Abstract
Sec7 domains (Sec7d) catalyze the exchange of guanine nucleotide on ARFs. Recent studies indicated that brefeldin A (BFA) inhibits Sec7d-catalyzed nucleotide exchange on ARF1 in an uncompetitive manner by trapping an early intermediate of the reaction: a complex between GDP-bound ARF1 and Sec7d. Using 3H-labeled BFA, we show that BFA binds to neither isolated Sec7d nor isolated ARF1–GDP, but binds to the transitory Sec7d–ARF1–GDP complex and stabilizes it. Two pairs of residues at positions 190–191 and 198–208 (Arno numbering) in Sec7d contribute equally to the stability of BFA binding, which is also sensitive to mutation of H80 in ARF1. The catalytic glutamic (E156) residue of Sec7d is not necessary for BFA binding. In contrast, BFA does not bind to the intermediate catalytic complex between nucleotide-free ARF1 and Sec7d. These results suggest that, on initial docking steps between ARF1–GDP and Sec7d, BFA inserts like a wedge between the switch II region of ARF1–GDP and a surface encompassing residues 190–208, at the border of the characteristic hydrophobic groove of Sec7d. Bound BFA would prevent the switch regions of ARF1–GDP from reorganizing and forming tighter contacts with Sec7d and thereby would maintain the bound GDP of ARF1 at a distance from the catalytic glutamic finger of Sec7d.
Sec7 domains (Sec7d) catalyze the exchange of guanine nucleotides on ARF, a family of small G proteins involved in vesicular traffic and signal transduction (1, 2). Sec7d displays a characteristic hydrophobic groove which forms the binding site for ARF (3–9). Almost all of the catalytic activity of Sec7d is borne by a single residue, the glutamic finger, which lies on the border of the groove, and which, through its carboxylate group, displaces the β-phosphate of ARF1-bound GDP (6–9). The penetration of the glutamic finger into the nucleotide-binding site of ARF1 is coupled to a dramatic conformational change of ARF1 (7). This conformational change is not limited to the region of ARF1 that interacts with Sec7d, but extends up to the N terminus of ARF1, which interacts with membrane lipids (7, 10).
In line with structural and kinetic studies on other G proteins and their nucleotide-exchange factors, it is likely that the nucleotide-exchange reaction proceeds through the formation of several intermediates (8–13). The first intermediate would be a “docking” complex, in which GDP-bound ARF1 binds through its protuberant switch regions to the hydrophobic groove of Sec7d (6). In this low-affinity complex, the glutamic finger, which lies at one edge of the Sec7d groove, would be too distant to displace the β-phosphate of GDP. In a second step, the switch regions of ARF1–GDP would reorganize to yield larger contacts with Sec7d (7). This reorganization would allow the glutamic finger to penetrate into the nucleotide-binding site of ARF1 and destabilize the bound GDP (7, 10). The nucleotide-free intermediate is very stable in the absence of added nucleotide (14) and its crystal structure has been determined (7). In contrast, because of its transient nature, attempts to isolate the first putative docking intermediate have been unsuccessful. A recent study suggested that the inhibitor brefeldin A (BFA), a small (Mr 280) fungal metabolite, stabilizes a close mimic of such a docking complex (15).
BFA inhibits the exchange activity of several ARF guanine nucleotide-exchange factors by acting on the Sec7d (2, 15–21). Based on sequence comparisons between Sec7d having different BFA sensitivities, key residues for the inhibition by BFA have been identified recently (15, 21). For simplification, these residues are numbered here according to the sequence of the mammalian nucleotide-exchange factor Arno (1). Peyroche et al. (15) studied a pair of amino acids at position 190–191, whose sequence is either YS in BFA-sensitive Sec7d (such as the yeast exchange factors Geap and Sec7p), or FA in BFA-insensitive forms (such as the mammalian exchange factor Arno). Exchanging a YS pair for an FA pair switches the sensitivity of Sec7d both in vivo and in vitro. In an independent study, Sata et al. (21) found another pair of critical residues. When residues S198 and P208 of the Sec7d of cytohesin, a close Arno homolog, were replaced by the cognate residues of the exchange factor p200 (Asp and Met, respectively; ref. 19), the Sec7d became sensitive to BFA in vitro (21).
Because residues 190, 191, and 198 line the groove of Sec7d—i.e., the ARF-binding site (3–5)—one model for the inhibitory effect of BFA could be that the drug competes with ARF for Sec7d. To our surprise, kinetics studies revealed a rare mechanism of inhibition, sometimes termed “uncompetitive,” that is distinct from competitive and noncompetitive mechanisms (15, 22). The key observation was that the extent of Sec7d inhibition by BFA increased with the concentration of ARF1–GDP, suggesting that the target of BFA is not isolated Sec7d but rather a complex between ARF1 and Sec7d (15, 23). Gel-filtration studies revealed the nature of this complex. In the presence of BFA, a stoichiometric complex between ARF1 and a BFA-sensitive Sec7d could be isolated in which GDP remains associated with ARF1 (15).
To test and study directly this unique model of G-protein inhibition, we used 3H-labeled BFA. Thus, instead of measuring the effect of BFA on the Sec7d/ARF1 interaction and the kinetics of guanine-nucleotide exchange, we assessed the effect of the Sec7d/ARF1 interaction on the binding of [3H]BFA. We show that the binding of BFA strictly requires two conditions: (i) Sec7d must be in complex with ARF; and (ii) in the complex, ARF must be in the GDP-bound form. Both pairs of residues at position 190–191 and 198–208 in Sec7d contributed to the stability of BFA in the abortive complex with ARF1–GDP. The binding of BFA was also sensitive to mutation of residue H80 in the switch II region of ARF1. The glutamic finger of Sec7d, which is strictly required for the destabilization of GDP from ARF1, was not essential for the binding of BFA.
Materials and Methods
Materials.
[3H]BFA (20 Ci/mmol; 1 Ci = 37 GBq) was from ICN. Alkaline phosphatase and unlabeled BFA were from Sigma.
Proteins.
The expression and purification of [Δ17]ARF1 and the Sec7d of Arno and Gea2p have been described elsewhere (3, 6, 14, 15). A large fraction of bacterially expressed [Δ17]ARF1 was in the GTP-bound form. To get pure GDP-bound [Δ17]ARF1, the bacterial lysate was incubated for 15 min with 100 μM GDP at room temperature and in a low Mg2+ buffer (20 mM Tris⋅HCl, pH 7.0/2 mM EDTA/1 mM MgCl2) before purification. Mutations were introduced into expression plasmids by PCR using appropriate primers and the quick-exchange site-directed mutagenesis kit of Stratagene. The full sequence and the presence of the desired mutation were verified by automated sequencing.
BFA-Binding Measurements.
The binding of [3H]BFA to proteins was determined by a fast filtration procedure. The small ligand is retained on a nitrocellulose filter when bound to a protein. All experiments were performed at 27°C in a buffer containing 50 mM Hepes (pH 7.5), 100 mM KCl, 1 mM MgCl2, and 1 mM DTT. In all cases, [3H]BFA (250 dpm/pmol), Sec7d, and [Δ17]ARF1–GDP were added sequentially from concentrated stock solutions to the sample buffer. Some experiments included the addition of unlabeled BFA, GDP, or alkaline phosphatase (see figures). The final amount of methanol resulting from the addition of BFA did not exceed 4%. At the indicated times, aliquots (15 μl) were withdrawn to determine [3H]BFA binding. The order of additions, concentration of reactants, and time schedule for taking aliquots are indicated in the figures. Each aliquot was immediately diluted into 2 ml of ice-cold buffer (50 mM Hepes, pH 7.5/100 mM KCl/10 mM MgCl2) and loaded onto a nitrocellulose filter (BA 85, Schleicher & Schuell) mounted on a vacuum pump. The filter was washed two times with 2 ml of cold buffer, and bound radioactivity was measured.
Results
All experiments were performed with various purified Sec7d and an N-terminal-truncated form of ARF1, [Δ17]ARF1, which was purified in the GDP-bound form. Because it lacks the N-terminal myristoylated helix that interacts with membrane lipids, [Δ17]ARF1 provides a convenient minimal model for studying the ARF1/Sec7d interface in solution (6, 7, 14). Previous studies have shown that the uncompetitive inhibition mechanism of BFA also applies when full-length ARF1 is used as a substrate of the Sec7d (10, 23). Most experiments were performed with mutated forms of the Sec7d of Arno, engineered to increase the sensitivity to BFA. Because all mutants carry two to five point mutations, the following abbreviations will be used: [YS]Arno-Sec7, the F190Y-A191S mutant of Arno-Sec7 described by Peyroche et al. (15); [DM]Arno-Sec7, the S198D-P208M mutant described by Sata et al. (21); [YSDM]Arno-Sec7, a quadruple mutant carrying both the F190Y-A191S and S198D-P208M mutations; [YSDM-E156K]Arno-Sec7, the same mutant with an additional mutation of the glutamic finger into a Lys. The mutants displayed similar exchange activity on [Δ17]ARF1 as the wild-type form (kcat/Km = 4 × 105 to 8 × 105 M−1⋅s−1; ref. 6), except [YSDM-E156K]Arno-Sec7, which was inactive because of the mutation of the glutamic finger (data not shown).
BFA Does Not Bind to ARF1–GDP Alone or Sec7d Alone, but Binds to a Complex Between ARF–GDP and Sec7d.
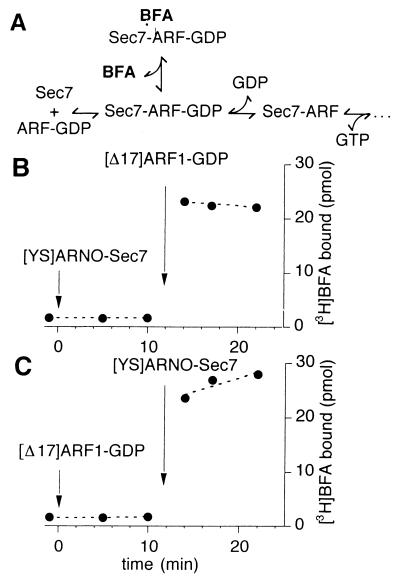
The uncompetitive mechanism of Sec7d inhibition by BFA predicts that the drug should bind to BFA-sensitive Sec7d only in the presence of ARF1–GDP (Fig. 1A; ref. 15). Fig. 1B shows that this is the case. At zero time, 5 μM [YS]Arno-Sec7 was added to a solution containing 25 μM [3H]BFA. No binding of the drug to [YS]Arno-Sec7 could be detected after 10 min of incubation at 27°C. However, the subsequent addition of 10 μM [Δ17]ARF1–GDP promoted a stable binding of [3H]BFA in less than 1 min. In a reverse experiment, [Δ17]ARF1–GDP was added before the addition of [YS]Arno-Sec7 (Fig. 1C). Again, the binding of [3H]BFA was detected only after the addition of the second protein. These results show that BFA binds specifically to a complex between [Δ17]ARF1 and [YS]Arno-Sec7.
Figure 1.
BFA binds to a complex between Sec7d and [Δ17]ARF1. (A) Proposed scheme for the inhibition of Sec7d by BFA as suggested by kinetic studies of Sec7d-catalyzed nucleotide exchange on ARF1 (15, 23). (B and C) Direct measurements of [3H]BFA binding. The sample (100 μl) contained initially 25 μM [3H]BFA (253 dpm/pmol) in buffer and was incubated at 27°C. At the indicated times, [YS]Arno-Sec7 (5 μM) and [Δ17]ARF1–GDP (10 μM) were added sequentially from concentrated stock solutions to the sample. Note the different order of protein additions between B and C. To determine the binding of [3H]BFA, 15-μl aliquots were withdrawn from the sample and filtered immediately onto cellulose filters, and the amount of [3H]BFA trapped on the filter was measured (see Materials and Methods). The amount of protein in each 15-μl aliquot was 75 pmol of [YS]Arno-Sec7 and/or 150 pmol of [Δ17]ARF1.
Similar experiments were performed with other Sec7d, including the wild-type forms of Gea2p-Sec7 and Arno-Sec7, and two mutated forms of Arno-Sec7, [DM]Arno-Sec7 and [YSDM]Arno-Sec7. For all Sec7d tested, the binding of BFA depended strictly on the presence of [Δ17]ARF1–GDP, suggesting that the uncompetitive mechanism applies for all Sec7d (data not shown). However, under the same experimental conditions (10 μM [Δ17]ARF1–GDP/25 μM [3H]BFA/5 μM Sec7d), the extent of BFA binding was clearly different. Thus, the binding of BFA in the presence of wild-type Arno-Sec7 was barely detectable (<0.1 pmol of BFA per pmol of Sec7d), whereas a significant binding was observed in the presence of Gea2p-Sec7 (≈0.25 mol of BFA per mol of Sec7d). This is in agreement with the difference between these two Sec7d for their sensitivity to BFA in vivo and in vitro (15). Interestingly, the two double mutants, [DM]Arno-Sec7 and [YS]Arno-Sec7, showed similar levels of BFA binding (0.4 ± 0.1 mol of BFA per mol of Sec7d), whereas the quadruple mutant [YSDM]Arno-Sec7 displayed roughly 2 times better binding of BFA than either of the two double mutants (≈0.7 mol of BFA per mol of Sec7d). This finding suggests that both pairs of residues Y190-S191 and D198-M208 of Sec7d contribute to the formation of the Sec7d–BFA–ARF1–GDP abortive complex.
Dissociation Rate of BFA from the ARF–GDP–Sec7d Complex.
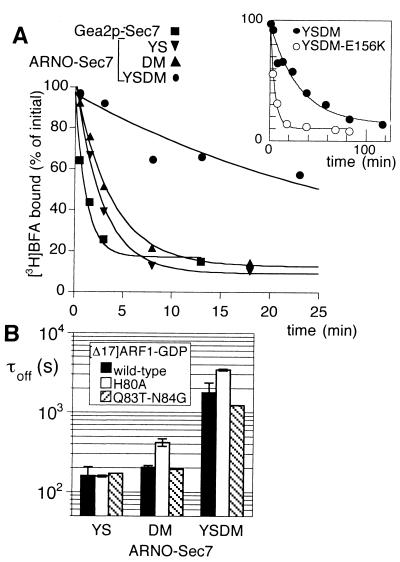
Because the target of BFA is not a single protein but a complex, the real affinity of BFA cannot be determined directly because it would require extrapolation of binding data at infinite concentration of ARF1–GDP—i.e., when Sec7d is saturated. As a more direct parameter of the tightness of the interactions among BFA, Sec7d, and [Δ17]ARF1–GDP, we assessed the dissociation rate of radiolabeled BFA from the Sec7d–[Δ17]ARF1–GDP complex. Each Sec7d (5 μM) was first mixed with a 2-fold excess of [Δ17]ARF1–GDP, 20 μM free GDP, and 25 μM [3H]BFA. After few minutes of incubation at 27°C, bound [3H]BFA reached a stable level, and [3H]BFA dissociation was monitored on the addition of 300 μM unlabeled BFA (Fig. 2A). [3H]BFA dissociated from the abortive complexes between [Δ17]ARF1–GDP and either [YS]Arno-Sec7 or [DM]Arno-Sec7 with a time constant (1/e) of about 200 s (τoff =160 ± 40 s and 203 ± 10 s, respectively; Fig. 2 A and B). For the abortive complex with the quadruple mutant [YSDM]Arno-Sec7, a 10-fold slower kinetics was observed (τoff = 1,800 ± 500 s; Fig. 2 A and B). This result suggests that the two pairs of residues Y190-S191 and D198-M208 in Sec7d contribute about equally to the stability of BFA in the complex with [Δ17]ARF1–GDP. Surprisingly, the dissociation rate of BFA from the complex between [Δ17]ARF1–GDP and Gea2p-Sec7 was about 20-fold faster than that observed for the complex with [YSDM]Arno-Sec7, despite the fact that the four critical “BFA-binding” residues are present in the Sec7d of Gea2p. However, Gea2p is a yeast exchange factor and a mammalian form of ARF1 was used here. Therefore, some differences at the level of the ARF1/Sec7d interface might explain this result.
Figure 2.
Effect of point mutations in Sec7d and [Δ17]ARF1–GDP on the dissociation rate of BFA from the abortive [Δ17]ARF1–GDP–Sec7d complex. (A) Time course of BFA dissociation. In a preliminary step, Sec7d (5 μM) was incubated for 10 min at 27°C with 10 μM [Δ17]ARF1–GDP/20 μM GDP/25 μM [3H]BFA. At zero time, 300 μM unlabeled BFA was added and the kinetics of [3H]BFA dissociation was followed by measuring the bound radioactivity in 15-μl aliquots. The Sec7d used were Gea2p-Sec7 (■); [YS]Arno-Sec7 (▾); [DM]Arno-Sec7 (▴); and [YSDM]Arno-Sec7 (●). (Inset) Time course of BFA dissociation from the abortive complex with [YSDM]Arno-Sec7 (●) on a different time scale as well as the dissociation of BFA from the abortive complex with [YSDM-E156K]Arno-Sec7 (○). (B) Conditional effect of the H80A mutation of [Δ17]ARF1–GDP on the dissociation rate of BFA. The time course of [3H]BFA dissociation from various abortive [Δ17]ARF1–GDP–Sec7d complexes carrying the indicated mutations in [Δ17]ARF1 and in Arno-Sec7 was measured as in A. The time constant, τoff(1/e), for BFA dissociation was determined by fitting the experimental points with a single exponential. Error bars show the standard error for two to four independent experiments. Note that the H80A mutation of [Δ17]ARF1–GDP slowed down the dissociation of BFA only when Sec7d bore the DM mutation.
A Mutation in the Switch II Region of ARF1 That Stabilizes BFA Binding.
The critical residues for BFA binding in Sec7d define a region that overlaps the binding site for the switch regions of ARF1 (3–9). Because BFA binds to a complex between Sec7d and ARF1, one might expect to find critical residues for the binding of BFA not only in or near the groove of Sec7d but also in the switch regions of ARF1. However, most mutations described in these regions affect the ability of ARF1 to be activated by Sec7d (6). Therefore, it could be difficult to distinguish between a direct effect of a mutation on the binding of BFA and an indirect effect caused by a decrease in the interaction of ARF1–GDP with Sec7d. These considerations prompted us to focus only on two mutations in the switch II regions of ARF1, H80A and Q83T-N84G, which have nearly no effect on the sensitivity of ARF1 to the exchange activity of Arno-Sec7 (6). We first noticed that introducing the H80A mutation in [Δ17]ARF1–GDP slowed down the dissociation of BFA from the complex with [YSDM]Arno-Sec7 by a factor of 2, whereas the Q83T-N84G mutations had no effect (Fig. 2B). We then examined separately the effect of the H80A mutation of [Δ17]ARF1 in the context of either the YS mutation or the DM mutation of Arno-Sec7. Interestingly, the H80A mutation of [Δ17]ARF1–GDP slowed down the dissociation of BFA from the abortive complex with [DM]Arno-Sec7 by a factor of 2, but had no effect on the dissociation of BFA from the complex with [YS]Arno-Sec7 (Fig. 2B). This finding suggests that some residues of one protein contribute to the binding of the drug only when the appropriate residues on the other protein are present.
BFA Does Not Bind to the Nucleotide-Free Complex Between [Δ17]ARF1 and Sec7d.
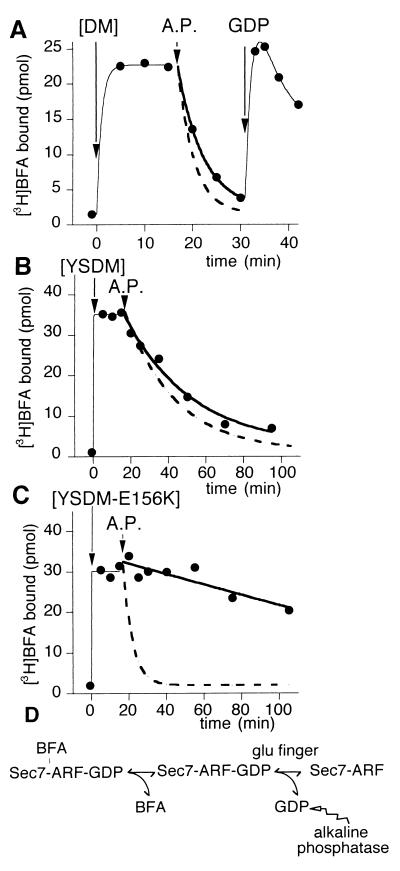
The observation by Peyroche et al. (15) that GDP remained bound to the complex between [Δ17]ARF1 and [YS]Arno-Sec7 induced by BFA was striking because the only complex that can usually be isolated between a G protein and its exchange factor is a nucleotide-free complex (7–9, 13, 14). To study the mutual influence of GDP and BFA on their binding to a [Δ17]ARF1–Sec7d complex, we first determined whether BFA could bind to the nucleotide-free complex between [Δ17]ARF1 and Sec7d. For this purpose, we prepared the nucleotide-free complex by incubating a BFA-sensitive Sec7d and [Δ17]ARF1–GDP with a catalytic amount of alkaline phosphatase, which hydrolyzes free GDP as it dissociates from [Δ17]ARF1 on Sec7d stimulation (10, 24). This pretreatment abolished the binding of [3H]BFA (data not shown), suggesting that the binding of BFA is specific for the GDP-bound complex. An experiment in which the reactants were added in a different order was even more informative (Fig. 3). In a first phase, BFA was incubated with [Δ17]ARF1–GDP and at zero time, the mutated Sec7d of Arno carrying either the DM pair (Fig. 3A) or both the YS and DM pairs (Fig. 3B) was added. When the binding of [3H]BFA reached a stable level, alkaline phosphatase was added to the sample. For both the DM and YSDM mutants of Arno-Sec7, this induced the complete dissociation of BFA from the complex with [Δ17]ARF1 (Fig. 3 A and B). Moreover, the dissociation could be temporarily reversed by the addition of a very large excess of GDP (1 mM) to overcome the alkaline phosphatase activity (Fig. 3A). Again, this shows the strict specificity of BFA for the GDP-bound complex as compared with the nucleotide-free complex. Strikingly, for each Sec7d mutant, the kinetics of release of [3H]BFA observed on alkaline phosphatase addition was close to the kinetics of [3H]BFA dissociation as measured by exchange with unlabeled BFA (compare Figs. 2 and 3), suggesting that the dissociation of BFA was the rate-limiting step for the liberation of GDP from ARF1 and its hydrolysis by alkaline phosphatase (Fig. 3D). Taken together, these results suggest the following interplay between BFA and GDP: (i) the binding of BFA to a Sec7d–ARF1 complex is strictly dependent on the presence of GDP in the nucleotide-binding site of ARF1; and (ii) as soon as BFA has bound to the Sec7d–ARF1–GDP complex, no significant release of GDP occurs.
Figure 3.
BFA does not bind to the nucleotide-free complex between [Δ17]ARF1 and Sec7d. The sample (200 μl) contained initially 10 μM [Δ17]ARF1–GDP and 25 μM [3H]BFA. The binding of 25 μM [3H]BFA was initiated at zero time by the addition of Arno-Sec7 (5 μM) carrying either the DM mutations (A), the YSDM mutations (B), or the YSDM + E156K mutations (C). At t = 17 min, alkaline phosphatase (200 units/ml) was added to the sample. In A, 2 mM GDP was further added at t = 31 min. Each experimental point corresponded to the bound radioactivity in a 15-μl aliquot. The continuous thick lines show an exponential fit of the data points after the addition of alkaline phosphatase. The dashed lines report the time course of [3H]BFA dissociation as measured by exchange with unlabeled BFA and measured in Fig. 2. Note the good correlation between the two fits (A and B) except for the complex with [YSDM-E156K]Arno-Sec7 (C). These experiments suggest that once BFA has bound to a Sec7d–ARF1–GDP complex, the release of GDP and its hydrolysis by alkaline phosphatase requires first the dissociation of BFA from the complex and then the destabilization of GDP by the glutamic finger of Sec7d (D).
Removal of the Glutamic Finger of Sec7d Does Not Prevent the Binding of BFA.
The key catalytic residue of Sec7d that promotes the dissociation of GDP from ARF1 is the glutamic finger (E156 in Arno; refs. 6 and 7). Because binding of BFA to the Sec7d–ARF1–GDP complex blocks the release of GDP, one might suggest that BFA binds directly to the glutamic finger, thereby preventing it from destabilizing bound GDP. However, the glutamic finger (E156) and the critical BFA-binding residues (190, 191, 198, and 208) are not on the same border of the Sec7d groove. We have generated a quintuple mutant of Arno-Sec7 carrying the four “BFA mutations” (YSDM) and the mutation of the glutamic finger (E156K). [YSDM-E156K]Arno-Sec7 exhibited, in the presence of [Δ17]ARF1–GDP, a level of [3H]BFA binding which was comparable to that of the YSDM mutant (compare Fig. 3 B and C). Thus, the glutamic finger is not essential for the binding of BFA. However, the E156K mutation accelerated by a factor of ≈8 the kinetics of [3H]BFA dissociation as measured by exchange with unlabeled BFA (Fig. 2A Inset). In contrast, almost no dissociation of [3H]BFA was observed on alkaline phosphatase addition (Fig. 3C). The first observation suggests that the E156K mutation reduces the stability of the abortive complex, whereas the second observation reflects the fact that the E156K mutation by itself—i.e., in the absence of BFA—abolishes the exchange activity of Sec7d (6). In that case, even after BFA dissociation, Sec7d could not promote the dissociation of GDP, which therefore remained inaccessible for hydrolysis by alkaline phosphatase (Fig. 3D).
The Slow Binding Kinetics of BFA Are Related to the Transience of Its Target Complex.
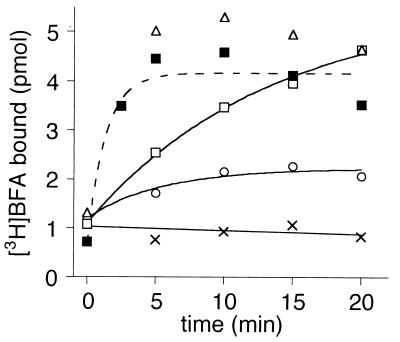
One of the first clues for the uncompetitive mechanism of BFA inhibition came from the observation that a preincubation of about 1 min of BFA with both ARF1–GDP and [YS]Arno-Sec7 was required to observe inhibition of GDP/GTP exchange as monitored by a real-time assay (15). This suggested that, although used at relatively high concentration (10−5 to 10−4 M), BFA exhibited slow binding kinetics. Under our standard conditions, the time course of [3H]BFA binding was too fast to be resolved (Figs. 1 and 3). Therefore, we decreased the concentrations of the reactants (Fig. 4). In the presence of 15 μM BFA/2 μM [Δ17]ARF1–GDP/0.5 μM [YSDM]Arno-Sec7, BFA bound with a time constant of about 1,000 s (□). For a simple interaction between a ligand and a protein, such kinetics would be several orders of magnitude slower than a diffusion-limited process. However, the target of BFA is not an isolated protein but a complex. In that case, the slow rate constant of binding should reflect the low abundance of this complex versus the other species, which are not competent for BFA binding. This possibility is illustrated by the fact that increasing the concentration of [Δ17]ARF1–GDP accelerated the kinetics of [3H]BFA binding (Fig. 4). Indeed, increasing the concentration of [Δ17]ARF1–GDP should increase the fraction of Sec7d in complex with [Δ17]ARF1–GDP at the expense of free Sec7d (Fig. 1A). In addition, mutagenesis of the glutamic finger also increased the apparent rate of BFA binding (Fig. 4). Because this mutation prevents the GDP-bound complex from switching to nucleotide-free form, it also favors statistically the binding of BFA.
Figure 4.
Mutation of the glutamic finger of Sec7d increases the rate of BFA binding. Association kinetics of [3H]BFA (15 μM) with 0.5 μM [YSDM]Arno-Sec7 (white symbols) or [YSDN-E156K]Arno-Sec7 (■), in the presence of 0 (×), 1 (○), 2 (■, □), or 5 μM (▵) [Δ17]ARF1–GDP. The reaction was initiated by the addition of Sec7d. The t = 0 point corresponds to the initial level of bound [3H]BFA just before the addition of Sec7d and represents the nonspecific binding of BFA. Bound radioactivity was determined on 15-μl aliquots (7.5 pmol of Sec7d).
Discussion
The fact that BFA acts as an uncompetitive inhibitor by forming an abortive complex with Sec7d and GDP-bound ARF1 was unexpected (2, 15, 25). First, an uncompetitive mechanism is a rare mechanism of inhibition. Second, the overlap in Sec7d between residues that are important for the BFA sensitivity and the ARF1-binding site (15, 21) might suggest a more classical competitive mechanism. Finally, ternary complexes between G proteins, guanine nucleotides, and exchange factors are very unstable (11, 12, 14). By using radiolabeled BFA, the binding of BFA to the Sec7d–ARF1–GDP complex was directly addressed here. As expected from the uncompetitive inhibition mechanism (15, 23), this binding depends not only on the presence of specific residues in Sec7d but also on the interaction of Sec7d with ARF1–GDP.
Binding of [3H]BFA was observed when both [Δ17]ARF1–GDP and a BFA-sensitive Sec7d were present (Fig. 1). In contrast, no binding of [3H]BFA could be detected on isolated ARF1–GDP or isolated Sec7d, even when residues that determined the sensitivity of Sec7d to BFA were present. Moreover, the binding of BFA was abolished when, in the Sec7d–ARF1 complex, the nucleotide-binding site of ARF1 was vacant (Fig. 3). Thus, it seems that a binding site for BFA is transiently formed when ARF1–GDP interacts with Sec7d.
What might be this transient BFA-binding site? Three parameters must be considered: (i) the localization of residues of Sec7d that define the sensitivity to BFA; (ii) the size and hydrophobicity of BFA; and (iii) the conformational change at the ARF1/Sec7d interface. In the structure of Sec7d (3–5, 7), residues at positions 190 and 191 line the characteristic hydrophobic groove (Fig. 5). Although at a distance in the primary sequence, residues 198 and 208 are exposed a few angstroms away (Fig. 5). Other critical residues for the effect of BFA have been identified by random mutagenesis (15). Together with residues 190, 191, 198, and 208, they form a patch at the surface of Sec7d, which is centered at M194, a critical residue for both ARF1 binding and BFA inhibition (3–5, 7, 15). The patch overlays the hydrophobic groove but also projects beyond the groove (Fig. 5). The size of the patch is compatible with the size of BFA, and future docking and structural studies should reveal the mode of interaction of BFA and explain the stereospecificity of this drug for the positioning of its hydroxyl groups (16, 17).
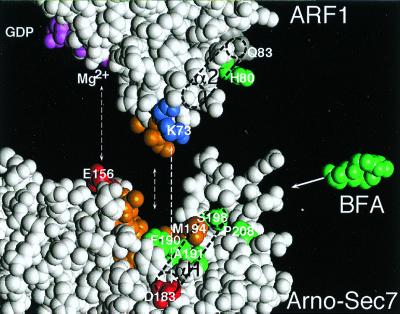
Figure 5.
Model for the binding of BFA to the Sec7d–ARF1–GDP complex. A docking approach of free ARF1–GDP (26) onto the Sec7d of Arno (3), based on previously identified contacts (dotted lines; ref. 6) is shown. Note the complementarity of shape between the interacting surfaces: the protuberant switch regions of ARF1–GDP and the groove of Sec7d. This might represent the first step of the nucleotide-exchange reaction, i.e., when ARF1–GDP contacts Sec7d. In a second step, a dramatic conformational change occurs in ARF1, which is coupled to the penetration of the glutamic finger (E156) of Sec7d into the nucleotide-binding site of ARF1 and the destabilization of GDP (7). BFA inhibits this second step by binding specifically to the Sec7d–ARF1–GDP complex. The positions of residues in ARF1 and Sec7d that are important for the binding of BFA are shown in green. This includes H80 in helix α2 of ARF1 and residues F/Y190, A/S191, S/D198, and P/M208 in helix αH of Sec7d (underlined amino acids favored the binding of BFA). In the nucleotide-free Sec7d–ARF1 complex, helix α2 of ARF1 interacts extensively with helix αH of Sec7d. In contrast, when ARF1–GDP contacts Sec7d, helix α2 of ARF1 would be at a distance from helix αH of Sec7d, and BFA could insert between the two helices like a wedge. This insertion would block the subsequent reorganization of the ARF1/Sec7d interface.
We cannot exclude a direct binding of BFA to isolated Sec7d, but the binding measurements shown here suggest that the binding, if any, must be of very low affinity (undetectable at concentrations of BFA and Sec7d in the 10−5 M range; Fig. 1). In contrast, the binding of BFA to complexes between GDP-bound ARF1 and BFA-sensitive Sec7d was remarkably stable, featuring half-times of dissociation in the range of a few minutes to half an hour (Fig. 2). This result is striking because the interaction between GDP-bound ARF1 and Sec7d, and more generally between small G proteins in the GDP-bound state and their exchange factors, is of very low affinity in contrast to the nucleotide-free complex (11, 12, 14). Therefore, the binding of BFA converts the weak and transient interaction between ARF1–GDP and Sec7d into a very stable interaction. It should be noted that although the koff of BFA from the Sec7d–ARF1–GDP complex can be determined (Fig. 2), it is more difficult to determine the kon and the corresponding equilibrium constant (Kd = koff/kon). Indeed, the target of BFA is not a stable entity but an intermediate of a reaction. Therefore, the rate of BFA binding depends not only on a bimolecular kinetic constant (kon) but also on the fraction of Sec7d in complex with ARF1–GDP (Fig. 4). Because this fraction is low, high concentrations of BFA are required to enhance the probability of “capture” of a versatile and rare target, but this does not mean that the intrinsic affinity of BFA for the complex is weak.
One key factor for the formation of the nucleotide-free complex between ARF1 and Sec7d is the flexibility of the switch regions of ARF1 (7–9). In the nucleotide-free complex with Gea2p-Sec7, ARF1 displays a conformation that is very different from isolated ARF1–GDP (7, 26, 27). Thus, when one docks the structure of ARF1–GDP onto the structure of Sec7d, the resulting interacting surface is much less well packed than in the nucleotide-free complex (Fig. 5; refs. 6 and 7). Notably, there are some spaces between the two proteins, which could be filled by BFA. In the nucleotide-free complex, helix α2 in the switch II region of ARF1 is packed against helix H of Sec7 (7). Helix H contains most residues of Sec7d identified as determinants of the BFA sensitivity, namely F/Y190, A/S191, M194, and S/D198 (refs. 15 and 21; this study). Some of these residues, such as F/Y190, are completely hidden by helix α2 of ARF1 in the nucleotide-free complex (7), so it is clear that in the nucleotide-free complex, BFA cannot bind to this region. However, before the formation of extensive interactions between helix α2 of ARF1 and helix H of Sec7d, one can imagine that BFA could intercalate, like a wedge, between the two helices (Fig. 5). The presence of some hydrophobic residues in helix α2 of ARF1 (I74, L77, and W78) could favor this intercalation. This is also suggested by the effect of the H80A mutation (helix α2 of ARF1) on the binding of BFA (Fig. 2). Although this effect is modest, it is interesting to note that it required the presence of the DM pair on Sec7d, which faces residue H80 in the nucleotide-free complex (7).
In this model, the inhibition of GDP release can be understood as an indirect consequence of the intercalation of BFA. Indeed, the penetration of the glutamic finger of Sec7d within the phosphate- and Mg2+-binding region of ARF1 is coupled to the extensive packing of the switch regions of ARF1 on the Sec7d surface (7–9). By hindering this packing, BFA would maintain the glutamic finger at a distance from the nucleotide. This would illustrate the emerging concept that the driving force for the destabilization of bound nucleotide by exchange factors is the formation of extensive contacts with the flexible switch regions of G proteins, which allow a few residues of the exchange factor to penetrate into the nucleotide-binding site (8, 9). Structural studies of the abortive complex between BFA, Sec7d, and ARF1–GDP will be important for our understanding of the nucleotide-exchange reaction on G proteins.
Acknowledgments
We thank Anne Peyroche for the gift of Gea2-Sec7 and Pierre Chardin, Jacqueline Cherfils, Cathy Jackson, Marcus Lee, and David Madden for discussions and comments on the manuscript. This work was supported in part by a grant from the Human Frontier Science Program.
Abbreviations
- ARF
ADP-ribosylation factor
- BFA
brefeldin A
- Sec7d
Sec7 domain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170290597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170290597
References
- 1.Chardin P, Paris S, Antonny B, Robineau S, Béraud-Dufour S, Jackson C L, Chabre M. Nature (London) 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 2.Jackson C L, Casanova J E. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 3.Cherfils J, Menetrey J, Mathieu M, Le Bras G, Robineau S, Béraud-Dufour S, Antonny B, Chardin P. Nature (London) 1998;392:101–105. doi: 10.1038/32210. [DOI] [PubMed] [Google Scholar]
- 4.Mossessova E, Gulbis J M, Goldberg J. Cell. 1998;92:415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- 5.Betz S F, Schnuchel A, Wang H, Olejniczak E T, Meadows R P, Lipsky B P, Harris E A S, Staunton D E, Fesik S W. Proc Natl Acad Sci USA. 1998;95:7909–7914. doi: 10.1073/pnas.95.14.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. EMBO J. 1998;17:3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg J. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 8.Cherfils J, Chardin P. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 9.Sprang S R, Coleman D E. Cell. 1998;95:155–158. doi: 10.1016/s0092-8674(00)81746-8. [DOI] [PubMed] [Google Scholar]
- 10.Béraud-Dufour S, Paris S, Chabre M, Antonny B. J Biol Chem. 1999;274:37629–37636. doi: 10.1074/jbc.274.53.37629. [DOI] [PubMed] [Google Scholar]
- 11.Klebe C, Prinz H, Wittinghofer A, Goody R S. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 12.Lenzen C, Cool R H, Prinz H, Kuhlmann J, Wittinghofer A. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 13.Boriack-Sjodin P A, Margarit S M, Bar-Sagi D, Kuriyan J. Nature (London) 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 14.Paris S, Béraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. J Biol Chem. 1997;272:22221–22226. doi: 10.1074/jbc.272.35.22221. [DOI] [PubMed] [Google Scholar]
- 15.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson C L. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 16.Donaldson J G, Finazzi D, Klausner R D. Nature (London) 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 17.Helms J B, Rothman J. Nature (London) 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 18.Peyroche A, Paris S, Jackson C L. Nature (London) 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 19.Morinaga N, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1997;94:12926–12931. doi: 10.1073/pnas.94.24.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sata M, Donaldson J G, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1998;95:4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sata M, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1999;96:2752–2757. doi: 10.1073/pnas.96.6.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999. pp. 112–114. [Google Scholar]
- 23.Mansour S J, Skaug J, Zhao X H, Giordano J, Scherer S W, Melancon P. Proc Natl Acad Sci USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, Goody R S. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 25.Chardin P, McCormick F. Cell. 1999;97:53–55. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- 26.Amor J C, Harrison D H, Kahn R A, Ringe D. Nature (London) 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- 27.Greasley S E, Jhoti H, Teahan C, Solari R, Fensome A, Thomas G M, Cockcroft S, Bax B. Nat Struct Biol. 1995;2:797–806. doi: 10.1038/nsb0995-797. [DOI] [PubMed] [Google Scholar]