P = 0.049). Taken together, our results strongly suggest that let-7i might be used as a therapeutic target to modulate platinum-based chemotherapy and as a biomarker to predict chemotherapy response and survival in patients with ovarian cancer.
Introduction
Epithelial ovarian cancer (EOC) is the most frequent cause of gynecologic malignancy-related mortality in women (1). Although advances in platinum-based chemotherapy have resulted in improved survival, patients typically experience disease relapse within 2 years of initial treatment and develop platinum resistance (2). Therefore, a better understanding of the mechanisms that underlie platinum resistance, including the discovery of robust predictive biomarkers which monitor the treatment and development of combination therapy that uses platinum with resistance modulators or new molecularly targeted drugs, should allow optimized therapy, such that substantial improvements in the outlook for women with this disease can be achieved (2, 3). Nevertheless, studies in the identification of druggable targets and biomarkers for ovarian cancer have thus far mainly focused on the role of protein-coding genes, whereas our knowledge of functional noncoding genomic sequences, such as microRNAs (miRNAs), is still in its infancy. miRNAs are ∼22-nucleotide noncoding RNAs, which negatively regulate gene expression in a sequence-specific manner (4–6). The potential regulatory circuitry afforded by miRNA is enormous (4). Increasing evidence indicates that miRNAs are key regulators of various fundamental biological processes (4). In EOC, we have generated evidence that miRNA exhibits high-frequency genomic alterations (7), and that its expression is remarkably deregulated (8), strongly suggesting that miRNA is involved in the initiation and progression of this disease. Indeed, recent studies have shown that miRNAs play a critical role in tumor cells by serving as either oncogenes or tumor suppressor genes (5, 6), as well as by offering resistance to cytotoxic anticancer therapy (9–11). The current rapid advances in oligonucleotide/nanoparticle therapy create realistic optimism for the establishment of miRNAs as a new and potent therapeutic target and/or chemoresistant modulator in cancer treatment.
Let-7 is among the founding and best understood miRNAs in the Caenorhabditis elegans genome. It times seam cell terminal differentiation, possibly by acting as a regulator of multiple genes required for cell cycle and proliferation (12–15). In other organisms such as mouse, rat, and human, the let-7 family is composed of multiple members with overlapping or distinct functions (16). Eleven members of let-7 have been identified in the human genome (16). Most importantly, the let-7 family is one of the first reported tumor suppressor miRNAs in cancer, which negatively regulates the RAS and is expressed at lower levels in lung tumors than in normal lung tissue (17, 18). Reduced expression of let-7 has also been associated with shortened postoperative survival in human cancer patients (18–21). In addition, forced expression of let-7 family members is able to suppress cancer cell growth both in vitro (22–24) and in vivo (25, 26). Finally, increasing evidence indicates that the let-7 family negatively regulates numerous well-characterized oncogenic proteins, such as RAS (17, 25, 27), HMGA2 (23, 24, 27, 28), c-Myc (29), CDC25A (22), CDK6 (22), and cyclin D2 (22). Although the let-7 family has been generally shown to be a tumor suppressor gene, there have been contradictory reports that it can serve an oncogenic function. For example, Brueckner and colleagues reported that let-7a-3 hypomethylation results in enhanced tumor phenotype in colon cancer (30).
In the present investigation of miRNA signatures of human EOC by microarray, we found that let-7i is significantly reduced in chemotherapy-resistant patients and lower let-7i expression is strongly associated with shorter progression-free survival of the patients. In vitro study using various ovarian and breast cancer cell lines further confirmed that let-7i is involved in the cancer cell response to cis-platinum. Therefore, our results strongly suggest that let-7i might be used as a therapeutic target to modulate platinum-based chemotherapy and as a biomarker to predict chemotherapy response and survival in ovarian cancer patients.
Materials and Methods
Patients and specimens
All frozen ovarian cancer specimens used in this study were collected at the University of Turin, Turin, Italy. Clinical characteristics were as previously defined (7, 8) and listed in Table 1. Optimal surgical debulking was ≤1 cm of residual individual tumor nodules. Front-line chemotherapy comprised platinum, platinum-cyclophosphamide, or (after 1995) platinum-paclitaxel. Complete response to therapy was defined by normalization of physical examination, abdomino-pelvic computerized tomography (CT) scan and serum CA-125. Noncomplete response included partial response (≥50% decrease in the sum of greater tumor dimensions by CT) and no response (<50% decrease or any increase in tumor). Progression-free survival was the time between completion of chemotherapy and first recurrence (if a complete response had been achieved) or progression of disease, defined as ≥50% tumor increase by CT scan or two increasing CA-125 values. All tumors were from primary sites, and were immediately snap-frozen and stored at −80°C. Tissues were obtained after patients' written consent under a general tissue collection protocol approved by the Institutional Review Board of the University of Pennsylvania and the University of Turin.
Table 1.
Patient characteristics (N = 72)
| Characteristic | No. (%) |
|---|---|
| Age | |
| 20–29 | 1 (0.01) |
| 30–39 | 3 (0.04) |
| 40–49 | 10 (0.14) |
| 50–59 | 23 (0.32) |
| 60–69 | 20 (0.28) |
| 70–79 | 14(0.19) |
| >80 | 1 (0.01) |
| Stage | |
| III | 61 (0.85) |
| IV | 11 (0.15) |
| Grade | |
| 0 | 1 (0.01) |
| 1 | 4(0.06) |
| 2 | 12 (0.17) |
| 3 | 55 (0.76) |
| Histologic subtypes | |
| Serous | 41 (0.57) |
| Endometrial | 6 (0.08) |
| Mucinous | 7 (0.10) |
| Clear cell | 4(0.06) |
| Others | 14(0.19) |
| Debulking status* | |
| Optimal (≤1 cm) | 23 (0.32) |
| Suboptimal (>1 cm) | 48 (0.67) |
| Chemotherapy response† | |
| Complete response | 42 (0.58) |
| Noncomplete response | 27 (0.38) |
One patient not available.
Three patients not available.
Cell lines and cell culture
Ovarian (SKOV3, 2008, OVCAR10, OVCAR3), cervical (HeLa), and breast (MCF7, MDA-MB-468) cancer cell lines were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotics (Invitrogen).
RNA isolation
Total RNA was isolated from 100 to 500 mg of frozen tissue or 1 × 106 cultured cells with TRIzol reagent (Invitrogen). The quality and quantity of the isolated RNA was analyzed using a Bioanalyzer 2100 system (Agilent).
miRNA microarray
miRNA microarray was performed as previously described (8). Briefly, 5 μg of total RNA was reverse-transcribed using biotin end-labeled random-octamer oligonucleotide primer. Hybridization of biotin-labeled complementary DNA was performed on the Ohio State University miRNA microarray chip (OSU_CCC version 3.0), which contains 1,100 miRNA probes, including 326 human miRNA genes, spotted in duplicates. Often, more than one probe exists for a given mature miRNA. Additionally, there are quadruplicate probes corresponding to most pre-miRNAs. The hybridized chips were washed and processed to detect biotin-containing transcripts by streptavidin-Alexa 647 conjugate and scanned on an Axon 4000B microarray scanner (Axon Instruments).
Microarray analysis
The normalized microarray data were managed and analyzed by GeneSpring (Agilent), GenePattern,10 BRB-ArrayTools version 3.6,11 and microarray software suite 4(TM4).12 Java Treeview 1.0 (Stanford University School of Medicine, Stanford, CA) was used for tree visualization.
Stem-loop real-time reverse transcription-PCR (TaqMan miRNA assay)
Expression of mature miRNAs was analyzed by TaqMan miRNA Assay (Applied Biosystems) under conditions defined by the supplier. Briefly, single-stranded cDNA was synthesized from 5.5 ng of total RNA in a 15 μL reaction volume using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). The reactions were first incubated at 16 °C for 30 min, then at 42°C for 30 min. The reactions were inactivated by incubation at 85 °C for 5 min. Each cDNA generated was amplified by quantitative PCR using sequence-specific primers from the TaqMan MicroRNA Assays Human Panel on an Applied Biosystems 7900HT sequence detection system (Applied Biosystems). The 20 μL PCR included 10 μL of 2× Universal PCR Master Mix (no AmpErase UNG), 2 μL of each 10× TaqMan MicroRNA Assay Mix and 1.5 μL of reverse transcription product. The reactions were incubated in a 384-well plate at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min.
Retroviral transduction and stable cell line generation
The retrovirus-based human miRNA expression vector was purchased from GeneService. Retroviral vector containing human let-7i or control vector was transfected into the packing cell line PT67 (Clontech) using FuGene6 Transfection Reagent (Roche). The medium was changed 48 h posttrans-fection and the medium containing retrovirus was collected 48 h later. Human tumor cells were infected with retrovirus in the presence of 8 μg/mL of of polybrene.
Transfection of inhibitor oligos
miRIDIAN inhibitors and negative controls were purchased from Dharmacon. Cells were seeded in a 96-well or 24-well plate in antibiotic-free medium to reach a 40% to 50% confluence the next day. Twenty-four hours later, the medium was replaced prior to transfection. Transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen) following the instructions of the manufacturer. For 24-well plates, the concentration used for inhibitors was 80 nmol/L, and for 96-well plates, the concentration used for inhibitors was 66 nmol/L. Cells were incubated in the medium containing the transfection mixture for 72 h until RNA extraction (from 24-well plate) or 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (in 96-well plates) was performed.
Cis-platinum treatment
Cells were seeded in a 96-well plate in antibiotic-free medium. cis-Diamineplatinum(II) dichloride (Sigma) or mock Dulbecco's PBS alone was added into the medium at various concentrations. The MTT assay was performed 72 h post-drug addition.
MTT assay
MTT assay was performed in a 96-well plate using the Cell Proliferation Kit (I) (Roche) following the manufacturer's instructions. Four to six wells were done for each sample and experiments were repeated twice. The resulting colored solution was quantified using an Emax precision microplate reader (Molecular Devices) at 570 nm with a reference wavelength of 650 nm.
Tissue microarray
The tissue microarray was constructed as described previously (8). In brief, tumors were embedded in paraffin and 5-μm sections were stained with H&E to select representative regions for biopsies. Four core tissue biopsies were obtained from each specimen. The presence of tumor tissue on the arrayed samples was verified on H&E-stained sections. The patient material consisted of 53 primary ovarian carcinomas with serous histology only. The patients were treated at the Helsinki University Central Hospital between 2000 and 2004. Patients who became disease-free after the primary treatment (surgery and platinum-taxane-based chemotherapy) were included in the study, and disease-free survival was the time from diagnosis to relapse of the disease.
miRNA in situ hybridization and image analysis
In situ detection of miRNA expression was performed on formalin-fixed paraffin-embedded tissue microarray sections. Slides were deparaffinized in xylene series and rehydrated through an ethanol series (100% to 25%). After proteinase K digestion (30 μg/mL; Roche) for 10 min and postfixation in 4% paraformaldehyde, slides were prehybridized in hybridization solution (50% formamide, 5× SSC, 500 μg/mL yeast tRNA, 1 × Denhardt's solution) for 1 h and hybridized overnight with digoxigenin-labeled miRNA-locked nucleic acid probe (Exiqon) in hybridization solution. After stringent washes (50% formamide, 2× SSC) at hybridization temperature, chromogenic detection of signals was performed using anti-digoxigenin antibody (Roche, 1:400 dilution) and PowerVision+ Poly-HRP IHC detection kit (ImmunoVision Technologies) according to the manufacturer's instructions. Occasionally, a nuclear signal was seen most likely representing nonspecific staining as it was also seen in the negative controls. Therefore, only cytoplasmic staining (mature miRNA) of the tumor cells was recorded and classified as positive or negative without knowledge of the patient outcome.
Array-based comparative genomic hybridization
BAC clones included in the “1 Mb array” platform were recently described (7). Briefly, 4,134 clones from the CalTech A/B and RPCI-11 libraries were collected from both commercial and private sources and were mapped to build 34 of the human genomes using either an STS-marker (29%), end sequences (68%), or full sequences (3%). A minimum of two replicates per clone were printed on each slide. One microgram of tumor and reference DNA was labeled with Cy3 or Cy5, respectively (Amersham) using the BioPrime random-primed labeling kit (Invitrogen). In parallel experiments, tumor DNA and reference DNA were labeled with the opposite dye to account for differences in dye incorporation and to provide additional data for analysis. A systematic protocol was used to analyze array-based comparative genomic hybridization (aCGH) data for copy number alterations. For quality control purposes, clones demonstrating an adjusted foreground-to-background intensity ratio of <0.8 in the reference channel were removed. With dye swap data merged as input, copy number breakpoints were estimated for each sample by the Circular Binary segmentation algorithm using breakpoint significance based on 10,000 permutations. Additional analyses and visualization of aCGH data were done using the CGHAnalyzer software suite described previously.
Statistical analysis
Statistical analysis was performed using the SPSS statistics software package (SPSS). All results were expressed as mean ± SD, and P < 0.05 was used for significance. Kaplan-Meier curves were used to estimate 5-year survival rates and were compared with the use of log rank statistics.
Results
Let-7i expression is significantly reduced in patients with chemotherapy-resistant EOC
To identify miRNA expression signatures associated with resistance to chemotherapy in patients with EOC, specimens from 72 late-stage (stage III and IV) patients were initially analyzed by miRNA microarray. A total of 69 patients with well-documented chemotherapy response information were included for further biomarker identification, and all (n = 72) were used for survival analysis. The clinical characteristics of those patients are listed in Table 1. First, differences in miRNA expression between the complete response (n = 42) and noncomplete response groups (n = 27, including partial response and no response) were analyzed. It was found that 34 miRNAs were statistically different (P < 0.05) between the groups, with 24 (70.6%) miRNAs higher in the noncomplete response group and 10 miRNAs (29.4%) higher in the complete response group (Supplementary Table S1). Importantly, nine miRNAs exhibited even greater statistical significance (P < 0.015) and of those, six were higher in the noncomplete response group and three were higher in the complete response group (Fig. 1A and B). In particular, let-7i, a tumor suppressor miRNA (17, 18), was the top differential miRNA between the two groups and expressed at remarkably lower levels in the non-complete response group (expression ratio of complete response group to noncomplete response group = 9.3, P = 0.003, n = 69; Fig. 1A). To further validate this finding, we examined let-7i expression in 62 randomly selected late-stage EOC specimens by stem-loop real-time reverse transcription-PCR. Consistent with the microarray data, let-7i expression was indeed significantly reduced in the noncomplete response patients (9.1 ± 1.5 relative expression unit; let-7i/U6; n = 25) as compared with their counterparts with complete response (4.3 ± 0.7 relative expression unit; let-7i/U6; n = 37, P = 0.015; Fig. 1C). In addition, this result was further confirmed in EpCAM-positive tumor cells isolated from the ascites of late-stage ovarian cancer patients (∼13.9-fold higher in enriched tumor cells from the complete chemotherapy response patients compared with those from the noncomplete chemotherapy patients; n = 8).13 Taken together, we found that there was a distinguishable miRNA expression signature between the chemotherapy-responsive and chemotherapy-resistant EOC patients, and expression of the tumor suppressor miRNA let-7i was significantly reduced in the chemotherapy-resistant EOC patients.
Figure 1.
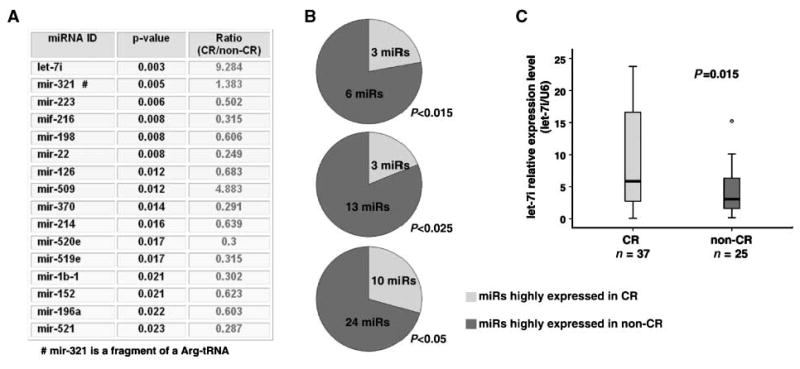
let-7i expression is significantly reduced in patients with chemotherapy-resistant EOC. A, microarray analysis of miRNA expression between complete response (CR) and noncomplete response (non-CR) ovarian cancer patients. B, differentially expressed miRNAs between complete response and noncomplete response patients at various statistical significance (P < 0.015, P < 0.025, and P < 0.05). C, validation of let-7i expression in complete response and noncomplete response patients by real-time reverse transcription-PCR.
Decreased let-7i expression increases the chemotherapy resistance of EOC cells
Studies from our group and other groups have demarcated that miRNAs are globally down-regulated in human cancers including EOC (8, 31). Those down-regulated miRNAs, such as the let-7 family (17, 18), might serve as tumor suppressor genes and their suppression can have an important effect on tumor cells, e.g., by rendering them more resistant to cytotoxic anticancer therapy (9–11). let-7i has been reported to be down-regulated in recurrent ovarian tumors compared with primary tumors (32). Therefore, to further investigate whether the above identified miRNAs are functionally involved in tumor resistance to chemotherapy, three miRNAs (let-7i, mir-321, and mir-509; Fig. 1A) that were significantly repressed in the chemotherapy-resistant tumors were focused on. mir-321, a fragment of Arg-tRNA, was excluded from our study, and both mature forms of mir-509 (mir-509-5p and mir-509-3p) were included. A total of three mature miRNAs, let-7i, mir-509-5p, and mir-509-3p were examined in EOC cell lines (2008 and SKOV3) in vitro. Endogenous miRNA expression was blocked by specific antisense oligonucleotide inhibitors. The effect on miRNA expression by the inhibitor was confirmed by stem-loop real-time reverse transcription-PCR. More than 90% of the endogenous miRNA expression was blocked by the inhibitor 48 hours posttransfection (Fig. 2). It was found that knockdown of the let-7i expression, but not that of mir-509-3p or mir-509-5p, significantly increased cell resistance to cis-platinum treatment in various EOC cell lines (2008, P = 0.004; SKOV3, P = 0.006; Fig. 2A). A similar result was also found in short-term primary cultured ovarian tumor cells (Supplementary Fig. S1). To complement this loss-of-function study, we also stably enforced let-7i expression in EOC (2008 and SKOV3) and breast (MCF7) cell lines via retroviral transduction before exposing them to serial concentrations of cis-platinum. Overexpression of let-7i in each of the above cell lines was confirmed by stem-loop real-time reverse transcription-PCR (Fig. 2C). Consistent with the loss-of-function study, overexpression of let-7i significantly increased the chemotherapy response sensitivity in vitro (Fig. 2C). Taken together, down-regulated or intrinsically reduced let-7i expression could render EOC cells more resistant to the cis-platinum treatment. Therefore, let-7i might serve as an important chemotherapy response modulator in cancer cells.
Figure 2.

let-7i expression regulates cis-platinum resistance of EOC cells. A, inhibition of let-7i, but not mir-509-3p or mir-509-5p, increased resistance to cis-platinum treatment in 2008 and SKOV3 cells. B, stem-loop real-time reverse transcription-PCR showed endogenous let-7i was significantly blocked by let-7i inhibitor. C, overexpression of let-7i by retroviral infection in 2008, SKOV3, and MCF7 cells increased their sensitivity to the cis-platinum treatment. Inset, stem-loop real-time reverse transcription-PCR showed that let-7i was stably overexpressed in EOC cell lines by retroviral transfection.
let-7i DNA copy number does not exhibit genomic alteration in human cancer
The molecular mechanism of let-7i down-regulation in patients with chemotherapy-resistant EOC is unclear. Our previous studies indicated that DNA copy number of miRNAs is highly altered in human cancer including EOC (7), and DNA copy number alteration significantly contributes to miRNA expression in cancer. For example, let-7a3 deletion was found in 31.2% of EOC specimens (n = 106), which significantly reduced let-7a3 expression in EOC (7). Therefore, we questioned whether DNA copy alteration of let-7i contributes to the reduced expression of let-7i in patients with chemotherapy-resistant EOC. In the 69 patients that were used for initial analysis of chemotherapy-associated miRNA markers, 30 were analyzed by aCGH (complete response patients, n = 20; and noncomplete response patients, n = 10). We first analyzed the genomic locus, Chr12_61-62 Mb, which contains the primary let-7i gene sequence, in these specimens. However, there was only one patient with a let-7i DNA copy number alteration in the chemotherapy response group (1/30, 3.3%), no patients with either deletion or application were found in the chemotherapy-resistant group. This indicates that unlike other let-7 family members, let-7i does not significantly exhibit DNA copy alteration in EOC. Therefore, DNA copy number alteration might not affect let-7i reduced expression in patients with chemotherapy-resistant EOC. For future confirmation of this conclusion, we expanded our aCGH study to a large collection of specimens with multiple cancer types including nine different types of human solid tumors (bladder breast, colon, lung, ovarian and pancreatic cancer, sarcoma, neuroblastoma, and melanoma; n = 1,315; Fig. 3 and Supplementary Table S2; ref. 33). Consistent with the first analysis, the DNA copy number of let-7i was found in only extremely low frequency alterations (gained three to five copies in 5% and heterogeneously deleted in 6%, <10% alteration was usually considered as the background signal of aCGH), which was significantly lower than other members of the let-7 family (e.g., let-7a-3 and let-7b deleted in 31.2% of EOC; ref. 7). These results suggested that other unknown mechanisms reduced let-7i expression in the chemotherapy-resistant patients, e.g., mutation, miRNA biogenesis pathway (34), epigenetic, or transcriptional regulation.
Figure 3.

let-7i DNA copy number does not exhibit genomic alteration in human cancer. Genomic locus harboring let-7i did not exhibit alteration in EOC (n = 106). Black, deletion; gray, amplification.
Low let-7i expression is significantly associated with shorter survival of patients with EOC
It has been reported that the expression of let-7 family is a strong prognostic marker for human cancer patients (18–21). In this study, we identified let-7i as an important predictor for chemotherapy resistance in patients with EOC. We further investigated whether let-7i could also serve as a prognostic marker in patients with EOC. To examine the correlation between let-7i expression and rapid recurrence of the disease, we first studied the let-7i expression in 72 late-stage EOC patient samples by miRNA microarray. Kaplan-Meier survival analysis indicated that low expression of let-7i was significantly associated with shorter progression-free survival of the patients as compared with the high let-7i expression group (P = 0.042, n = 72; Fig. 4A). We then validated this result by a more accurate mature miRNA quantitative method in the same sample set. Consistently, a similar result was also observed in the 62 randomly selected EOC patient samples analyzed by stem-loop real-time reverse transcription-PCR (n = 62, P = 0.001; Fig. 4B). Finally, we analyzed let-7i expression in an independent sample set using a completely different methodology—in situ hybridization. Again, we found that lower let-7i expression was significantly associated with shorter disease-free survival in 53 samples examined by in situ hybridization of tissue array (n = 53, P = 0.049; Fig. 4C). In conclusion, the above data strongly suggests that the expression level of let-7i could serve as a novel prognostic and prediction biomarker for the survival of patients with EOC.
Figure 4.

Low let-7i expression is significantly associated with shorter survival of patients with EOC. Correlation between let-7i expression and survival of EOC patients analyzed by microarray (A, progression-free survival), real-time reverse transcription-PCR (B, progression-free survival), and tissue array (C, disease-free survival).
Discussion
EOC is the most lethal gynecological malignancy in western countries (1). The role of miRNAs in ovarian cancer has recently been proposed and investigated (7, 8, 20, 21, 32, 35–37), which might offer novel strategies for prevention, early detection, diagnosis, and treatment of this disease. Here, we used miRNA microarray on 69 ovarian tumor specimens to identify the miRNA signature associated with chemotherapy response in ovarian cancer. Let-7i, a let-7 family member, was found to be an important miRNA, differentiating ovarian cancer patients with complete response or noncomplete response to chemotherapy. Further investigation using a variety of cultured cancer cell lines confirmed that let-7i is as functionally involved in the tumor cells response to cis-platinum. The present study may therefore provide a novel prognostic biomarker and therapeutic target for ovarian cancer.
The role of let-7 in cancer was first shown by the Slack group when they found that the let-7 family negatively regulates let-60/RAS in C. elegans by binding to the multiple let-7 complementary sites in its 3′-untranslated region (17). Moreover, having found that let-7 expression is lower in lung tumors than in normal lung tissue, whereas RAS protein is significantly higher in lung tumors, they proposed let-7 as a tumor suppressor gene (17), which is consistent with previous clinical observation in lung cancer (18). The inhibitory function of the let-7 family in cancer has been corroborated by a number of groups and in various types of tumors (9–11, 17, 18, 20–23, 25–29, 34, 35, 38). Let-7 probably performs those functions by targeting various genes. First, let-7 inhibits several well-characterized oncogenic proteins such as K-RAS (17, 25, 27), H-RAS (17, 25, 27), HMGA2 (23, 24, 27, 28), c-Myc (29), and NF2 (11). In addition, let-7 may target multiple cell cycle–associated genes, e.g., CDC25A (22), CDK6 (22), CDK4(39), and cyclin A (39), cyclin D1 (39), cyclin D2 (22), and cyclin D3 (39). Finally, let-7 regulates a panel of oncofetal genes, e.g., IMP-1/CRD-BP (40), and Toll-like receptors, e.g., TLR4 (38). Consistent with other investigators' reports (17, 23–25, 27, 28), we also found that overexpression of let-7i was able to remarkably down-regulate oncogenic proteins such as H-RAS and HMGA2, in EOC cell lines (data not shown). Up to one-third of human mRNAs seem to be miRNA targets (41). Each miRNA can target hundreds of transcripts directly or indirectly (42, 43), and more than one miRNA can converge on a single transcript target (44). Thus, the potential regulatory circuitry afforded by the let-7 family is enormous. We would not expect that one or even a few target proteins play a key role in let-7i's function of chemotherapy sensitivity. Instead, we believe that there is a complex molecular network involved in this function (Fig. 5A), which indicates that restoring let-7i might be a more efficient strategy compared with only targeting one protein-coding gene to modulate chemotherapy because multiple pathways will be affected by let-7i–based therapy.
Figure 5.

Illustration of the potential mechanism of let-7i regulating chemotherapy sensitivity in human cancer.
Recently, an interesting role of the let-7 family in self-renewing progenitor cells has been reported (27, 45). Ibarra and colleagues found that let-7 is depleted in the mouse mammary epithelial cell line, comma-Dβ, which contains a population of self-renewing progenitor cells that can reconstitute the mammary gland, suggesting its role in the regulation of progenitor maintenance (45). Self-renewing tumor-initiating cells (T-IC) or cancer stem cells have been identified and implicated to give rise to cancer. By comparing miRNA expression in self-renewing and differentiated cells from breast cancer lines and in breast T-IC (BT-IC) and non–BT-IC from first-degree breast cancers, Yu and colleagues found that the let-7 family was markedly reduced in BT-IC and increased with differentiation (27). They also showed that the let-7 family regulates multiple BT-IC stem cell–like properties and tumorigenicity of breast cancer cells by silencing more than one target including H-RAS and HMGA2 (27). Those findings are particularly important to our understanding of the role of let-7 family in cancer, and especially in patient response to chemotherapy, because chemotherapy selectively enhances the proportionate survival of BT-IC (27). In addition, those results strongly suggest that let-7– based targeted therapy might more efficiently differentiate the chemotherapy-resistant cancer stem cell population (Fig. 5B).
Although the let-7 family has been generally shown as a tumor suppressor gene, there have been contradictory reports that certain members of the let-7 family could also serve an oncogenic function. For example, let-7a-3 has been reported to be located in a CpG locus by two groups (21, 30), and we also reported that the chromosome region harboring this miRNA is highly deleted in EOC (7). However, Lu and colleagues reported that hypermethylation of let-7a-3 in EOC is associated with a favorable prognosis (21), whereas Brueckner and colleagues made the opposite discovery that its hypomethylation results in enhanced tumor phenotype in cultured HCT 116 colon cancer cells (30). Those results indicated that the highly homologous let-7 family members may play contradicting functions in different cancer types or cellular context. Most recently, Vasudevan and colleagues documented that let-7 induces translation up-regulation of target mRNAs on cell cycle arrest, yet it represses translation in proliferating cells (46). Therefore, the function of let-7 family in cancer might be more complex than we previously expected before. Further studies are needed on the function of individual let-7 family members in human cancer.
miRNAs provide a therapeutic target for cancer treatment (5). Modified antisense oligonucleotides complementary to miRNAs are used by many groups to inhibit miRNAs with oncogenic properties. To supplement and/or enhance the function of tumor suppressor miRNAs, enforced expression of a short hairpin RNA from a polymerase II or III promoter in a nonviral or viral vector, which can be further processed into mature miRNAs, has been tested (47–50). In addition, in vivo delivery of double-stranded miRNA mimics has been reported (47–50). The current rapid advances in oligonucleotide/nanoparticle therapy create realistic optimism for the establishment of the let-7 family as a new and potent therapeutic target and/or chemoresistant modulator in cancer treatment.
Supplementary Material
Acknowledgments
Grant support: Ovarian Cancer Research Fund (L. Zhang and G. Coukos), NCI Ovarian Specialized Programs of Research Excellence P50-CA083638 (Career Development Award and Project 7, L. Zhang), American Cancer Society IRG-78-002-30 (L. Zhang), the Mary Kay Ash Charitable Foundation (L. Zhang), and the Italian Association for Cancer Research (D. Katsaros).
We thank Saundra Ehrlich for her excellent editing assistance on the manuscript.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Unpublished observation.
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3:502–16. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 3.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–9. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blower PE, Chung JH, Verducci JS, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- 11.Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256–64. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- 12.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 13.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 14.Lin SY, Johnson SM, Abraham M, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–50. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 15.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–30. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 17.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 19.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–22. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 22.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 23.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Y, Laser J, Shi G, et al. Antiproliferative effects by let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–73. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 25.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 27.Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 28.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 30.Brueckner B, Stresemann C, Kuner R, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–23. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 32.Laios A, O'Toole S, Flavin R, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greshock J, Nathanson K, Martin AM, et al. Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Res. 2007;67:3594–600. doi: 10.1158/0008-5472.CAN-06-3674. [DOI] [PubMed] [Google Scholar]
- 34.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 36.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 37.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 38.Chen XM, Splinter PL, O'Hara SP, Larusso NF. A Cellular micro-RNA, let-7i, regulates toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–57. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 40.Boyerinas B, Park SM, Shomron N, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–91. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 41.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 43.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 44.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 45.Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–43. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 47.Esau CC, Monia BP. Therapeutic potential for microRNAs. Adv Drug Deliv Rev. 2007;59:101–14. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Grimm D, Kay MA. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? J Clin Invest. 2007;117:3633–41. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117:3623–32. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corey DR. Chemical modification: the key to clinical application of RNA interference? J Clin Invest. 2007;117:3615–22. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


