A 57-year old female was referred to our hospital because of flu like symptoms, and extensive bilateral pleural and pericardial effusion. However, the patient showed neither lymphadenopathy nor hepatosplenomegaly. Examination of cells in her pleural effusion revealed atypical cells with convoluted nuclear contour, dense chromatin, and bizarre mitotic figures. Numerous atypical cells had “flower shape” nuclei (Figure 1A).. She was negative for HTLV-I virus. Similar abnormal cells were also detected in the peripheral blood and bone marrow.
Fig. 1. NUP214-ABL1 gene in T-cell leukemic cells.
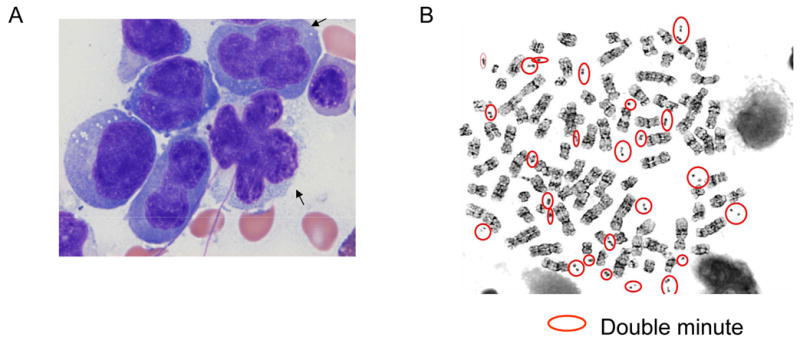
A: Morphology of leukemic cells. Atypical cells show convoluted nuclear contour, dense chromatin, and bizarre mitotic figures. Cells with “flower shape” nuclei are also observed (arrows).
B: Metaphase spreads. Double minute chromosomes are circled.
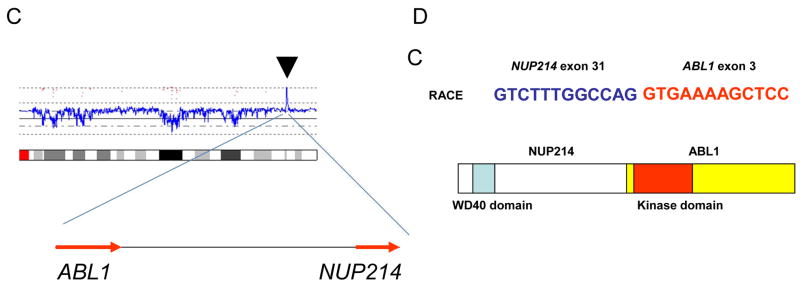
C: High copy number amplification of NUP214 and ABL1. The result of SNP-chip analysis (chromosome 9) revealed high copy number amplification of chromosome 9q34 region (arrow head) which contained NUP214 and ABL1 genes (arrows indicate the direction of transcription of the genes).
D: NUP214-ABL1 fusion was detected in T-cell leukemia. Rapid amplification of cDNA ends (RACE) method and nucleotide sequencing showed the fusion of exon 31 of NUP214 and exon 3 of ABL1. Schematic representation of the structure of NUP214-ABL1 fusion protein is shown.
Flow cytometry analysis revealed that the abnormal cells were TdT−, CD2 +, CD4+, CD5+, CD7+, CD10−, CD20−, CD33− and CD34−. Loss of CD3 and CD8 was also noted. She was diagnosed as T-cell acute lymphoblastic leukemia (T-ALL), although the differential diagnosis, based on morphologic and immunophenotypical features of the leukemic cells, included T-cell prolymphocytic leukemia (T-PLL).
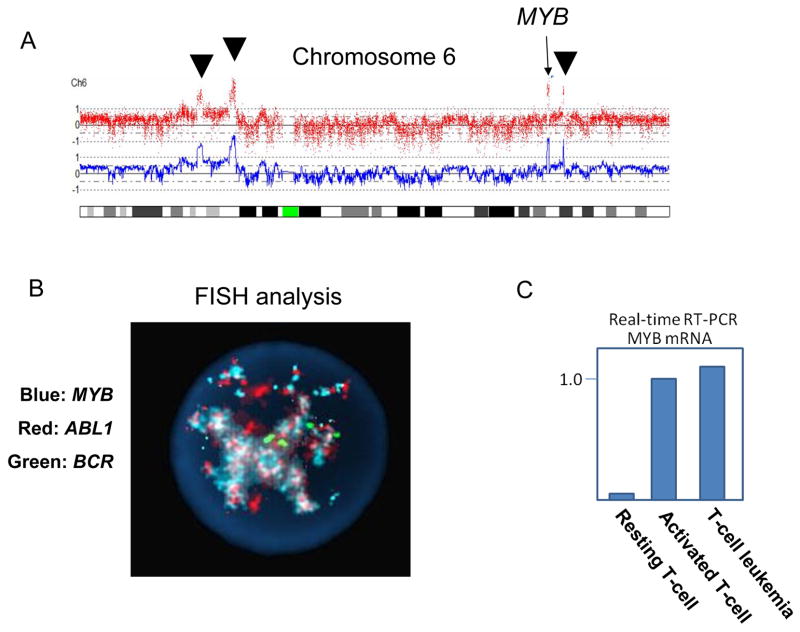
The karyotype of her leukemic cells was pseudotetraploid with a deleted copy of chromosome 1, numerous marker chromosomes and the presence of multiple double minute chromosomes: 89,XXXX,+del(1)(q25), +2, +3, +4×2, +5×2, +7×2, +8×4, +9, −10, +11, +12×2, +13, +14, 15×3, +16×2, +17×2, +18×2, +19×2, +20×2, +21×2, +22×2, +Mar1,+Mar2, +Mar3, +Mar4, +Mar5, +Mar6, +dms (Fig. 1B). To further characterize the double minutes, we performed single nucleotide polymorphism DNA microarray (SNP-chip) analysis on the leukemic cells using Affymetrix GeneChip Nsp 250K as previously reported [1]. The analysis demonstrated high copy number gene amplification of several chromosomal regions, including 6q and 9q (Figs. 1C and 2A). One of the amplified regions contained the NUP214 and ABL1 genes located at 9q34.1 (Fig. 1C). Rapid cloning of 5’end (RACE) was used to clone the gene fused to ABL1 and demonstrated that exon 31 of NUP214 was fused to exon 3 of ABL1 gene (Fig. 1D). Furthermore, we determined that the MYB gene was involved in one of the amplified region on chromosome 6 (Fig. 2A). FISH using ABL1, MYB, and BCR probes (Abbott Molecular Des Plaines, IL). showed that the ABL1 and MYB genes were extensively amplified with co-localization in the nucleus (Fig. 2B). Consistent with the amplification of MYB, real-time RT-PCR showed that the MYB gene was highly expressed in the leukemic cells (Fig. 2C). FISH and SNP-chip analysis also revealed a homozygous deletion of the p15/p16 genes at 9p21 (not shown).
Fig. 2. MYB and NUP214-ABL1 genes are components of double minute chromosomes in T-cell leukemic cells.
A: The result of SNP-chip analysis (chromosome 6). High copy number amplifications of four chromosomal regions (arrow heads and an arrow) are detected. One of the amplified regions contains MYB gene (arrow).
B: FISH analysis. MYB (aqua), ABL1 (red) and BCR (green) genes were examined by FISH analysis. MYB and ABL1 show co-amplification. BCR shows four signals in this cell, consistent with four copies of chromosome 22 (see text).
C: MYB gene is over expressed in the leukemic cells. The expression levels of MYB gene were measured in resting and activated normal T-cells (stimulated with PHA) as well as the patient’s leukemic cells by real-time RT-PCR.
Double minute chromosomes, which are cytological manifestations of gene amplification, are rare abnormalities in leukemic cells. It can be difficult to define the genomic regions involved in their formation. In this study, SNP-chip analysis provided insight into the composition of the double minute chromosomes, while FISH analysis provided confirmation. NUP214-ABL1 gene fusion has been reported in about 6% of T-acute lymphoblastic leukemia (ALL), usually presenting as cryptic episomes, and has been associated with a poor prognosis [2]. The present case is the first in which NUP214-ABL1 fusion was found in larger double minute chromosomes. The increased size of the amplified structures may reflect the presence of other genomic material included in the double minutes. SNP-chip analysis had revealed amplification of MYB which was also confirmed by FISH. Co-localization of NUP214-ABL1 and MYB was observed by the FISH analysis. Translocations and duplications leading to increased expression of the MYB gene have recently been reported in about 8% of T-ALL patients [3, 4]. It has been reported that homozygous deletion of p15/p16 genes (9p21), as seen here, are detected in at least 65% of T-ALL [5]. The molecular features of this case, including presence of NUP214-ABL1 fusion, over expression of MYB and deletion of the p16/p15 genes, supported the diagnosis of T-ALL.
There have been several reports of amplification of large genomic regions associated with complex translocations (complicons), especially in B-cell malignancies, involving IGH/MYC, IGH/BCL2, and IGH/CCND1 [6–8]. This is the first report of a complicon in a T-cell disorder, and also the first demonstration of nonsyntenic co-amplification of a complicon with two distinct oncogenes (NUP214-ABL1 and MYB). This complex phenomenon would not have been suspected without the information provided by the SNP-chip analysis.
Acknowledgments
We thank the Parker Hughes Fund and NIH grants for supporting this study. NK is supported by the fellowship from The Tower Cancer Research Foundation. HPK holds the Mark Goodson Chair in Oncology Research at Cedars Sinai, and is a member of the Jonsson Cancer Center and the Molecular Biology Institute of UCLA. This work was also supported by grant-in-aid from Department of Health, Welfare and Labor and from MEXT of the Japanese government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–84. doi: 10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 3.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–61. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 4.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–5. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi S, Bartram CR, Seriu T, Miller CW, Tobler A, Janssen JW, et al. Analysis of a family of cyclin-dependent kinase inhibitors: p15/MTS2/INK4B, p16/MTS1/INK4A, and p18 genes in acute lymphoblastic leukemia of childhood. Blood. 1995;86:755–60. [PubMed] [Google Scholar]
- 6.Gruszka-Westwood AM, Atkinson S, Summersgill BM, Shipley J, Elnenaei MO, Jain P, et al. Unusual case of leukemic mantle cell lymphoma with amplified CCND1/IGH fusion gene. Genes Chromosomes Cancer. 2002;33:206–12. doi: 10.1002/gcc.1216. [DOI] [PubMed] [Google Scholar]
- 7.Martín-Subero JI, Odero MD, Hernandez R, Cigudosa JC, Agirre X, Saez B, et al. Amplification of IGH/MYC fusion in clinically aggressive IGH/BCL2-positive germinal center B-cell lymphomas. Genes Chromosomes Cancer. 2005;43:414–23. doi: 10.1002/gcc.20187. [DOI] [PubMed] [Google Scholar]
- 8.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–21. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]