Abstract
Purpose
The discovery of effective biomarkers is a fundamental goal of molecular medicine. Developing a systems-biology understanding of radiosensitivity can enhance our ability of identifying radiation-specific biomarkers.
Methods and Materials
Radiosensitivity, as represented by the Survival Fraction at 2 Gy (SF2) was modeled in 48 human cancer cell lines. We apply a linear regression algorithm that integrates gene expression with biological variables including: ras status (mut/wt), tissue of origin (TO) and p53 status (mut/wt).
Results
The biomarker discovery platform is a network representation of the top 500 genes identified by linear regression. This network was reduced to a 10-hub network that includes: c-Jun, HDAC1, RELA (p65 subunit of NFKB), PKC-beta, SUMO-1, c-Abl, STAT1, AR, CDK1 and IRF1. Nine targets associated with radiosensitization drugs link to the network, demonstrating clinical relevance. Furthermore, the model identifies four significant radiosensitivity clusters of terms and genes. Ras was a dominant variable in the analysis along with TO and their interaction with gene expression but not p53. Overrepresented biological pathways differed between clusters but included: DNA repair, cell cycle, apoptosis and metabolism. The c-Jun network hub was validated using a knockdown approach in 8 human cell lines representing lung, colon and breast cancers.
Conclusions
We developed a novel radiation-biomarker discovery platform using a systems biology modeling approach. We propose this platform will play a central role in the integration of biology into clinical radiation oncology practice.
Keywords: Radiosensitivity, systems biology, gene expression, linear regression, mathematical modeling
Introduction
The discovery of novel biomarkers to better define treatment and disease outcome in oncology are central tenets of the molecular medicine era 1. However, an efficient and coordinated strategy to identify radiation-specific biomarkers has been lacking. Thus, in spite of significant effort, few biomarkers have become routine in clinical radiation oncology practice.
The generation of high-throughput datasets in the “omics” era provides an opportunity to address biomarker discovery from a different perspective. For example, gene expression signatures have been shown to be prognostic in breast, lung, head and neck and colon cancer 2–5. Furthermore, these high-throughput technologies are central to the development of a systems-view of complex biological systems6. In systems biology, regulatory pathways are proposed to be organized as complex interacting networks similar to the world-wide web7, 8. Thus, the first step in understanding a regulatory network is defining its components and organization.
One important feature of systems biology is that it integrates biological scales (molecular, regulatory network, cellular, tissue, organism) when modeling disease, thus representing a more global approach to modeling 6–8. Further, it may provide insights into the central function of a biological system by considering all scales involved. We hypothesized that developing a radiosensitivity systems model could provide significant biological/clinical insights in our understanding of intrinsic radiosensitivity.
Previously, we developed a linear regression algorithm to correlate gene expression (molecular scale) and intrinsic cellular radiosensitivity (cellular scale) in a 35 cancer cell line database 9. The model correctly predicted cellular radiosensitivity (SF2) in 22/35 cell lines (p=0.002). Importantly, we showed that the algorithm led to biological discovery. It identified four known genes (topoisomerase 1, rbapa48, rgs19, r5pia) that were highly correlated with radiosensitivity. We showed that RbAp48-overexpression led to radiosensitization in three cancer cell lines tested as predicted. Further it led to a higher proportion of cells in the G2/M phase of the cell cycle and to de-phosphorylation of Akt, consistent with a mechanistic role for RbAp48-overexpression in radiosensitization. Furthermore, Topoisomerase 1 was validated by others as a target for radiosensitization 10. Thus we concluded that the linear regression algorithm was a valid strategy to the discovery of novel radiosensitivity biomarkers.
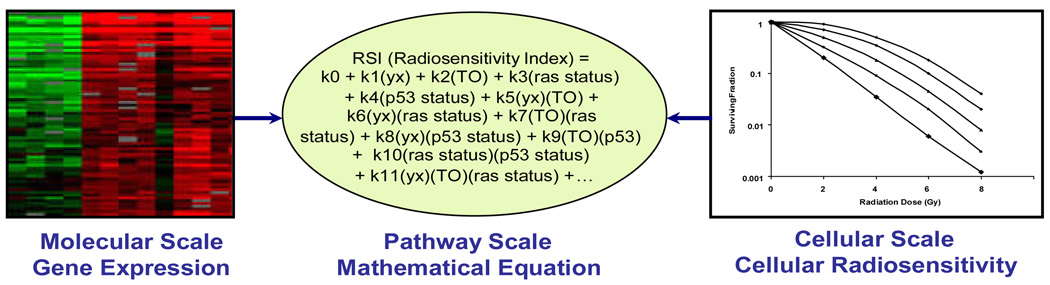
Based on this success and the establishment of linear regression as a valid approach to relate biological scales within the radiosensitivity biological continuum, we reasoned that we could use a similar approach to define the pathway/regulatory network scale, in an expanded 48 cancer cell line database (Figure 1). We hypothesized that this could serve as a strategy to the discovery of radiation-specific biomarkers.
Figure 1. Defining the pathway scale by mathematical modeling.
A linear regression algorithm is used to model the pathway/network scale in the radiosensitivity continuum. Biological variables (ras status, p53 status and TO) known to influence radiosensitivity along with gene expression are included in the model
Methods and Materials
Cell lines
Cell lines were obtained from the NCI and cultured in RPMI-1640 supplemented with glutamine (2 mM), penicillin/streptomycin (10 U/ml) and Heat-inactivated Fetal Bovine Serum (10%) at 37°C (5% CO2).
Radiation Survival Assays (SF2)
SF2s were obtained from the literature in 23/48 cell lines (Table 1). For literature-based SF2s, we used papers (published before 2004) that reported on clonogenic assays performed without any substrate (i.e. agar) and that irradiated cells in log phase. We also required at least two SF2 values reported in the literature by different laboratories. We determined the mean SF2 and used it for model generation. The remaining 25 SF2s were determined in our lab as previously described9.
Table 1.
SF2 values for 48 cell lines in the database
| Cell Line | Recorded SF2 | Cell Line | Recorded SF2 |
|---|---|---|---|
| BREAST_HS578T | 0.79 | COLON_COLO205 | 0.69 |
| BREAST_MDAM B231 | 0.82 | COLON_HCC-2998 | 0.44 |
| COLON_HCT116 | 0.38 | COLON_HT29 | 0.79 |
| COLON_HCT15 | 0.4 | COLON_KM12 | 0.42 |
| COLON_SW620 | 0.62 | MELAN_LOXIMVI | 0.68 |
| LEUK_CCRFCEM | 0.185 | MELAN_M14 | 0.42 |
| LEUK_HL60 | 0.315 | MELAN_MALME3M | 0.8 |
| LEUK_MOLT4 | 0.05 | MELAN_SKMEL28 | 0.74 |
| MELAN_SKMEL2 | 0.66 | MELAN_SKMEL5 | 0.72 |
| NSCLC_A549ATCC | 0.61 | MELAN_UACC257 | 0.48 |
| NSCLC_H460 | 0.84 | MELAN_UACC62 | 0.52 |
| NSCLC_HOP62 | 0.164 | NSCLC_EKVX | 0.7 |
| NSCLC_NCIH23 | 0.086 | NSCLC_HOP92 | 0.43 |
| OVAR_OVCAR5 | 0.408 | OVAR_OVCAR3 | 0.55 |
| RENAL_SN12C | 0.62 | OVAR_OVCAR4 | 0.29 |
| BREAST_BT549 | 0.632 | OVAR_OVCAR8 | 0.6 |
| BREAST_MCF7 | 0.576 | OVAR_SKOV3 | 0.9 |
| BREAST_MDAM B435 | 0.1795 | PROSTATE_DU145 | 0.52 |
| BREAST_T47D | 0.52 | PROSTATE_PC3 | 0.484 |
| CNS_SF268 | 0.45 | RENAL_7860 | 0.66 |
| CNS_SF539 | 0.82 | RENAL_A498 | 0.61 |
| CNS_SNB19 | 0.43 | RENAL_ACHN | 0.72 |
| CNS_SNB75 | 0.55 | RENAL_CAKI1 | 0.37 |
| CNS_U251 | 0.57 | RENAL_UO31 | 0.62 |
siRNA transfection
Cells were plated overnight (antibiotic-free medium) and transfected with either a pool of 4 negative control siRNAs (25 nM) or c-jun siRNA (25 nM) (dharmaFECT transfection protocol, Dharmacon, Inc., Lafayette, CO). 72 hours after transfection, cells were irradiated (2 Gy). Clonogenic survival was assessed 2–3 weeks post-irradiation.
Microarrays
Gene expression profiles were from Affymetrix HU6800 chips from a previously published study 11. The gene expression data was preprocessed using the Affymetrix MAS 5.0 algorithm.
Gene Expression Microarray Analysis: Identification of Systems Model
A linear model was created to correlate radiosensitivity with gene expression and biological variables (ras status, (TO) and p53 status) for each probeset in the cell line dataset using the R software (equation in text). The model consisted of all non-singular terms (28 terms) including gene expression, p53 mutation status, ras mutation status, TO and all possible interactions among terms (Supplemental Table 5). TO, p53 mutation and ras mutation status are categorical variables and were coded as dummy variables. This analysis is performed on a gene by gene basis, totaling 7,168 probesets. The 500 gene-based models with the smallest sum of squared residuals were selected for further analysis.
Systems Model: Network Representation
This was generated by GeneGO™ MetaCore™ software (Encinitas, CA). The software interconnects all 500 selected genes based on literature-based annotations. Only direct connections between identified genes were considered. Major hubs were defined as having more than 5 connections and less than 50% of edges hidden within the network.
Evaluation of each term in the model: Ranking of terms
Term impact was compared using clustering, by obtaining p-values for each coefficient of the variables within the models. These p-values were –log2 transformed and clustered using complete linkage and uncentered correlation as the similarity metric in Cluster 3.0 and visualized using Java Treeview.
Pathway Analysis
The 500 genes selected were uploaded into the GeneGO™ MetaCore™ software and p-values for overrepresented pathways were calculated. Only significant (p<=0.05) maps were considered further. Additional lists were generated for each of the three radiosensitivity clusters defined by the clustering method described earlier.
Predictive Model Development
A model was developed in the 48 cell line database using gene expression from the 10 gene (hub) system. The model is based on gene expression ranks from the highest (10) to the lowest expressed gene (1)12. The coefficients were determined by fitting the model in the 48 cell line dataset (equation in text).
Predictive Model Test Set and Permutation Analysis
The pre-defined predictive model, was tested in a independent set, consisting of the 12 remaining cell lines from the NCI (60) whose SF2s were recently reported 13. To assess the likelihood of identifying 5 /12 cell lines within 10% of the reported SF2 value by chance, we computed 10,000 permutations of 12 random numbers generated from a uniform distribution. We calculated the number of permutations in which 5 or more random values were correct.
Results
A linear regression algorithm to model the radiosensitivity network: Defining the pathway/network scale using mathematics
Since we had previously been successful in establishing a correlation between the cellular and molecular scale for radiosensitivity, and we had biologically-validated the linear regression approach (supplemental data figure 1), we reasoned we could use a similar approach to model radiosensitivity at the pathway/network scale (figure 1). Further we reasoned that within this biological scale would reside pathways/molecules that could serve as potential clinical biomarkers. Thus, we modeled the radiosensitivity network scale using a systems biology approach with the following linear regression equation:
Single gene expression was combined with key biological variables that have been reported to perturb the radiosensitivity network: TO 14, 15, ras status 16, 17 and p53 status 18–20. Thus, we focused on identifying common elements of the radiosensitivity network across the molecular diversity introduced by multiple cell lines and biological conditions.
A gene-based linear model was constructed for each gene (7168 probesets), correlating expression and biological parameters with the measured SF2 using a least-squares fit. We compared the sum squared error of the gene expression based linear models to the null model, consisting of biological parameters and no expression (SSE = 1.2). The 500 genes with the smallest sum squared error were considered in further analysis (max SSE = 0.54).
The linear model identifies a 10 gene network to represent the pathway scale: A radiosensitivity biomarker discovery platform?
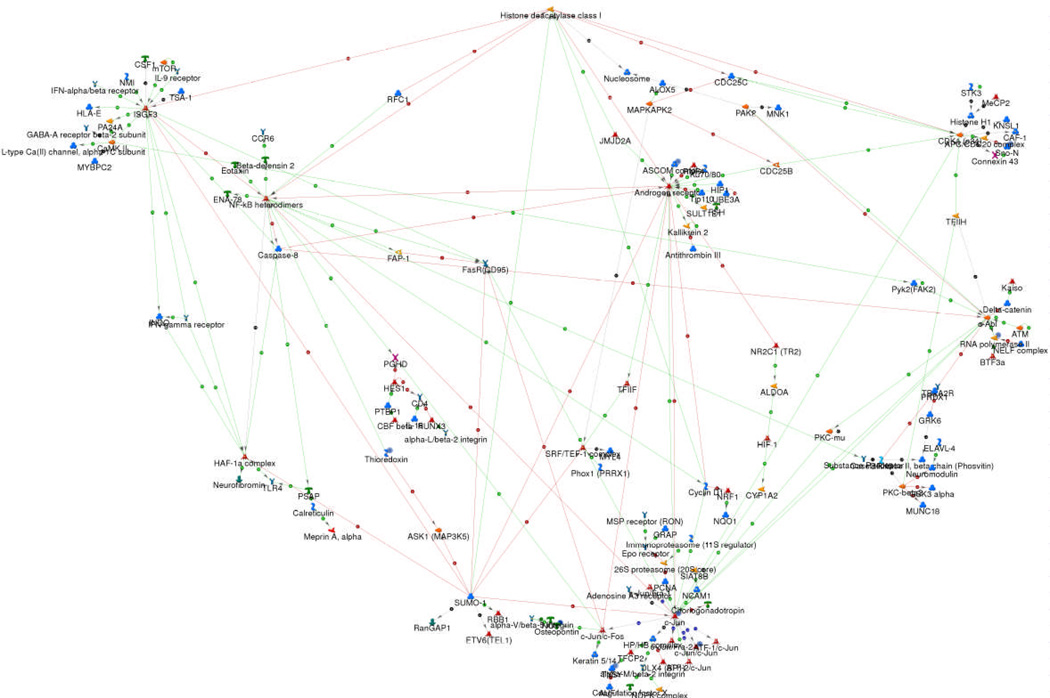
The biomarker discovery platform is a network-based representation of the 500 identified radiosensitivity genes. To create this network, genes (nodes) and literature-based connections (edges) were plotted using GeneGo MetaCore (Figure 2). The network architecture is consistent with a scale-free network and represents interactions between individual targets.
Figure 2. The radiosensitivity network.
GeneGO™ MetaCore™ was used to generate a network of direct connections between the 500 genes selected for analysis. Red, green and gray arrows indicate negative, positive and unspecified effects.
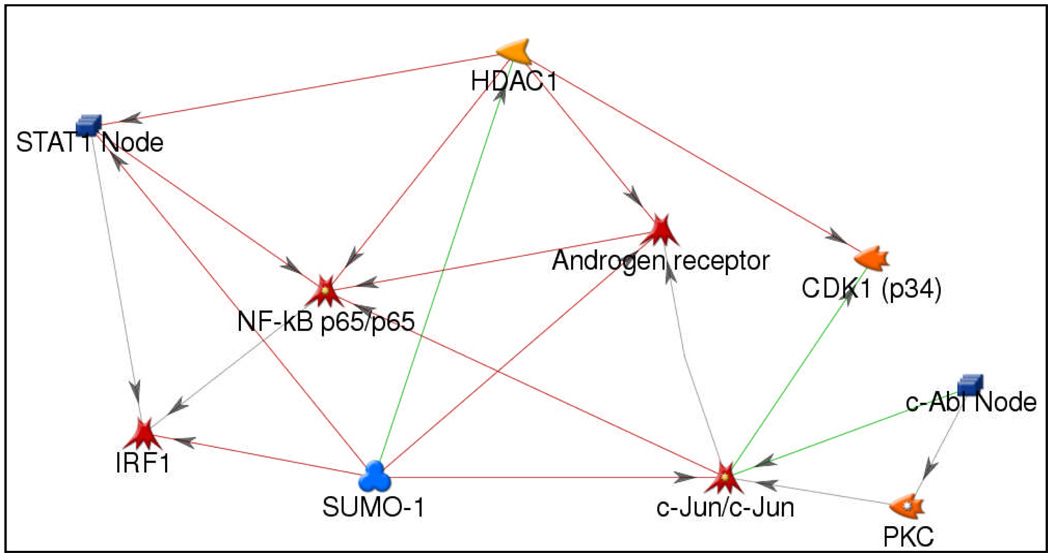
Since targets with high degrees of connectivity are considered to be the most important components of a network 21, we examined hubs with more than 5 connections. The ten hubs are shown in Figure 3: c-Jun, HDAC1, RELA (p65 subunit of NFKB), PKC-beta, SUMO-1, c-Abl, STAT1, AR, CDK1 and IRF1. Remarkably, several of these hubs have been previously reported to be involved in radiation signaling 22–31and 7/10 (HDAC1, PKC-beta, NFKB, c-Abl, STAT1, AR, CDK1) have been studied as targets for radiosensitizer development 32–37. Further, a recent study identified an interferon-related signature that includes STAT1 (one of the ten hubs) and correlated signature predictions to clinical outcome in breast cancer patients treated with chemotherapy and/or radiotherapy 38.
Figure 3. A hub-based network view of the radiosensitivity model.
Hubs were identified as having more than 5 connections within the network. STAT1, IRF1, NFKB, AR, and c-Jun are indicated as transcription factors while HDAC1, CDK1, PKC and c-Abl are annotated as enzymes. SUMO1 is annotated as a protein.
The 10 Gene Systems Model as a Biomarker Panel
The model proposes a 10 gene network/system to define the pathway scale within the radiosensitivity biological continuum. Thus we explored whether the 10 gene system could function as a biomarker panel. To determine whether this was reasonable to pursue, we investigated connections between known radiosensitizer drug targets and the network. If the biomarker platform models radiosensitivity then these drug targets should be connected to the platform. We tested 9 targets associated with drugs in clinical development or routine clinical use. All are linked by primary interconnection to at least one of the hubs (Table 2), arguing for the biological/clinical relevance of the model. Interestingly, each target interferes with a minority of the hubs, suggesting that our current clinical approach to radiosensitization might be inefficient at disrupting the proposed radiosensitivity network.
Table 2.
Radiosensitization targets link to the network.
| Target | Incoming Edge (Network to Target) | Outgoing Edge (Target to Network) |
|---|---|---|
| TOP1 | c-AblPKC-betaSUMO-1 | c-Jun |
| Ras/Raf/Mek/Erk | ARCDK1STAT1 | c-JunAR |
| EGFR | ARPKC-beta | c-AblSTAT1 |
| COX2 | c-JunSTAT1NFKB | --- |
| DNAPK | c-Abl | c-Abl |
| PARP1 | HDAC1 | NFKB |
| BIRC5 | CDK1NFKB | --- |
| HSP90 | STAT1 | AR |
| TGFB1, TGFB2 | c-JunARNFKBHDAC1 | --- |
The biomarker discovery platform represents the biological diversity of the network: Ras and TO play a key role but not p53
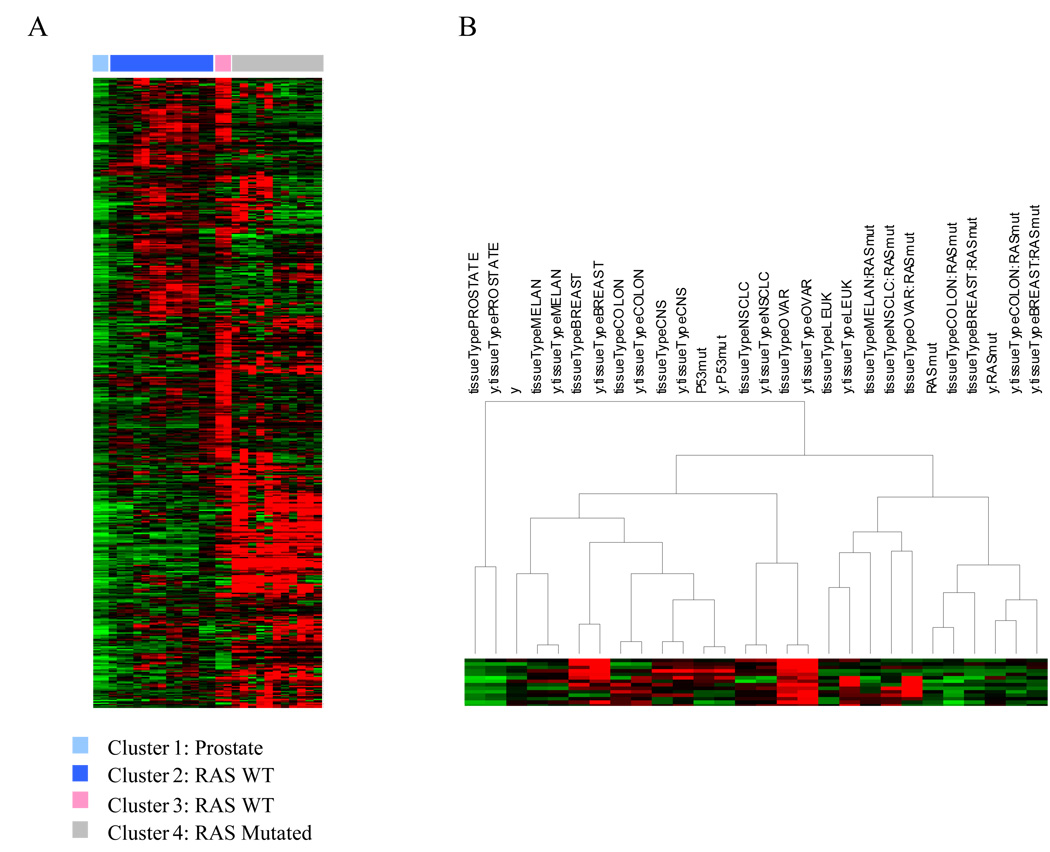
This platform provides an opportunity to study the molecular diversity within the radiosensitivity network (500 gene network). Figure 4 is a cluster heatmap of the impact of each biological variable considered across the genes in the network. Thus, we tested all 28 terms within the model and ranked them in importance. The model identifies four significant radiosensitivity clusters of terms and genes. TO, ras status and their interaction with gene expression proved to be key variables in defining the four clusters. Interestingly, the prostate cancer term grouped separately. Further, p53 did not have a large impact in the analysis. The ras wt population was divided into two groups (lung and ovarian vs. other TO). The ras term was dominant. Cell lines with mutated ras grouped closer than cell lines from the same TO, as exemplified by breast cancer cell lines.
Figure 4. Integration of biological parameters in the gene expression/SF2 model.
500 gene-based linear models of radiosensitivity were clustered based on the impact of each term within the model. Each spot in the heatmap represents a p-value from a single coefficient within each individual gene-based model. Terms include gene expression (y), TO (tissueType), ras status (RASmut) and p53 status (P53mut). The combination of two terms (e.g. y:RASmut) indicates an interaction term.
Radiosensitivity clusters in the platform are biologically distinct
To explore the functional difference in the radiosensitivity clusters, we performed pathway analysis using the genes identified within each cluster. An AP-1 regulated pathway was the only commonality between the three main clusters suggesting a key role for c-Jun in the network. Table 3 shows key biological differences across clusters in the network. For example, genes in cluster 2 represented pathways in metabolism, hypoxia and Akt. Genes in cluster 3 represented 29 pathways, 11 of which were cell-cycle related. Finally, genes in cluster 4 were the most functionally diverse, representing pathways in DNA repair, cell cycle regulation, adhesion, apoptosis, immune response and protein kinase cascades. Importantly, while many of these pathways have been implicated in the regulation of radiation response, our model suggests that the importance of each pathway depends on the biological context that defines network dynamics.
Table 3.
Clusters in the radiosensitivity network are biologically distinct.
| Cluster | Map | Cell Process | p value |
|---|---|---|---|
| transcription, proteolysis, | 0.00059 | ||
| Role of Akt in hypoxia induced HIF1 activation | protein kinase cascade | ||
| 2 | Glycolysis and gluconeogenesis | Carbohydrates metabolism | 0.00066 |
| 0.02093 | |||
| Role of AP-1 in regulation of cellular metabolism | transcription | ||
| Role SCF complex in cell cycle regulation | cell cycle | 0.00349 | |
| 3 | Role of AP-1 in regulation of cellular metabolism | transcription | 0.00428 |
| Role APC in cell cycle regulation | cell cycle | 0.00769 | |
| transcription, transcription, protein | 0.00010 | ||
| Role ASK1 under oxidative stress | kinase cascade | ||
| Role of Brca1 and Brca2 in DNA repair | cell cycle | 0.00033 | |
| Role of IAP-proteins in apoptosis | cell death, apoptosis | 0.00188 | |
| Role of AP-1 in regulation of cellular metabolism | transcription | 0.00364 | |
| 4 | Chemokines and adhesion | cell adhesion | 0.00435 |
| Regulation of G1/S transition (part 2) | cell cycle | 0.01252 | |
| ATM/ATR regulation of G1/S checkpoint | cell cycle | 0.02392 | |
| FAS signaling cascades | cell death, apoptosis | 0.02701 | |
| IL2 activation and signaling pathway | immune response | 0.03031 | |
Validation of c-Jun Hub
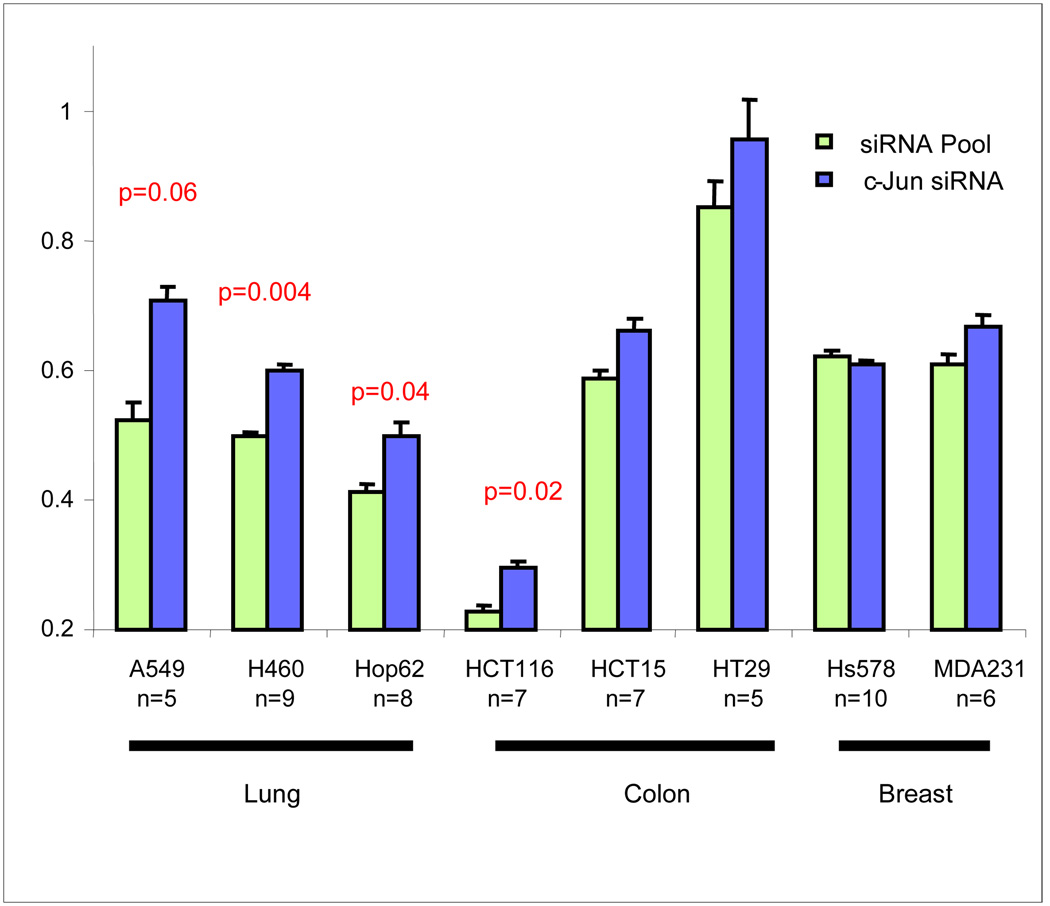
We examined the hub-based model and chose to validate the effect of c-Jun knockdown on radiosensitivity. We chose c-Jun because it was identified as a central hub and it had the largest number of network connections. Although c-Jun has been identified as an early response gene in radiation response 23, 39, its functional role in radiosensitivity is still in debate. The model ranked TO as the most important variable influencing the overall importance of c-Jun to the network. Therefore we tested the effect of c-Jun knockdown on eight cell lines from three different TO: lung, colon and breast cancer (Figure 5). An overall significant trend towards radioresistance was observed across cell lines (p=0.016, paired Wilcoxon rank-sum test, n=8). However, at the individual level significant changes were observed in 3/8 cell lines (H460 p=0.004; Hop62 p=0.04; HCT116 p=0.02) with the main effect being observed in lung cancer cell lines (H460, p=0.004; Hop62, p=0.04; A549, p=0.06), thus confirming TO as an important variable influencing this hub. Interestingly, consistent with these results, our analysis identified a direct correlation between gene expression and radiosensitivity in lung cancer cell lines (Figure 5B). Additionally, since the network model indicates signal redundancy and feedback among central hubs, perturbing c-Jun alone may not uniformly result in changes to radiosensitivity.
Figure 5. Biological validation of c-Jun.
A. c-Jun was knocked down in eight cell lines using siRNA and SF2 was determined. The mean and standard errors from at least five independent experiments in triplicates are represented. Down-regulation of c-Jun was verified by Western blot.B. C-Jun gene expression is directly proportional to radiosensitivity in lung cancer cell lines. Graphic representation of c-Jun gene expression and SF2 in lung cancer cell lines in the 48 cell line dataset (A549, H460, HOP62, NCIH23, HOP92, EKVX).
A Predictive Model of Cellular Radiosensitivity
We developed a linear regression algorithm to predict radiosensitivity in the 48 cell line database, using gene expression rank of the 10 hubs. This rank-based model is given below:
This model which predicts a continuous variable (SF2) was independently tested in the 12 remaining cell lines from the NCI (60) using SF2s recently reported 13. We defined a correct prediction as values within ±10% of the measured value 9. This accounts for the reported average variability of the clonogenic assay 40. A comparison of SF2s in our dataset compared with Amundson (supplementary figure 3), shows a mean difference of ± 17% with only 17/48 cell lines falling within ±10% between the two datasets. Thus, this definition is on the conservative side of the mean SF2 variability ranges observed. The model predicted 5/12 cell line SF2 correctly (p=0.07, compared to chance, Table 4). Since this is not a binary prediction (e.g. radiosensitive vs. radioresistant), the likelihood that a prediction will be correct by chance is about 20% .
Table 4.
Model Predictions vs. Measured Value for independent test set
| Cell Line | Predicted | Reported |
|---|---|---|
| SF2 | SF2 | |
| BREAST_MCF7ADRr | 0.57 | 0.56 |
| BREAST_MDN | 0.45 | 0.70 |
| CNS_SF295 | 0.23 | 0.73 |
| LEUK_K562 | 0.82 | 0.05 |
| LEUK_RPMI8266 | 0.64 | 0.10 |
| LEUK_SR | 0.73 | 0.07 |
| NSCLC_NCIH226 | 0.79 | 0.63 |
| NSCLC_NCIH332M | 0.56 | 0.65 |
| NSCLC_NCIH522 | 0.52 | 0.43 |
| OVAR_IGROV1 | 0.49 | 0.39 |
| RENAL_RXF393 | 0.57 | 0.67 |
| RENAL_TK10 | 0.71 | 0.52 |
Interestingly, the model is most inaccurate in predicting leukemia cell lines, a finding consistent with our previously published model 9. If leukemia cell lines are excluded, the results are significant (5/9 correct, p=0.02).
Discussion
The identification of novel biomarkers is fundamental to the development of biologically-guided treatment strategies in radiation oncology. In this paper we present a discovery platform that we propose as a rational strategy to the identification of novel radiation-specific biomarkers. The platform applies a systems biology approach to modeling the radiosensitivity network in a database of 48 human cancer cell lines. It proposes a highly interconnected radiosensitivity network with ten central hubs and significant signal redundancy, a key feature of complex and robust biological systems 41.
The applicability of the platform as an approach to biomarker discovery is supported by several observations. First, the platform identified 10 hub genes that are proposed to play a central role in determining radiophenotype. Indeed, these hubs are known to play a mechanistic role in radiation response 22–31 or have been identified as potential targets for development of radiosensitizers 32–37. Further, we confirmed the influence on radiophenotype of one hub in the 10 gene system: c-Jun. Interestingly, the model ranked TO as the most important variable for c-Jun in the system. We confirmed this analysis by showing that the induction of radioresistance by c-Jun knockdown is primarily observed in lung but not breast or colon cancer cell lines. Furthermore, we developed a ranked-based linear regression model of cellular radiosensitivity based on the 10 gene systems model. This was independently validated in the remaining 12 cell lines in the NCI (60) and SF2 was accurately predicted in 5/12 cell lines (p=0.07). However, the model was inaccurate when predicting leukemia cell lines and when these cell lines were excluded the predictive test was significant (5/9, p=0.02). A possible explanation for this finding is that radiation-induced death occurs via different mechanisms between epithelial and leukemia cells (mitotic cell death vs. apoptosis) 42. Since the majority of cell lines in the database are of epithelial origin, it is possible the model is more accurately representing the biological networks of epithelial cells. One strategy to improve accuracy would be to exclude leukemia cell lines from model development. However since leukemia cell lines only represent 3/48 cell lines in the database, we think its likely individual impact across the model is small. Another possibility would be to include the observed SF2 heterogeneity in model development.
Recently, an interferon-related signature was identified as a predictive marker of outcome in breast cancer patients treated with chemotherapy and/or radiation therapy. This signature was cross-validated against our published cell line dataset, to determine whether there was any association with radioresistance. Indeed 36/49 signature genes were in the top 25% of all genes ranked by their correlation to SF2 with Stat1 being the most highly correlated. 38. Finally, we have demonstrated that the gene expression model is an accurate predictor of response or prognosis after concurrent chemoradiotherapy in three independent cohorts of 118 patients with rectal, esophageal or head and neck cancer (see companion paper).
Although network hubs can be used as biomarkers of radiosensitivity, they can also be investigated as potential drug targets. Since these hubs are the most highly connected nodes, disrupting them may lead to the most significant downstream signaling changes. Consistent with this, we found that nine radiosensitizing targets are linked to at least one network hub by primary interconnection. However, the platform suggests that targeting single hubs would be an inefficient clinical strategy for radiosensitization/radioprotection since the network has significant signal redundancy. For example, we observed that c-Jun knockdown resulted in induction of radioresistance in lung cancer cell lines but not in breast cancer cells lines
Therapeutically targeting a single hub could lead to inconsistent system outputs, since phenotypic response could be driven by competing signals. The radiosensitivity platform provides a systems-level view of the signaling networks that could be used to identify competing signaling networks. For example, a model could be developed by generating experimental data of radiophenotypic changes as a result of hub gene expression perturbations (siRNA-knockdown). This could serve as a framework to identify clinical strategies to more effectively target the network.
Although biological targets can be identified within the network, the importance of these targets may vary depending on clinical context. For instance, genes important in RAS-mutated cell lines were more biologically diverse than in RAS wild-type. The platform can be utilized to develop a RAS-specific radiosensitivity network, thus allowing the integration of biological context into rational targeting strategies. Additionally, ras was the dominant term in the model over TO and p53. This suggests that it is reasonable to consider ras status in clinical trial design. This is supported by the demonstration that wild-type Kras is required for panitunumab efficacy 43.
There are inherent biases to our analysis that are important to consider. First, the analysis will favor well-studied genes, since literature-based primary interconnections are key criteria utilized to build the network. We took this approach since hub connectivity is correlated with biological importance in yeast studies 21. Thus, the current model may not be as effective at identifying orphan genes that function as central hubs in the network. Second, there is the notable absence of microenvironment-related genes in the identified hubs. Although clusters 2 and 4 represented a hypoxia-related pathway, these genes were dropped when primary interconnections were considered. This is not surprising since the data was generated in tissue culture under normoxic conditions. Further, none of the genes in the 10 hub system is involved in double-strand break (DSB) repair, an important process mediating radiosensitivity 44. To address this concern we identified all 31 DSB repair genes present in the HU-6800 chip and mapped their relationship to the 10 gene system. Almost all DSB genes considered interact with the network either by direct connection or by an intermediary (supplemental section table 13 and supplemental section figure 2). Thus changes in DSB repair could influence the network and vice-versa.
In conclusion, we developed a discovery platform for the identification of novel radiation-specific biomarkers. The platform identifies a biologically diverse radiosensitivity network with ten central hub genes. We propose this platform may play a central role in the integration of biology into clinical radiation oncology practice.
Supplementary Material
Acknowledgments
Support: NCI 5K08CA108926-03, NFGC – DAMD 170220051, Moffitt GU Foundation and Mr. Arnold VanZanten
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: SE and JTR are named as inventors in a patent application for the technology described.
References
- 1.Dalton WS, Friend SH. Cancer biomarkers--an invitation to the table. Science. 2006;312:1165–1168. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 2.van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 4.Chung CH, Parker JS, Karaca G, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 5.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 6.Hood L, Perlmutter RM. The impact of systems approaches on biological problems in drug discovery. Nat Biotech. 2004;22:1215–1217. doi: 10.1038/nbt1004-1215. [DOI] [PubMed] [Google Scholar]
- 7.Kitano H. Systems Biology: A Brief Overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 8.Hood L, Heath JR, Phelps ME, et al. Systems Biology and New Technologies Enable Predictive and Preventative Medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 9.Torres-Roca JF, Eschrich S, Zhao H, et al. Prediction of Radiation Sensitivity Using a Gene Expression Classifier. Cancer Res. 2005;65:7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 10.Chen AY, Okunieff P, Pommier Y, et al. Mammalian DNA topoisomerase I mediates the enhancement of radiation cytotoxicity by camptothecin derivatives. Cancer Res. 1997;57:1529–1536. [PubMed] [Google Scholar]
- 11.Staunton JE, Slonim DK, Coller HA, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Tan AC, Winslow RL, et al. Merging microarray data from separate breast cancer studies provides a robust prognostic test. BMC Bioinformatics. 2008;9:125. doi: 10.1186/1471-2105-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amundson SA, Do KT, Vinikoor LC, et al. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 14.Fertil B, Malaise EP. Intrinsic radiosensitivity of human cell lines is correlated with radioresponsiveness of human tumors: analysis of 101 published survival curves. Int J Radiat Oncol Biol Phys. 1985;11:1699–1707. doi: 10.1016/0360-3016(85)90223-8. [DOI] [PubMed] [Google Scholar]
- 15.Malaise EP, Fertil B, Chavaudra N, et al. Distribution of radiation sensitivities for human tumor cells of specific histological types: comparison of in vitro to in vivo data. Int J Radiat Oncol Biol Phys. 1986;12:617–624. doi: 10.1016/0360-3016(86)90071-4. [DOI] [PubMed] [Google Scholar]
- 16.Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22:8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- 17.Kim IA, Fernandes AT, Gupta AK, et al. The influence of Ras pathway signaling on tumor radiosensitivity. Cancer and Metastasis Reviews. 2004;23:227–236. doi: 10.1023/B:CANC.0000031763.95152.09. [DOI] [PubMed] [Google Scholar]
- 18.Cuddihy AR, Bristow RG. The p53 protein family and radiation sensitivity: Yes or no? Cancer and Metastasis Reviews. 2004;23:237–257. doi: 10.1023/B:CANC.0000031764.81141.e4. [DOI] [PubMed] [Google Scholar]
- 19.El-Deiry WS. The role of p53 in chemosensitivity and radiosensitivity. Oncogene. 2003;22:7486–7495. doi: 10.1038/sj.onc.1206949. [DOI] [PubMed] [Google Scholar]
- 20.Gudkov AV, Komarova EA. The Role of p53 in Determining Sensitivity to Radiotherapy. Nature Reviews Cancer. 2003;3:117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 21.Jeong H, Mason SP, Barabasi AL, et al. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 22.Deng X, Hofmann ER, Villanueva A, et al. Caenorhabditis elegans ABL-1 antagonizes p53-mediated germline apoptosis after ionizing irradiation. Nat Genet. 2004;36:906–912. doi: 10.1038/ng1396. [DOI] [PubMed] [Google Scholar]
- 23.Hallahan DE, Dunphy E, Kuchibhotla J, et al. Prolonged c-jun expression in irradiated ataxia telangiectasia fibroblasts. International Journal of Radiation Oncology*Biology*Physics. 1996;36:355–360. doi: 10.1016/s0360-3016(96)00327-6. [DOI] [PubMed] [Google Scholar]
- 24.Kao GD, McKenna WG, Muschel RJ. p34(Cdc2) kinase activity is excluded from the nucleus during the radiation-induced G(2) arrest in HeLa cells. J Biol Chem. 1999;274:34779–34784. doi: 10.1074/jbc.274.49.34779. [DOI] [PubMed] [Google Scholar]
- 25.Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappa B through two distinct mechanisms. PNAS. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Yang D, Minemoto Y, et al. NF-[kappa]B Is Required for UV-Induced JNK Activation via Induction of PKC[delta] Molecular Cell. 2006;21:467–480. doi: 10.1016/j.molcel.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Mårten Fryknäs SDFÖLRMRHGMGUPPNRLAI. STAT1 signaling is associated with acquired crossresistance to doxorubicin and radiation in myeloma cell lines. International Journal of Cancer. 2007;120:189–195. doi: 10.1002/ijc.22291. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima T, Yukawa O, Azuma C, et al. Involvement of protein kinase C-related anti-apoptosis signaling in radiation-induced apoptosis in murine thymic lymphoma(3SBH5) cells. Radiat Res. 2004;161:528–534. doi: 10.1667/rr3176. [DOI] [PubMed] [Google Scholar]
- 29.Pamment J, Ramsay E, Kelleher M, et al. Regulation of the IRF-1 tumour modifier during the response to genotoxic stress involves an ATM-dependent signalling pathway. Oncogene. 2002;21:7776–7785. doi: 10.1038/sj.onc.1205981. [DOI] [PubMed] [Google Scholar]
- 30.Terzoudi GI, Jung T, Hain J, et al. Increased G2 chromosomal radiosensitivity in cancer patients: the role of cdk1/cyclin-B activity level in the mechanisms involved. Int J Radiat Biol. 2000;76:607–615. doi: 10.1080/095530000138268. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Meng A, Lang H, et al. Activation of Nuclear Factor {kappa}B In vivo Selectively Protects the Murine Small Intestine against Ionizing Radiation-Induced Damage. Cancer Res. 2004;64:6240–6246. doi: 10.1158/0008-5472.CAN-04-0591. [DOI] [PubMed] [Google Scholar]
- 32.Wang QE, Zhu Q, Wani G, et al. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell JS, Brady K, Burgan WE, et al. Gleevec-Mediated Inhibition of Rad51 Expression and Enhancement of Tumor Cell Radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 34.Ma BBY, Bristow RG, Kim J, et al. Combined-Modality Treatment of Solid Tumors Using Radiotherapy and Molecular Targeted Agents. J Clin Oncol. 2003;21:2760–2776. doi: 10.1200/JCO.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 35.Cerna D, Camphausen K, Tofilon PJ. Histone deacetylation as a target for radiosensitization. Curr Top Dev Biol. 2006;73:173–204. doi: 10.1016/S0070-2153(05)73006-4. [DOI] [PubMed] [Google Scholar]
- 36.Milas L, Mason KA, Liao Z, et al. Chemoradiotherapy: Emerging treatment improvement strategies. Head & Neck. 2003;25:152–167. doi: 10.1002/hed.10232. [DOI] [PubMed] [Google Scholar]
- 37.Kaminski JM, Hanlon AL, Joon DL, et al. Effect of sequencing of androgen deprivation and radiotherapy on prostate cancer growth. Int J Radiat Oncol Biol Phys. 2003;57:24–28. doi: 10.1016/s0360-3016(03)00539-x. [DOI] [PubMed] [Google Scholar]
- 38.Weichselbaum RR, Ishwaran H, Yoon T, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallahan DE, Gius D, Kuchibhotla J, et al. Radiation signaling mediated by Jun activation following dissociation from a cell type-specific repressor. J. Biol. Chem. 1993;268:4903–4907. [PubMed] [Google Scholar]
- 40.Peters LJ. The ESTRO Regaud lecture Inherent radio sensitivity of tumor and normal tissue cells as a predictor of human tumor response. Radiotherapy and Oncology. 1990;17:177. doi: 10.1016/0167-8140(90)90202-8. [DOI] [PubMed] [Google Scholar]
- 41.Albert R, Jeong H, Barabasi A-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 42.Brown JM, Wouters BG. Apoptosis, p53, and Tumor Cell Sensitivity to Anticancer Agents. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- 43.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 44.Willers H, Dahm-Daphi J, Powell SN. Repair of radiation damage to DNA. Br J Cancer. 2004;90:1297–1301. doi: 10.1038/sj.bjc.6601729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.