Abstract
Single molecule-based protocols have been gaining popularity as a way to visualize DNA replication at the global genomic and locus-specific levels. These protocols take advantage of the ability of many organisms to incorporate nucleoside analogs during DNA replication, together with a method for displaying stretched DNA on glass for immunostaining and microscopy. We describe here a microfluidic platform that can be used to stretch and capture labeled DNA molecules for replication analyses. This platform consists of parallel arrays of 3-sided, 3 or 4 μm high, variable width capillary channels fabricated from polymethyl siloxane (PDMS) by conventional soft lithography, and silane-modified glass coverslips to reversibly seal the open side of the channels. Capillary tension in these microchannels facilitates DNA loading, stretching and glass coverslip deposition from μL-scale DNA samples. The simplicity and extensibility of this platform should facilitate DNA replication analyses using small samples from a variety of biological and clinical sources.
INTRODUCTION
DNA replication lies at the heart of biology: accurate and complete genome replication ensures genetic continuity and cell viability at each division, and species continuity between generations. Replication is well-understood in outline 1,2. Five ‘classical’ DNA polymerases replicate the bulk of genomic DNA, and are aided by an additional group of ‘specialized’ polymerases that replicate specific chromosomal regions such as telomeres or synthesize DNA on damaged templates to complete genome replication 3-7. This central role of replication in biology has made DNA replication a high value therapeutic target. Drugs that target and disrupt human DNA replication are widely used as cancer chemotherapeutic and immunosuppressive agents. Replication inhibitors that selectively target bacterial or viral replication, such as DNA gyrase inhibitors and nucleoside analog chain terminators, have also gained wide use as antibacterial and antiviral agents 8-10.
Assays available to analyze DNA replication
A wide variety of assays have been developed to analyze DNA replication by examining genomic DNA, chromosomes or individual genes. These complementary assays have been used to gain mechanistic insight and to identify genetic, environmental and pharmacologic perturbations that alter DNA replication. Replication assays that focus on single replicating DNA molecules have recently received renewed interest 11. This reflects the ability of these assays to simultaneously reveal both global replication dynamics and altered replication in subsets of replicating molecules.
Current single molecule replication assays exploit the ability of many organisms to incorporate halogenated pyrimidine nucleoside analogs into replicating DNA. The incorporated BrdU, CldU and IdU are then detected by immunostaining DNA. Sequential labeling with pairs of these analogs followed by detection with analog-specific antisera provides a powerful way to reveal replication dynamics 12,13. These methods, recently reviewed in detail 11, represent facile and more readily applicable extensions of earlier 3H-thymidine incorporation/autoradiography protocols that were used to visualize replicating bacterial and mammalian DNA molecules 14,15.
Several single molecule replication protocols have been developed to visualize replication tracks in DNA or chromatin fibers 13,16-19. All of these protocols share common labeling and detection protocols, though differ in their solution to the problematic step of reproducibly stretching and capturing long DNA fragments for immunostaining and fluorescence microscopy. We recently developed a facile new solution to this problem in which microfluidic capillary channels are used to stretch and capture replicating DNA molecules 20. This protocol builds on earlier work in which a similar approach was used for restriction and methylation site mapping of prokaryotic and eukaryotic genomes 21-25.
Advantages and disadvantages of our protocol
Our single molecule replication analysis protocol, termed microfluidic-assisted replication track analysis or ‘maRTA’, provides a simple, inexpensive and extensible platform for single molecule DNA replication analyses. Though rigorous comparison of maRTA with other available single molecule technologies (SMARD, Dynamic combing, DIRVISH) will require side by side testing, several suggestions can be made regarding its comparative advantages and disadvantages. MaRTA is based on cheap, easily fabricated polydimethyl siloxane (PDMS) micro-capillary channels that require no specialized equipment to use yet achieve orderly deposition of DNA over a fixed grid-like pattern. Thus, subsequent scanning of fibers is easy and automatable. MaRTA offers a simple protocol for preparing silanized glass coverslips that are stable and storable; we have not noticed any between-batch differences in the quality of glass generated by this protocol. Our tests show that similar to other single molecule technologies, maRTA platform is compatible with fiber FISH. Also of note is that maRTA uses small (μL scale) sample volumes. These features should allow maRTA to be further developed to permit multiplexed, microfluidic processing and analysis of DNA replication in small biological or clinical samples.
On the other hand, the front-end requirement to manufacture a negative master for making microchannels, and the fact that PDMS requires an additional treatment to be rendered hydrophilic, can be considered disadvantages of maRTA.
Microfabrication and glass treatment for maRTA experiments
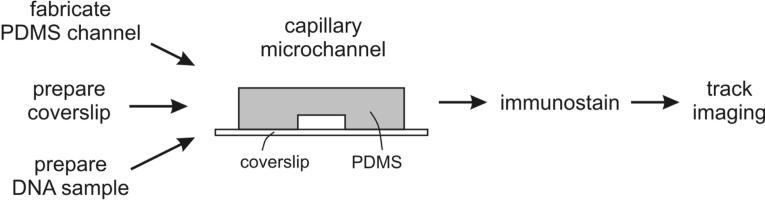
The flow diagram in Figure 1 depicts the steps needed to set up microchannel-assisted replication track analysis (maRTA). A negative master is fabricated using standard photolithography (figure 2, steps 1-9 of the protocol) and it serves as a template for polydimethylsiloxane (PDMS) microchannels which are cast using a soft lithography procedure (figure 3, steps 10-14). Generating a negative master requires access to a clean room with specialized equipment. As microfabrication for biomedical applications becomes more and more common, our first suggestion for those interested in implementing this protocol is to do a survey of your colleagues and departments to identify existing facilities and expertise in photolithography.
Figure 1. Overview of a setup for microchannel-assisted Replication Track Analysis (maRTA).
PDMS microchannels are fabricated and glass coverslips are silanized prior to the procedure. High molecular weight DNA is isolated out of nucleoside analog-labeled cells. A microchannel patch is reversibly sealed onto a coverslip and a μL-scale droplet of DNA solution is placed at one opening of microchannels. Capillary action draws DNA solution into microchannels and a combination of flow force and attachment to silanized glass stretches DNA. PDMS patch is then removed and DNA on glass can be stained for nucleoside incorporation.
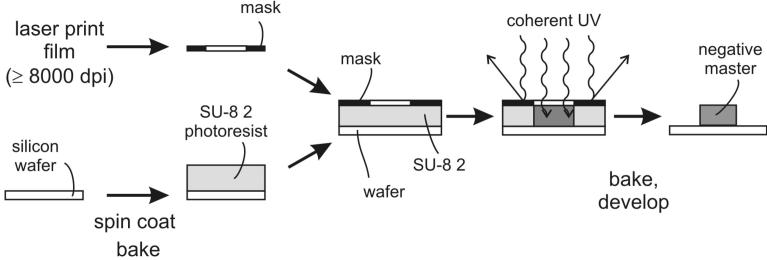
Figure 2. Fabrication of a ‘negative master’ for casting PDMS microchannels using standard photolithography procedures.
A pattern for microchannels is printed on a photomask. A layer of SU-8 2 photoresist is spun onto silicon wafer, then the photomask is layered on top of SU-8 2 and the sandwich is exposed to UV. SU-8 2 polymerizes and hardens only in areas that are not covered with the photomask. After development, unpolymerized SU-8 2 is washed away, leaving a negative master pattern of raised lines on a wafer.
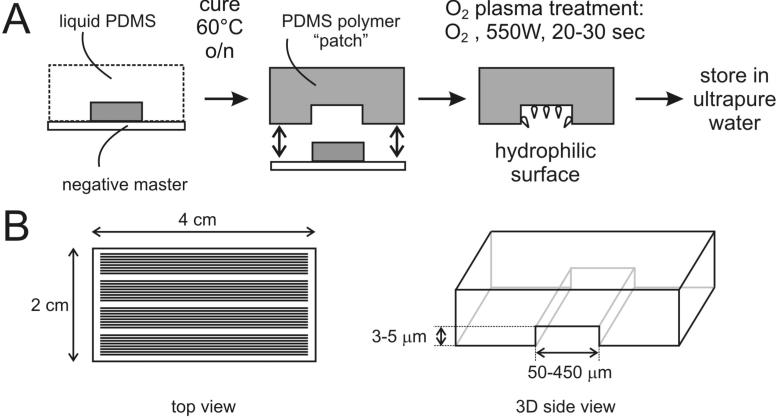
Figure 3. Fabrication of PDMS capillary microchannels by soft lithography.
A. A liquid mixture of a monomer and curing agent is poured onto the negative master. Baking at 65°C speeds up PDMS polymerization. Once polymerized, a PDMS patch is lifted off the master. Oxygen plasma treatment is the used to render it hydrophilic. B. Size parameters for PDMS patches we commonly use. One patch contains several clusters of microchannels.
Fabrication of a master can be also contracted out to a microfabrication foundry. Many of these facilities now exist worldwide, and can be found by a simple web search. They will make the SU-8 master and PDMS replicas (see below) for a fee to your design specifications.
One master can be used to mold over a hundred PDMS replicas. Replica molding does not require any specialized equipment and can be done in a regular lab. However, PDMS microchannels are hydrophobic as they come off the master and need to be treated with ionized oxygen to render them hydrophilic (steps 15-17). Hydrophilicity is needed for passive capillary filling of PDMS microchannels which is used in this protocol. Treated microchannels are stable for up to 2 months and they can be used for DNA stretching twice if rinsed and retreated with oxygen between uses. Oxygen plasma treatment of PDMS microchannels is a second aspect of microfabrication that is most easily addressed by identifying a local fabrication facility and colleagues. Many researchers already use such equipment as Plasma cleaner or UV-ozone cleaner to generate ultra-clean glass surfaces, and this equipment may be used to treat microchannels.
An alternative approach that does not require plasma treatment is to modify the microfluidic platform to include a sample injection port and feed line that can be used to actively load DNA. For additional background on microfluidics, the properties of microscale liquids, and different microfabrication procedures and options including the ‘soft lithography’ procedure used here, see the following reviews 26-30.
Glass coverslips are used both to seal the open face of PDMS patches to create capillary microchannels, and to provide an optical surface for DNA deposition and imaging (figure 4). These coverslips need to be derivatized with a mixture of two silanes (steps 18-23). Once silanized, coverslips can be stored for at least 1 month. DNA is stretched by a combination of forces of capillary tension that pulls DNA solution into the channels, and of binding to reactive moieties of silanes on glass (step 25).
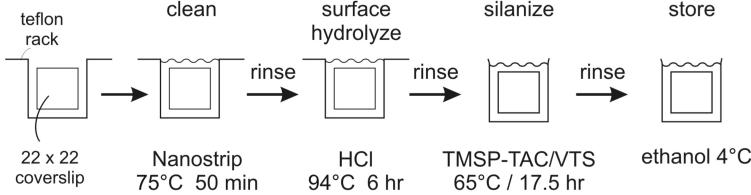
Figure 4. Derivatizing glass coverslips for DNA capture, immunostaining and microscopy.
Glass surface is cleaned and enriched for hydroxyl groups either by sequential incubation with Nanostrip and HCL, as shown, or by incubation with freshly mixed HNO3 and HCL. After extensive rinsing in water and then ethanol, glass is incubated in a silane-water solution, then rinsed again and stored in ethanol.
Experimental design for maRTA experiments
Whilst the specific design of a maRTA experiment will depend on the question(s) being addressed, it must always include pulse-labeling of cells in culture with CldU and IdU or BrdU. Labeling intervals in mammalian cells typically range from 10 to 40 min, or up to 1 hr if a replication-slowing drug is used. The optimal labeling duration should be determined experimentally and optimized for the question being addressed and the organism being examined (see Table 1 Quality controls for additional discussion). The DNA isolation procedure and experimental outline described below are tailored to mammalian cells. When working with yeast, note that genetically engineered strains are required to enable incorporation of halogenated nucleoside analogs (reviewed in 11). Consecutive labeling with two analogs has not yet been reproducibly successful for yeast, particularly in an unperturbed S phase. Isolation of chromosome length yeast DNA suitable for maRTA analyses can be done as described for pulsed-field gel electrophoresis (see, e.g., 31). In this case it is important to incubate cells for 30-60 min after labeling and prior to harvesting in order to ensure that all chromosomes are fully replicated and therefore will be able to enter a pulse-field gel. Cell cycle synchronization can be used to enrich human cell populations for S phase cells prior to pulse labeling, but this is not required. A sample experimental design to address the effects of DNA damage on DNA replication in human fibroblasts is given in Box #1 as an example.
Table 1. Quality controls.
| What to do | What it checks | What to expect |
|---|---|---|
| Pulse-labeling conditions: when labeling replicating cells add the second label for different times, e.g., for 10 and 20 min. | Whether track lengths of ongoing forks accurately reflect the speed of elongation and are not limited by replicon boundaries. Whether stretched DNA length is at least replicon-sized. |
For ongoing forks the rate of growth of second label segments should be similar when derived using length measurements for two different periods of time. |
| Add the 1st and 2nd labels consecutively for the same periods of time | Whether lengths of tracks laid by ongoing forks are not limited by replicon boundaries. Whether stretched DNA is sheared to lengths no smaller than replicon size. Whether replication is not under stress. |
Under normal conditions the ratio of lengths of labeled segments in ongoing forks should be distributed around 1-1.5 |
| Reverse the order in which CldU and IdU are added | Whether antibody staining is influencing the results | Results should not depend on the order of label addition |
| Stain a sample of DNA with anti total DNA antibody and determine DNA fragment lengths | Whether DNA is too sheared | The lengths of molecules should be longer than the lengths of replication tracks |
| Collect two subsets of data: viewing the fields through a filter to one fluorescent label, then to another, e.g first in green then in red. | Whether scanning process is biasing data collection | Calculated fractions of track types may depend on the way the images are collected |
BOX 1. Sample EXPERIMENTAL DESIGN.
A sample experimental design is outlined below to address effects of MMS on DNA replication elongation.
Plate SV40 transformed human fibroblasts at a density of 200,000 cells per 60mm plate.
The next day incubate cells for 12 hours with 0.5mM mimosine to arrest in late G1.
Release from mimosine by changing media, incubate for 8-10hrs. Cells are now in the mid S phase.
Pulse-label with 50μM of 1st label (IdU or CldU) for 20 min.
Wash away the 1st label, add the 2nd label (CldU or IdU, respectively) at 50μM for 20min (untreated control) or add 0.015-0.02% MMS for 20 min. Wash away MMS and add the 2nd label for 20 or 40 min.
Harvest cells or perform an optional step: wash away the 2nd label and incubate untreated and MMS-treated cells without label for additional 30-60min. This step allows replication forks to move away from labeled tracks. Junctions between replicated and unreplicated DNA can be more susceptible to breakage during DNA isolation and stretching.
Harvest cells as described in Preparation of high molecular weight DNA.
Anticipated results
Inhibition of replication elongation by MMS will cause the tracks labeled by the 2nd label to become shorter than in untreated controls. In particular, in ongoing forks the 1st label to 2nd label segment ratios will be higher after MMS treatment (Figure 7D).
MATERIALS
REAGENTS
-
·
silicon wafers (3-inch diameter, P/Boron; Silicon Senses Inc.)
-
·
SU-8 2 photoresist (MicroChem Corporation), Used to fabricate a ‘negative master’ mold for casting PDMS microcapillary channels. SU-8 (formulated in gamma-butyrolactone) and SU-8 2000 (formulated in cyclopentanone) are epoxy based, negative photoresists that have high functionality and high optical transparency. Negative photoresists of this type become polymerized and developer-resistant upon exposure to coherent UV light. Standard formulations of these photoresists allow a wide range of film thicknesses to be generated (from < 1 μm to > 200 μm). The thickness of the finished photoresist is determined primarily by the photoresist formula and the spin speed used to coat silicon wafers. SU-8 2 is used in this protocol to give finished feature heights of 3 to 5 μm. SU-8 5, SU-8 2002 and SU-8 2005 can also be used for negative mastering by modifying process parameters (see: http://www.microchem.com/products/su_eight.htm for additional details).
Δ CRITICAL See manufacturer’s data sheets for process guidelines.
! CAUTION SU-8 is highly sensitive to UV light/ambient room light. Store and process in a room equipped with UV filters.
-
·
SU-8 developer (1-methoxy-2-propyl acetate; MicroChem Corporation), used for removing unexposed SU-8. ! CAUTION Toxic. Wear protective gear and dispose according to your EHS regulations.
-
·
Tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (TFOCS; United Chemical Technology), used for treating the SU-8 masters to facilitate PDMS replica release.
!CAUTION Corrosive and toxic. Wear protective gear and dispose according to your EHS regulations.
-
·
isopropyl alcohol
-
·
Sylgard 184 polydimethylsiloxane (PDMS) polymer components (Dow Corning).
-
·
ΔCRITICAL PDMS consists of two components: silicone elastomer base and Sylgard 184 elastomer curing agent. PDMS is a bio-compatible transparent elastomeric polymer that can be inexpensively replica-molded and is widely used in soft lithographic applications. PDMS surfaces reversibly self-seal against smooth surfaces and can be rendered hydrophilic by a brief exposure to oxygen plasma.
-
·
Nano-Strip solution (Cyantek Corporation 539200) ! CAUTION Highly caustic. Wear protective gear and dispose according to your EHS regulations.
-
·
N-trimethoxysilylpropyl-N,N,N-trimethylammonium chloride (Gelest SIT8415.0)
-
·
vinyltrimethoxysilane (Gelest SIV9220.0)
-
·
CldU, chlorodeoxyuridine (Sigma c6891)
-
·
IdU, iododeoxyuridine (Sigma i7125)
-
·
mimosine (Sigma m0253)
-
·
MMS, methyl methane sulfonate (Sigma m4016) ! CAUTION Toxic and mutagenic. Wear gloves and handle in a ventilated hood. Solutions of MMS in water inactivate over time and can be disposed.
-
·
β-agarase (New England Biolabs m0392S)
-
·
proteinase K powder (Invitrogen 25530-015)
-
·
YOYO-1dye (Invitrogen Y3601)
-
·
rat anti-CldU/BrdU (bromodeoxyuridine) antibody (Serotec MA2060 or Accurate Chemical OBT0030G)
-
·
normal goat serum (NGS; Accurate Chemical ACL1200-100)
-
·
BSA, bovine serum albumin (Sigma a3294)
-
·
goat anti-rat Alexa 594-conjugated antibody (Invitrogen Molecular Dynamics A1100)
-
·
mouse anti-IdU/BrdU antibody (Becton Dickinson 347580)
-
·
goat anti-mouse Alexa 488-conjugated antibody (Invitrogen Molecular Dynamics A11001)
-
·
mouse anti-DNA antibody (anti-deoxythymidine) (Chemicon MAB3034).
-
·
low gelling temperature (LGT) agarose (any brand, but must be DNAse-free)
-
·
EDTA, disodium salt (any high quality supplier can be used for this and the chemicals below)
-
·
deoxycholic acid, sodium salt
-
·
N-lauroylsarcosine, sodium salt
-
·
NaCl
-
·
Tris base
-
·
Tween 20 non-ionic detergent
-
·
phosphate-buffered saline (PBS)
-
·
HCL, concentrated
-
·
‘Hard-as-Nails’ clear nail polish (Sally Hansen 2103, from your local drugstore)
REAGENT SETUP
-
·
CldU Make up as a 10mM solution in water. Store frozen at -20°C in the dark for up to several months.
-
·
IdU Make up as 2mM solution in water. Store frozen at -20°C in the dark for up to several months. Both CldU and IdU can also be made as 50mM solutions in DMSO and stored as above.
-
·
Mimosine Make up as 10mM solution in growth medium (typically Dulbecco-modified Eagle’s medium; DMEM). Filter sterilize and store at +4°C for up to several months.
-
·
LGT agarose Make up as 2% (w/v) solution in water. Store at room temperature (defined here and elsewhere as 20°C) for up to several months. Reheat as needed. CRITICAL Any brand of LGT agarose can be used, as long as it is DNAse-free, i.e. certified for recovery of DNA fragments
-
·
Sodium deoxycholate Make up as 10% (w/v) solution in water. Store at room temperature, indefinitely.
-
·
Sodium lauryl sarcosine Make up as 10% (w/v) solution in water. Store at room temperature, indefinitely.
-
·
Agarose Insert Buffer Make up as 10 mM Tris 7.5, 20 mM NaCl, 50 mM EDTA solution in water. Store at room temperature, indefinitely.
-
·
Lysis buffer Make up fresh as 100mM EDTA pH 8.0, 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1mg/ml Proteinase K solution in water, out of stock solutions (see above) and dry proteinase K powder.
-
·
Block buffer: Make up as 5% BSA, 0.5% Tween20 solution in PBS. Filter sterilize and store at +4°C for up to several months.
-
·
Wash buffer Make up as 1% BSA, 0.1% Tween2 solution in PBS. Can be made by diluting Block buffer 1:5 with PBS. Store at +4°C for up to several months.
EQUIPMENT
-
·
Photoresist spinner (Headway Research Inc. PWM32)
-
·
UV mask aligner (Neutronix-Quintel, Quintel Model 2001 3 inch)
-
·
Two hot plates (preferably programmable)
-
·
Surface Profilometer (KLA-Tencor P-15)
-
·
Fume hood
-
·
Balance
-
·
Rocking platform with variable tilt and rotation speed controls
-
·
Two desiccators (one for PDMS only, one for TFOCS only). CAUTION Due to the toxicity and corrosiveness of TFCOS, a dessicator for TFCOS use only should be set up in a fume hood for silicon wafer silanization.
-
·
Plasma processing reactor center (Plasmatic Systems Inc. Plasma Preen II-973) Also referred as a “plasma etcher”, a vacuum chamber where oxygen radicals are generated. CRITICAL We use a microwave-based plasma etcher (Plasma-Preen II) to increase the hydrophilicity of PDMS surfaces. Other types of plasma etcher can also be used.
-
·
Small oven that can maintain 65 °C setting to cure PDMS
-
·
Water bath that can maintain 65 °C for silanization of coverslips
-
·
Sand bath or a heating block with removable rack units. The receptacle of a heating block can be filled with dry, heat conducting medium such as sand in order to accommodate polypropylene containers (see below)
-
·
Agarose gel insert (‘plug’) molds (Bio Rad 1703706)
-
·
Glass coverslips (Fisher Finest 12-548-B)
-
·
Glass microscope slides (any brand)
-
·
Teflon racks (Invitrogen C14784)
-
·
Kimwipes
-
·
Parafilm
-
·
Polypropylene containers (All-Pak PLC03696)
-
·
Fluorescent Microscope (We use a Zeiss Axiovert confocal microscope equipped with 40x, 63x or 100x objectives, filter sets to red and green fluorescence, and a digital camera. Note that the ability to do confocal imaging is not required for this protocol).
PROCEDURE
Design of the photomask for the SU-8 negative master
• TIMING - variable
A pattern for a negative master can be generated using design software (Corel Draw, Corel Corporation; or AutoCAD, Autodesk, Inc.). Send the resulting file to a commercial printing service (CAD/Art services, http://www.outputcity.com/) to print the pattern at high (≥8000 DPI) resolution on a transparency photomask. Our pattern has clusters of 35 mm long lines ranging from 50 to 450 μm wide and spaced 450 μm apart.
Fabrication of the SU-8 negative master
• TIMING - 4 to 5 hrs
1. Clean silicon wafers by immersing new wafers in Nano-Strip for 20 min, followed by a deionized water rinse. Dehydrate the surface by baking at 200°C for 10 min on a contact hot plate or 30 min in an oven.
! CAUTION Nano-Strip is highly caustic.
Δ CRITICAL STEP Perform steps 1 through 7 in a dust-free ‘clean room’. A dust-free environment is critical because dust particles can be larger than the desired microstructure feature size and will interfere with photoresist coating. To obtain maximum process reliability and promote photoresist adhesion, silicon wafers should be clean and dry prior to applying the SU-8 photoresist. Use new wafers only.
2. Coat wafers with photoresist by spin coating clean silicon wafers on a photoresist spinner. This ensures that the SU-8 photoresist will be of uniform thickness. Use the manufacturer’s guide for setting up coating spins (see http://www.microchem.com/products/su_eight.htm for instructions). For example, for SU-8 2 photoresist a “spread” spin speed at 500 rpm for 10 s (set acceleration ramp rate to 100 rpm/s and hold for 5 sec) will allow photoresist to cover the wafer and spread from the center. A second spin at 1000 to 2000 rpm for 30 s (holding time) is then used to generate the desired finish height. Acceleration and deceleration rates for this second spin should be set to 500 and 1000 rpm/s respectively. Quickly place and center the wafer on a photoresist spinner vacuum holder. Turn on the vacuum, dispense ∼1 mL photoresist (SU-8 2) onto the center of the wafer, and start the “spread” spin. Finish with a second spin to generate a uniform, thin coating.
3. Preset two hot plates at 65 °C and 95 °C respectively. Transfer the coated wafer to the 65°C hot plate and bake for 1 min, then immediately transfer the wafer to the 95 °C hot plate and bake for an additional 3 min.
Δ CRITICAL STEP After coating, the photoresist must be soft baked to evaporate the solvent and solidify the film. Two steps of contact hot plate baking are typically used for thin (< 100 μm thick) SU-8 photoresists.
4. Place the SU-8-coated wafer onto the vacuum chuck of a UV aligner, and bring the photomask (emulsion side facing the wafer) into contact with the photoresist. Use the exposure dose recommendations from MicroChem based on photoresist film thickness and UV power output. 200 mJ/cm2 is used in this protocol to overexpose the photoresist film. This is done to ensure that the negative master pattern we are using, with 35 mm long lines ranging from 50 to 450 μm wide, adheres to the wafer substrate and is not damaged or lost during post-baking and developing.
Δ CRITICAL STEP SU-8 is optimized to polymerize and harden when exposed to near UV (350-400nm) light. Excessive UV doses at wavelengths below 350nm may lead to over-polymerization of the top portion of the photoresist, resulting in exaggerated negative sidewall profiles or ‘T-topping’ upon development. When using a broad spectral output source, filter out excessive energy below 350nm to ensure the straightest channel sidewalls. Under-exposure, in contrast, often results in catastrophic adhesion failure and excessive cracking. Thus if feature size is not absolutely critical, overexposure is preferred.
5. Place the wafer onto a 65 °C hot plate for 1 min, and then onto a 95 °C hot plate for 1 min. Then ramp down the 95 °C hotplate temperature to 25 °C (or your preferred room temperature) at 4 °C /min for better adhesion.
Δ CRITICAL STEP After UV polymerization a second bake must be performed to selectively cross-link the exposed portions of the film.
6. Develop the negative master by immersing the wafer in SU-8 developer for 5 min. Avoid strong agitation. Although the recommended time is 1 min, the actual developing time depends on pattern features, agitation speed, temperature, etc. Cross-linked SU-8 is very stable in developer, thus prolonged immersion causes little harm to the SU-8 mold. We use 5 min immersion with no agitation to ensure development and minimize the possibility of wafer adhesion failure.
TROUBLESHOOTING
7. Following development, rinse the wafer briefly with isopropyl alcohol, then air dry. When completely dry, a raised SU-8 master pattern of long lines should be visible on the wafer surface. All other unexposed area of the wafer should be completely free of photoresist and uniformly shiny (no streaks or shadows). Inspect the patterns under a microscope.
Δ CRITICAL STEP: a 5 min development time should be adequate to remove all unhardened photoresist. If a white film forms on the wafer after isopropyl alcohol rinsing, return the wafer to developer (step 6) for an additional 5 min or until fully developed.
8. Measure the height of the SU-8 patterns using a surface profilometer. This step is recommended though not obligatory because, according to MicroChem guidelines using the aforementioned spin speed will produce a negative master with the desired features, e.g. 2 to 5 μm tall. We are using a master with 3 - 4 μm tall features (e.g. it will cast microchannels that are 3 - 4 μm deep). Deeper microchannels (e.g., 10 μm), lead to higher flow rates and inefficient DNA deposition.
■ PAUSE POINT Store the finished SU-8 master mold in a dust-free environment for up to several years.
9. Surface silanize by vapor-coating with TFOCS in a vacuum chamber. A convenient way to do this is to attach a desiccator jar (containing a small piece of absorbent paper towel in place of the drying pellets) to a vacuum source. Then add a drop of TFOCS to the paper and evacuate air from the chamber. Apply vacuum for 1 min, close off the vacuum line and allow 30 min for deposition.
Δ CRITICAL STEP: To prevent bonding of oxidized silicon to PDMS and to facilitate release, the SU-8 master needs to be silanized prior to PDMS replica molding.
! CAUTION The desiccator chamber must be located inside a chemical fume hood owing to the toxicity and corrosiveness of TFOCS vapor.
■ PAUSE POINT The negative master for PDMS molding is complete at this point. A single silanized master can be used for many cycles (>100) of PDMS replica molding. Store the master for up to several years in a closed container to protect it from dust.
Replica molding of PDMS devices from the SU-8 master
• TIMING : ≥4 hrs
10. Weigh PDMS pre-polymer out in a weigh boat. Next add curing agent at a 10:1 polymer:curing agent weight ratio. A ∼ 20 - 30 gm mixture is needed to cover the surface of each 3 inch silicon wafer. Mix PDMS prepolymer and curing agent thoroughly to ensure uniform cross-linking. Thorough mixing can be accomplished by stirring the PDMS mixture with a wooden tongue depressor for 5 min.
11. Use a vacuum chamber (a desiccator without drying pellets) to degas the PDMS mixture for 10-15 min until bubbles clear.
12. Slowly pour PDMS onto the SU-8 master (from step 9) placed in a weigh boat or a P-100 petri dish to a height of ∼ 2-3 mm. A weigh boat is advantageous for preventing wafer breakage if the wafer needs to be released or transferred from the container at the end of molding. De-gas again for 10-15 min or until bubbles clear.
13. After de-gassing is complete, place PDMS into a 60-65°C oven (> 2 hr) for curing.
14. Remove the cured PDMS-covered master from the oven. Using a clean razor blade or a sharp scalpel, cut a 5 cm × 3 cm rectangle through the PDMS around the line pattern, then carefully peel the PDMS center piece off. A replica pattern of lines should be visible on the PDMS patch surface. The harvested patch then needs to be cleanly trimmed so that the pattern runs end to end and the channels are open. These PDMS microchannel patches form 3 of the 4 walls of the microcapillary channels that will be used for DNA stretching. PDMS replica molding is now complete. Additional replicas can be made from the same master by repeating steps 10-13. Cured PDMS outside of the pattern region can be left on the wafer, thus next time liquid PDMS mixture needs only to be poured into the center region.
Δ CRITICAL STEP: When cutting a PDMS patch out, avoid cutting across the lines of a negative master as this will destroy the master.
■ PAUSE POINT: Keep cured PDMS patches and the SU-8 master in a dust-free environment between casting.
Surface treatment of PDMS microchannels
•TIMING: 30 minutes.
15. If dust particles are visible on the PDMS microchannel (from step 14) surface, clean it with a piece of scotch tape. Gently apply the tape to the surface of the PDMS patch and lift to remove dust particles.
Δ CRITICAL STEP: A clean dust-free PDMS surface is critical for good sealing to glass substrates.
16. Place the PDMS microchannels inside a Plasma Preen II oxygen plasma etcher with the microchannel side up. Set the power to 550W, oxygen pressure at 30 psi and flow rate 3 - 5 SCFH. Treat for 20-30 sec. Optimal system parameters such as oxygen pressure, flow rate, plasma power and treatment time should be established empirically using the test described in step 17. Generally, the settings should be the lowest that produce the desired effect, e.g. a completely hydrophilic PDMS surface.
Δ CRITICAL STEP: To facilitate DNA stretching, hydrophobic PDMS microchannels need to be rendered hydrophilic using oxygen plasma.
17. Test PDMS surface. A simple test of PDMS surface treatment efficacy is to see how well the treated surface can be wetted by water. After oxygen plasma treatment place the PDMS microchannels in a petri dish channel side up and leaning against the edge of the dish. Drop ∼0.5 mL de-ionized water onto the PDMS surface. Water should easily wet the whole surface, leaving a uniform film if the plasma treatment was successful. After the test, immediately immerse PDMS devices in deionized water channel side down. If left dry, PDMS surfaces will revert to a hydrophobic state within 15 min and will not be useable.
■ PAUSE POINT: Treated PDMS microchannel patches can be stored submerged in sterile deionized water at 4°C without losing surface properties for up to 2 months. The PDMS surface can be re-treated with oxygen plasma if it is not sufficiently hydrophilic or is inadvertently dried.
Preparation of silanized glass coverslips
• TIMING: 24 hrs.
18. Clean and surface hydrolyze coverslips using either option A (nano-strip and hydrochloric acid) or option B (nitric and hydrochloric acid).
- Nano-strip and hydrochloric acid method.
-
Place coverslips in Teflon racks in a polypropylene container and immerse in Nano-Strip for 50 minutes at 75° C in a sand bath to maintain a uniform temperature.! CAUTION Nano-Strip is a toxic and caustic mixture of sulfuric acid and hydrogen peroxide. Wear protective gear and handle in fume hood, dispose properly.
- Remove Nano-Strip and rinse coverslips 4 to 5 times with ultrapure water.
- Add concentrated hydrochloric acid to coverslips in a polypropylene container and incubate at 95° C for 6 hours in a sand bath to maintain a uniform temperature.
-
- Nitric and hydrochloric acid method
-
Incubate coverslips 8 hr - overnight in a freshly made 2:1 (v/v) mixture of nitric and hydrochloric acid.! CAUTION: This alternative reagent is extremely corrosive and emits chlorine gas for several hours upon mixing. Incubation should be performed only in a well-ventilated fume hood. Dispose by neutralizing with baking soda.
-
19. Remove surface treatment and rinse coverslips 7-8 times with ultrapure water.
20. Wash coverslips in pure ethanol twice, then air dry in clean, dust-free air.
21. Make up a solution of 12.4 μL of N-trimethoxysilylpropyl-N,N,N-trimethylammonium chloride and 6 μL of vinyltrimethoxysilane in 50 mL of sterile, deionized water. Shake the solution vigorously by hand or on a shaker for 5 min to start silane hydrolysis. Vortex can be used as long as the mixture is moved briskly.
Δ CRITICAL STEP: The solution must be made fresh immediately prior to use and thorough mixing is important to initiate silane hydrolysis.
22. Incubate coverslips with a silane mixture from step 21 at 65° C for 17.5 hrs. Carry out incubation in glass containers (standard histology staining jars) with the coverslips laid flat on the bottom with no overlaps. Use water bath with a lid to minimize evaporation of silane solution.
Δ CRITICAL STEP After silanization it is important to keep track which side of the coverslip was facing up as this is the side to use for capturing DNA.
23. Rinse the silanized coverslips 5 times in ultrapure water and twice in ethanol.
PAUSEPOINT: The finished coverslips can be stored in Teflon racks in ethanol at 4° C for at least 1 month.
Preparation of high molecular weight DNA
• TIMING: 24-48 hrs
24. Harvest 200-400,000 nucleoside analog-labeled cells by trypsinization and wash once with 0.5mL of Agarose Insert buffer.
25. Resuspend cells in 50 μL Agarose Insert buffer and add an equal volume of melted 2% LGT (Low Gelling Temperature), DNAse-free agarose in water. Pipet to mix and immediately place in a well of an agarose insert mold taped off one end.
Δ CRITICAL STEP: Use a wide-bore tip for mixing and transfer. The resulting 100 μL volume of the mixture is enough for one insert of specified size (see Equipment section). Other insert sizes and fewer cells can be used, but adjust volumes accordingly. Some cell types may require adjusting cell concentration in plugs.
26. Let the gel inserts solidify at 4°C for 10 min or longer. Then remove tape from the bottom of the mold and push inserts out using a “tooth” that comes with the mold into flat-bottomed microtubes containing 1mL of Agarose Insert Buffer.
■ PAUSE POINT: At this stage gel inserts can be stored for up to several months at 4° C in the dark.
27. Aspirate off Agarose Insert buffer and add 0.5mL of Lysis buffer per 100μl gel insert. Incubate in a 50-55°C water bath overnight. This lyses embedded cells to free DNA.
Δ CRITICAL STEP: for MMS-treated DNA’s, incubate at 30° C for 36 hrs to avoid breakdown of methylated DNA (see 32).
28. Wash lysed agarose inserts at room temperature with four x 1-2mL changes of TE buffer for 30 min each. Rock or rotate gently to facilitate buffer exchange.
29. Add 0.3-0.5mL 1x agarase buffer per 100μl gel insert. Heat at 75°C for 5 min then inspect the tubes to ensure that the agarose inserts have melted. If needed, heat for another 2-5 min.
Δ CRITICAL STEP: From this point on, minimize agitation to avoid shearing the high MW DNA that is now in solution.
30. Transfer tubes to 42°C and let the temperature equilibrate for 5 min. Add 2-4 μL β-agarase to each tube containing melted agarose and mix by gently inverting once, then incubate for 3-4 hrs at 42°C. Alternatively, resuspend agarase in 50μl of buffer and add to melted agarose solution without further inversion.
Δ CRITICAL STEP: for MMS-treated DNAs, incubate at 37°C for 4-5 hrs.
31. Adjust pH of the DNA solution by adding 1/10 volume of 10xTE pH 8.0 to neutralize the β-agarase buffer (pH6.5).
Δ CRITICAL STEP: Proper stretching of DNA on glass is pH-dependent.
■ PAUSE POINT: After β-agarase digestion DNAs can be stored at 4°C in the dark for at least a month.
Assembly and loading of capillary microchannels
• TIMING: ≥1.5 hrs.
32. Remove PDMS microchannel patch stored in water from step 17, and blot dry against a clean glass surface.
Δ CRITICAL STEP: Microchannels must be absolutely dry for proper use. Residual liquid between channels will result in a failed seal and entry of DNA into space between channels. “Plugs” of liquid inside channels will impede capillary flow. However, once dry, microchannels should be used within 15 min (see step 17 above).
33. Remove silanized coverslip from ethanol and air dry. Position a PDMS microchannel patch on a silane-coated glass substrate from step 23 (channel side down). Microchannels will seal onto the surface spontaneously or after light tapping with forceps. Do not press hard on the microchannel patch, as this will result in flattening of the channels. A good seal should be visible as a striped pattern due to a difference in light reflection between sealed areas (between channels) and open areas (inside channels). A seal is more easily observed against a dark background. It is OK to break the seal to reposition microchannels on a coverslip if needed.
34. Using a wide-bore tip, load no more than 2 μL of DNA solution from step 31 at one end of the microchannels. The DNA solution should enter and progress only along the channels at a rate of 1cm/3-5 sec. Individual speeds may vary between channels of different widths. Exceedingly uniform and fast filling of all channels usually means the seal is imperfect. See steps 32 and 33.
TROUBLESHOOTING.
35. Insert a razor blade at a shallow angle between a microchannel patch and a coverslip at the front end of the patch (i.e., the end from which DNA entered the channels). Slowly push the razor blade forward, separating the patch from the glass, taking care that the seal breaks evenly and its front recedes perpendicular to the axis of channels. Use forceps to pick up the patch after it is separated from the glass. Note that PDMS patches can be recycled once. Rinse and store used PDMS patches in ultrapure water at room temperature for up to several months. Dry completely and retreat with oxygen plasma as described for new patches before reuse. Reusing more than once is not recommended because PDMS deteriorates after the third oxygen plasma treatment. This will show as increased stickiness of PDMS: it will begin adhering to glass.
Δ CRITICAL STEP: Do not drop or slide the PDMS patch on the coverslip or allow it to reseal, as this will result in disorganized deposition of DNA.
TROUBLESHOOTING
36. Lay coverslips flat with the DNA side up in a dust-free place and allow water to evaporate until no liquid remains.
37. Flood dried coverslips with a freshly made mixture of 3:1 methanol: acetic acid for 3 min to fix. Drain gently and air dry coverslips for 1 hr to overnight. Stretched DNA can be stained with YOYO-1 (1μM in TE with 20% β-mercaptoethanol) at this point to inspect the quality of stretching. If staining with YOYO-1, proceed to step 38 after microscopy. DNA can also be stained with YOYO-1 in gel inserts, prior to agarase digestion. Alternatively you can use an antibody against total DNA to examine the quality of stretching (see Reagents and step 50 note below).
Δ CRITICAL STEP: Minimize exposure of YOYO-1-stained DNA to the light beam of the microscope as excessive illumination will break DNA.
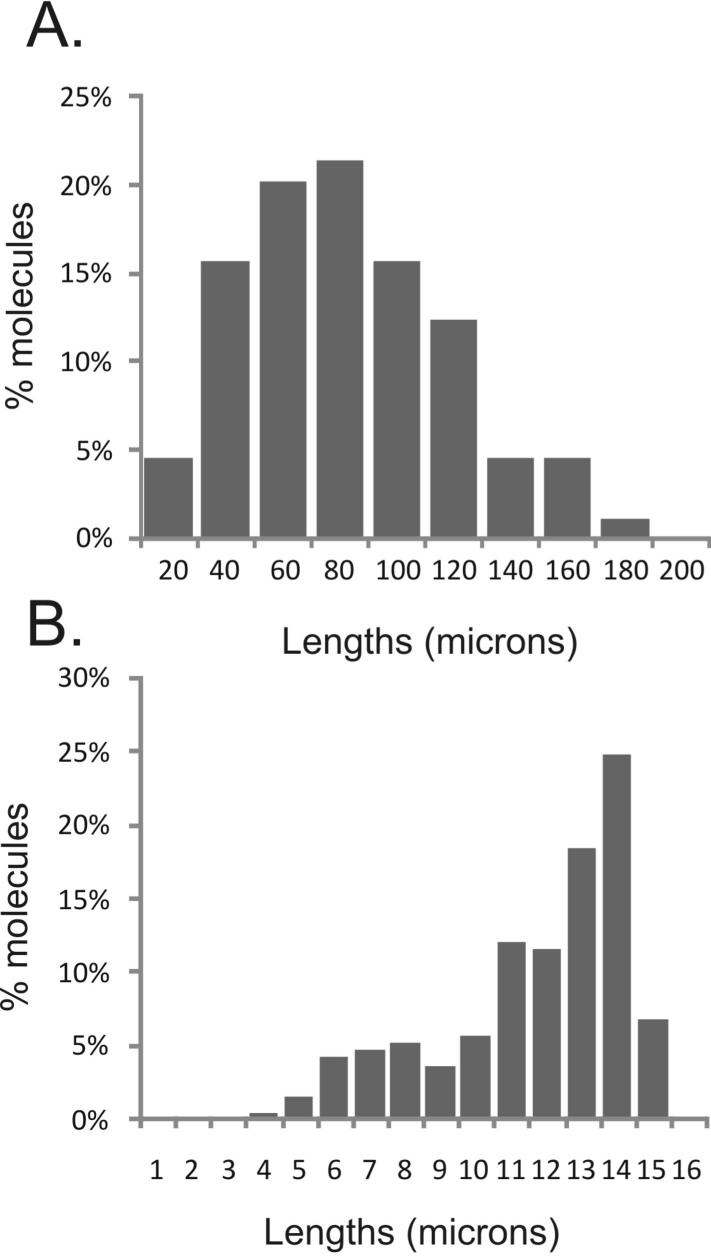
Note that DNA stretched as described above will be sufficiently concentrated to detect plenty of IdU and CldU tracks but too dense to measure lengths of individual DNA molecules. To achieve the latter, DNA should be carefully diluted 1:200-500 in TE. Figure 5A shows a distribution of DNA molecule lengths in microns obtained from a sample prepared and stretched as described above. Median and maximum lengths observed are 80 and 180 μm, respectively. To convert these lengths into microns, one can use a length standard, such as bacteriophage lambda DNA. Figure 5B shows a distribution of molecule lengths measured after stretching a stock solution of phage lambda DNA. The majority of intact molecules in Figure 5B measure around 13-14 μm in length, with the mean of 12.3μm. Thus, the conversion factor may be estimated as 48.5Kb (the length of phage lambda) divided by 12.3, e.g. 1μm=3.9Kb. This estimate is within the range of the original evaluation published for microchannel-assisted stretching on the two-silane platform 21. According to this estimate, median and maximum lengths of stretched DNA molecules shown in Figure 5A are approx. 300 and 700Kb.
Figure 5. Stretching parameters.
A. An example of a length distribution of DNA molecules isolated from human fibroblasts and stretched. DNA was stained with YOYO-1. B. A standard preparation of bacteriophage lambda DNA was stretched and stained with YOYO-1, and the lengths of molecules were measured and plotted. The smaller peak corresponds to broken molecules in the DNA sample. The observed lengths of intact molecules can be used to estimate the conversion, or stretching factor (see protocol).
TROUBLESHOOTING
Immunostaining of stretched DNA on coverslips
• TIMING: 7 hrs
38. Incubate fixed coverslips in 1-2 mls of 2.5 M HCl for 40 min to denature the DNA.
Δ CRITICAL STEP: This and subsequent steps are performed at room temperature unless otherwise noted. For this and other steps that involve washing coverslips can be kept DNA-side up in a six-well tissue culture plate.
39. Wash coverslips twice with 0.1 M Na Borate pH 8.5, once with PBS, and once with Wash Buffer to remove HCl.
40. Incubate coverslips in 1 ml Block buffer for 30 min.
41. Drain excess blocking buffer by touching the edge of a coverslip against a Kim wipe. Incubate blocked coverslips with rat anti-CldU/BrdU (primary antibody) freshly diluted 1:6 in Block buffer supplemented with 10% Normal Goat Serum (NGS) for 1 hr in the dark. Incubation with antibodies can be done with coverslips laid DNA side down into 10-20μl of antibody solution spotted onto parafilm. Use forceps to transfer coverslips between parafilm and a six-well plate. TROUBLESHOOTING.
42. Remove primary antibody by washing 3 times for 5 min. each time with 1 mL Wash buffer.
43. Incubate with goat anti-rat Alexa 594-conjugated antibody (secondary antibody) diluted 1:1000 in Block buffer supplemented with 10% NGS for 1 hr in the dark. Note that diluted secondary antibody in Block buffer can be kept at 4°C for up to 1 week.
44. Remove secondary antibody by washing 3 times for 5 min. each time with 1 mL Wash buffer.
45. Incubate coverslips for 20 min in 1 mL Block buffer supplemented with 10% NGS prior to staining with a second anti-analog antibody.
46. Incubate blocked coverslips with mouse anti-IdU/BrdU antibody (2nd primary antibody) diluted 1:6 in Block buffer supplemented with 10% NGS for 1 hr in the dark.
47. Remove 2nd primary antibody by washing 3 times for 5 min. each time with 1 mL Wash buffer.
48: Incubate coverslips with goat anti-mouse Alexa 488 conjugated antibody (2nd secondary antibody) diluted 1:1000 in Block buffer supplemented with 10% NGS.
49. Remove secondary antibody by washing 3 times for 5 min. each time with 1 mL Wash buffer. Rinse once with PBS.
50. Place in a clean, dust-free place and dry until all liquid evaporates (usually 5-7 min). Mount coverslips face down on microscope slides, affixing them with ‘Hard-as-Nails’ clear nail polish at the corners.
Δ CRITICAL STEP: We do not recommended the use of mounting medium because at least some brands, such as Vectashield, can cause deterioration of staining within hours after preparation. However, other brands, such as Prolong Gold antifade (Invitrogen) are reported to be compatible with staining. If using Prolong Gold, store coverslips at -20°C in the dark for up to several months.
■ PAUSE POINT: Doubly stained slides can store for weeks at 4°C in the dark.
51. If you so wish, also stain stretched DNA with mouse anti-deoxythymidine antibody. To do this, perform steps 46-50 above but using mouse anti-deoxythymidine antibody in place of the primary antibody in step 46 at a 1:2000 dilution in Block buffer supplemented with 10% NGS, followed by goat anti-mouse Alexa-conjugated secondary antibody in step 48. We recommend that beginners perform this staining to verify that the key elements of DNA isolation and stretching protocol are working.
Fluorescence microscopy of labeled and immunostained DNA on coverslips
• TIMING: variable, depending on nature of track analysis and number analyzed.
52. Observe stretched and immunostained DNA using either a confocal or regular fluorescent microscope. We use a confocal Zeiss Axiovert microscope equipped with a 63x and 100x objective and a digital camera. Briefly inspect the whole coverslip for DNA abundance, the quality of stretching and staining. Locate the areas with optimal DNA density and alignment (the density of DNA molecules goes down the farther they are from the loading end). See ‘Anticipated Results’ for further guidance on locating these areas.
TROUBLESHOOTING
Data acquisition, analysis, and quality control
• TIMING: 2 to 4 hrs/coverslip for image collection and track measurement
52. Collect images from the areas with optimal DNA density and alignment for subsequent analysis. We typically acquire 50-80 digital images from 3-4 microchannels for each sample, with the goal of collecting data on 300-400 replication tracks. This total number may have to be higher if a specific small subset of all tracks is sought after, for example tracks containing three segments (e.g., red-green-red). The only criteria for image acquisition should be the proper density and alignment of tracks. Photograph fields consecutively in two fluorescent colors to generate merged two-color images of tracks. See ‘Anticipated Results’ for further guidance regarding data analysis.
Troubleshooting
See Table 2 for troubleshooting guidance.
Table 2. Troubleshooting.
| Step number | Problem | Likely reasons | Solution |
|---|---|---|---|
| Step 6 | Patterns lift off in developer during photolithography | Photoresist adhesion problem | Reduce surface moisture on silicon wafer surface by drying wafer on a 200 °C hotplate for 10 min prior to coating; Increase exposure time and post exposure baking time; Ramp down temperature slowly after post UV exposure bake; Avoid agitation during developing |
| Steps 33-34 | DNA stretching is disorderly: DNA is not confined to channels | Residual moisture on PDMS surface has resulted in a poor seal and leaking of DNA into space between channels PDMS was not peeled off the cover slip properly PDMS patch surface is buckled, resulting in a poor seal and leaking of DNA into space between channels |
Make sure PDMS channels are completely dry by blotting several times against a glass surface before applying them onto coverslips. Remove PDMS from glass as described in Step 35. PDMS patches are too thick and thus rigid. Prepare thinner, more flexible patches for a better seal |
| Step 34 | DNA solution is not flowing into microchannels when loading | Blocked channels Hydrophobic PDMS surface |
Check if both ends of the microchannels are cut open Do not allow channels to remain dry for over 15 minutes prior to DNA stretching Re-treat PDMS with oxygen plasma |
| Step 37 | DNA is not fully attached to glass: “flapping” molecules can be seen | Suboptimal glass properties: vinyl silane concentration may be too low. | Remake coverslips according to the recipe; if needed, increase vinyl silane concentration Increase coverslip drying time after DNA capture |
| Step 37 | DNA is stretched poorly: unevenly stretched, curled, and clumped molecules are abundant | Suboptimal pH of DNA solution | Make sure DNA solution has a pH of 8.0 |
| Step 52 | DNA staining is too beady, impossible to identify tracks, beady background in channels | DNA concentration my be too high Proteinase K and/or agarase digests may be incomplete |
Dilute DNA solution used for loading 1:10 in TE Treat DNA samples with more agarase; reduce cell concentration in agarose gel inserts |
| Step 52 | Staining against CldU generates bright granular background throughout the cover slip | Some batches of rat anti-CldU antibody may be prone to generating aggregate background | Change the antibody batch, reduce antibody dilution, centifuge antibody prep to eliminate aggregates; Preincubate antibody dilution for 15min with a silanized coverslip that has no DNA but has been treated with methanol-acetic acid, dried, and subjected to HCL-neutralization-blocking steps |
Anticipated Results
Visual appearance of DNA
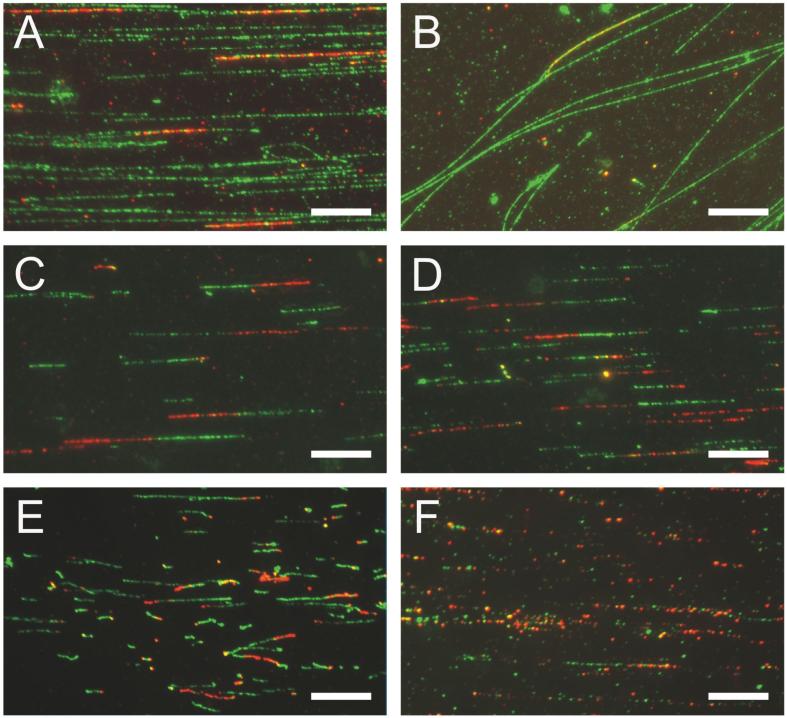
DNA stretched inside microchannels should appear as bands composed of molecules oriented parallel to the microchannel axis. Figure 6A shows a field within a microchannel, which displays a desired quality of stretching and reasonable density and length of DNA molecules. DNA is pulse-labeled with CldU and stained with antibodies against total DNA (anti thymidine, green) and anti CldU/BrdU (red). Note that some of the CldU tracks are located at the ends of molecules. It is expected that DNA at a replication fork may be more fragile to breakage during stretching than regular duplex DNA.
Figure 6. Examples of stretched DNA.
Scale bars are at 10μm. A) A fragment of area which was inside a microchannel when DNA was loaded. Flow inside microchannels results in deposition of highly aligned DNA molecules. Here, human fibroblasts were labeled for 30 min with CldU and harvested after another 30 min. DNA was prepped, stretched and stained with antibody against total DNA (anti-thymidine, green) and against CldU (red). Note that a subset of tracks is located at the ends of DNA molecules. B) A fragment of area outside a microchannel. Staining is as in A. C-F) Areas inside microchannels. C, D) Examples of good quality staining and stretching. Human fibroblasts were labeled with IdU then CldU for 30 min each and DNA was stained with antibodies against CldU (red) and IdU (green). E) An example of a less than optimal stretching resulting in “curling” of a subset of tracks. Human fibroblasts were labeled with IdU then CldU for 40 min each and DNA was stained with antibodies against CldU (red) and IdU (green). F) An example of suboptimal staining resulting in “beady” appearance of tracks. Human fibroblasts were labeled with CdU then IdU for 20 min each and DNA was stained with antibodies against CldU (red) and IdU (green).
Typically, DNA is also deposited and stretched outside the channels (Figure 6B). These molecules are usually unaligned or oriented at an angle to microchannel axes. Collecting data on this DNA should be avoided since it is often more sheared than DNA inside channels. When DNA is labeled consecutively with CldU and IdU (or BrdU and any one of the above nucleoside analogs) and stained with antibodies to these analogs, the anticipated patterns should be as in Figures 6C and D, in which densities of DNA molecules and staining quality are acceptable for data collection. The staining is optimal when tracks appear largely continuous. However, some discontinuity is unavoidable. At close inspection, tracks appear as chains of beads of about 1-1.5μm in diameter and it is common that some beads in this chain are missing or are dimmer than others. As a general rule of thumb, when discontinuities are larger than two beads width, the interpretation of track lengths and boundaries can be problematic.
Typically, double-labeled replication tracks comprise a fraction of all tracks labeled with the first label. Note that double-labeled tracks in Figure 6 have red and green segments of similar length. This should be expected if labels were added for the same periods of time (See Table 1 Quality Controls and Figure 7C).
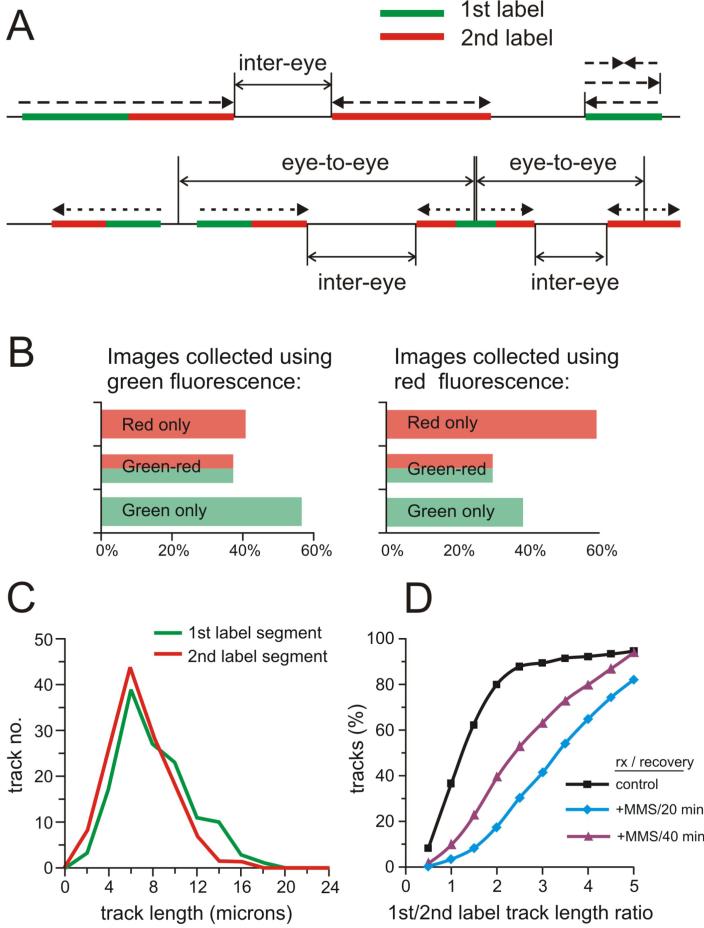
Figure 7. Replication track types and measures that can be derived from track images.
A) A schematic representation of the types of tracks and intra- and inter-replicon measurements. B) An example of scoring for the types of tracks in human fibroblasts labeled with IdU (green) then CldU (red) for 30 min each. The observed fraction of red only tracks appears higher if the images were collected looking for red fluorescence than when looking for green fluorescence. Approx. 250 tracks total was scored for each graph. C) Plotting of the track data as binned distributions of track lengths. Bins are in 2 micron increments, e.g. contain tracks that are 2-4, 4-6, etc. micron long. Human fibroblasts were labeled consecutively with two labels for 20 min each. Lengths of 1st and 2nd label segments in each two-segment track were measured and plotted. Note that these lengths are similar, as expected. D) Plotting of the track data as cumulative distributions. Human fibroblasts were labeled consecutively with IdU (green) then CldU (red) for 20 min. each (control), or treated with 0.015% MMS for 20 min. between the two labels (as described in more detail in Box1). In this case, the second, post-MMS label was added either for 20 or for 40 min. Ratios of 1st to 2nd label segment lengths in two-segment tracks were plotted (control, n=134; two segment tracks: 20min, n=118; 40 min, n=83). The slowing of replication elongation post MMS manifests as an increase in values of these ratios. Also, the ratios are lower the longer the post-MMS labeling time.
Figures 6E and F show examples of the most frequent problems encountered. In Figure 6E, immunostaining against CldU and IdU is good, but DNA is not well-stretched, as revealed by the variable thickness and buckling of some replication tracks. In Figure 6F, stretching is fair but immunostaining has not been successful: a majority of tracks are extremely discontinuous.
TROUBLESHOOTING
Determination of track types
This is the simplest kind of information that can be derived from replication track images, and it is useful for evaluating overall replication profiles. There are three basic types of tracks one can observe after consecutive pulse labeling with two labels (Figure 7A). Tracks containing the first label only represent one or two replication forks that terminated before the second label was added. Tracks containing consecutive segments of the first then second label represent ongoing forks; their direction can be determined from the temporal order of addition of each label. Tracks containing the second label only are origin firing events that occurred during the second labeling period. Note that these are the simplest interpretations of the track appearances and represent the most likely classes of event that could give rise to the specified track appearances. Finally, additional types of tracks may be observed: three-segment tracks of the 1st-2nd-1st label type are most likely two converged forks, and those of the 2nd-1st-2nd label type are origin firing events that occurred during the first labeling period. All these types can be scored manually, using any image display software such as Microsoft Picture Viewer or Adobe Photoshop.
It is important to realize that the process of image acquisition can and will introduce a bias into the estimated track type distribution (Figure 7B), especially when tracks are not very dense (2-5 per image). Scanning of coverslips for image acquisition is done in one color (i.e. with a filter set to fluorescence of one of the two labels), and the presence or absence of the second color in the field should not factor into a decision to acquire the image. Using first label color-only scanning may result in a data set that underestimates the proportion of the second label-only tracks among all tracks (Figure 7B). This bias can be avoided by collecting an additional set of images scanned using the second label color (see Table 1: Quality Controls). Use a consistent color scanning strategy in all experiments.
Measurement of replication track lengths
This measure provides information on replication elongation speeds and is particularly powerful when exploring genotype- or genotoxic drug-specific effects on replication. We measure lengths in microns for all types of tracks listed in the previous section using the AxioVision image analysis software supplied with the Zeiss Axiovert microscope, however, any software that has the capability to measure linear distances in a microscopic image can be used. The resulting sets of values can be processed in Microsoft Excel (or any graphing software). For tracks containing consecutive segments of each color, it is particularly informative to collect data on each segment and to determine mutual ratios of segment lengths (see Table 1: Quality controls). As mentioned before, our approximate conversion factor is 1μm=3.9Kb of DNA.
Inter-replicon measurements
This is a more demanding task than the intra-replicon characterization described above, as it requires identifying more than one adjacent replicon on one DNA molecule. When collecting inter-replicon data, it is helpful to use total DNA staining with an anti-thymidine antibody to identify replication tracks contained in single DNA molecules. If mouse anti-thymidine antibody is used for this purpose, a sheep or a goat antibody against IdU/BrdU should be used. Absolute length of DNA molecules becomes critical for these types of measures, since ability to identify more than one replicon on a single molecule is dependent on the presence of intact molecules of ≥0.5 Mb range.
Inter-origin or eye-to-eye distance is a distance between the centers of symmetry of two tracks that represent origin firing events (2nd-1st-2nd label or 2nd label only), and gives an estimate of the density of active origins and thus replicon size (Figure 7A). Pairs of closely spaced double-labeled tracks that represent diverging forks are also included into this measure: in this case an origin is implied to be at a halfway point between diverging forks. The acceptable spacing between such tracks should be less than the lengths of tracks.
The less stringent but more easily measured distance between converging forks, or Inter-Eye distance, may be affected by the density of origins as well as by fork processivity and speed. Origin density can also be calculated as a number of origin firing events per unit of total DNA length, per specified time. Note that all of these track measures may be biased by the position of cells in S phase at the time of labeling and harvesting.
When performing an experiment to take inter-replicon measurements, include a chase step after pulse labeling with nucleoside analog(s). This will allow replication forks to clear the vicinity of labeled segments of DNA and complete replication. Fully replicated DNA is thought to be more resilient to shear that incurs during stretching.
Analysis of track length, inter-origin distance or segment ratio data
We use two ways of displaying numerical values obtained from length or segment ratio measurements. Values are grouped into bins (usually at 2 μm resolution, e.g., 0-2, 2-4, 4-6, 6-8 μm etc.) and the number of tracks that fall within each length class or bin is plotted to give a frequency distribution as a function of length (Figure 7C). This type of graph readily visualizes the mode of the distribution; it also shows that the data are not distributed normally. Mean values may, as a result, be misleading and non-parametric significance tests should be used to compare data sets. We use a χ2 test on the binned distributions described above to assess differences between track length distributions. Cumulative distributions provide another useful and more compact way of plotting track length data (Figure 7D). This type of plot is of the fraction of track length values that are equal to or smaller than a given bin value. We use the Kolmogorov-Smirnov (K-S) test to determine p-values of different cumulative distributions.
Acknowledgements
This work was supported by NIH Seattle Cancer and Aging Program grant P20 CA103728-04, Subaward 000618167 to J.S., an NHGRI R01 (HG000225) to D.C.S., an R21/R33 EB0003307 award to A.F. and NCI PO1 CA88752 to R.J.M.,Jr. We would also like to thank Dhalia Dhingra and Paolo Norio for advice during early phases of this work, Amelia Gallaher for technical assistance and Alden Hackmann for graphics support.
REFERENCES
- 1.Alberts B. DNA replication and recombination. Nature. 2003;421:431–435. doi: 10.1038/nature01407. [DOI] [PubMed] [Google Scholar]
- 2.Kelly T, Stillman B. Duplication of DNA in eukaryotic cells. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbor Press; Cold Spring Harbor: 2006. pp. 1–29. [Google Scholar]
- 3.Loeb LA, Monnat RJ. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 4.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Research. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proceedings of the National Academy of Sciences. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi A, Shore D. How Telomerase Reaches Its End: Mechanism of Telomerase Regulation by the Telomeric Complex. Molecular Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg A, Baker TA. DNA replication. W.H. Freeman; New York: 1991. [Google Scholar]
- 9.Pommier Y, Diasio RB. Pharmacologic agents that target DNA replication. In: DePamphilis ML, editor. DNA replication and human disease. Cold Spring Harbort Press; Cold Spring Harbor: 2006. pp. 519–546. [Google Scholar]
- 10.Berdis AJ. DNA Polymerases as Therapeutic Targets†. Biochemistry. 2008;47:8253–8260. doi: 10.1021/bi801179f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DNA replication Methods and Protocols. Humana Press; New York: 2009. [Google Scholar]
- 12.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 13.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. Journal of Cell Biology. 1998;140:1285–95. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huberman JA, Riggs AD. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. Proc Natl Acad Sci U S A. 1966;55:599–606. doi: 10.1073/pnas.55.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns J. Cold Spring Harbor Symposia on Quantitative Biology. Vol. 28. Cold Spring Harbor, NY: 1963. The chromosome of Escherichia Coli; pp. 43–46. [Google Scholar]
- 16.Bensimon A, et al. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–8. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 17.Yokota H, et al. A new method for straightening DNA molecules for optical restriction mapping. Nucl. Acids Res. 1997;25:1064–1070. doi: 10.1093/nar/25.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrick J, Bensimon A. Single molecule analysis of DNA replication. Biochimie. 1999;81:859–71. doi: 10.1016/s0300-9084(99)00210-2. [DOI] [PubMed] [Google Scholar]
- 19.Norio P, Schildkraut CL. Visualization of DNA Replication on Individual Epstein-Barr Virus Episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- 20.Sidorova JM, Li N, Folch A, Monnat RJ., Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimalanta ET, et al. A Microfluidic System for Large DNA Molecule Arrays. Anal. Chem. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 22.Zhou S, Herschleb J, Schwartz DC. A single molecule system for whole genome analysis. In: Mitchelson KR, editor. New High Throughput Technologies for DNA Sequencing and Genomics. Elsevier; Amsterdam: 2007. pp. 265–302. [Google Scholar]
- 23.Zhou S, et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8:278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ananiev G, et al. Optical mapping discerns genome wide DNA methylation profiles. BMC Molecular Biology. 2008;9:68. doi: 10.1186/1471-2199-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 27.Beebe DJ, Mensing GA, Walker GM. PHYSICS AND APPLICATIONS OF MICROFLUIDICS IN BIOLOGY. Annual Review of Biomedical Engineering. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 28.Tegenfeldt J, et al. Micro- and nanofluidics for DNA analysis. Analytical and Bioanalytical Chemistry. 2004;378:1678–1692. doi: 10.1007/s00216-004-2526-0. [DOI] [PubMed] [Google Scholar]
- 29.Atencia J, Beebe DJ. Controlled microfluidic interfaces. Nature. 2005;437:648–655. doi: 10.1038/nature04163. [DOI] [PubMed] [Google Scholar]
- 30.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 31.Herschleb J, Ananiev G, Schwartz DC. Pulsed-field gel electrophoresis. Nat. Protocols. 2007;2:677–684. doi: 10.1038/nprot.2007.94. [DOI] [PubMed] [Google Scholar]
- 32.Lundin C, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucl. Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]