Abstract
This study was done to improve efficiency and islet specificity of the rat insulin promoter (RIP). Various rat insulin promoter lengths were prepared and tested in vitro to drive luciferase reporter gene expression in INS1-cells, alpha-cells, acinar cells, ductal cells, and fibroblasts. The CMV promoter was used as a positive control. In addition, the DsRed reporter gene was administered in vivo to rat pancreas by ultrasound-targeted microbubble destruction (UTMD). Confocal microscopy was used to detect the presence and distribution of DsRed within the pancreas after UTMD. A modified RIP3.1 promoter, which includes portions of the insulin gene after its transcription start site is 5-fold more active in INS-1 cells than the full length RIP promoter or the CMV promoter. RIP3.1 is regulated by glucose level and various islet transcription factors in vitro, and exhibits activity in alpha-cells, but not exocrine cells. In vivo delivery of RIP3.1-DsRed resulted in expression of DsRed protein in beta-cells, and to a lesser extent alpha cells under normal glucose conditions. No DsRed signal was present in exocrine pancreas under RIP3.1. A modified rat insulin promoter, RIP3.1, efficiently and specifically directs gene expression to endocrine pancreas.
Keywords: Rat Insulin Gene Promoter, Pancreatic islets, Gene Delivery, Islets Transcriptional Factors, Microbubble, Ultrasound
Introduction
Novel therapeutic strategies, including islet transplantation and gene therapy, are being sought to treat diabetes, which is increasing in prevalence worldwide (1–3). Gene therapy requires direct targeting of genes to the islets in vivo. So far, adenovirus, adeno-associated virus, lentivirus, and herpes simplex virus-1 vectors have enabled efficient gene transfer to islets, but suffer from host immune responses and vector cytotoxicity (4–10). Non-viral gene delivery systems, including naked DNA and DNA complexes, induce islet cell transfection at much lower levels and with only transient transgene expression. However, non-viral vector systems will more easily satisfy biosafety concerns in clinical trials.
We have recently shown that ultrasound-targeted microbubble destruction (UTMD) is able to target gene delivery to pancreatic islets in vivo (11–12). Gas-filled microbubbles, containing plasmids, are injected intravenously and destroyed within the pancreatic microcirculation by ultrasound, thereby delivering the plasmids through the vascular endothelial barrier. This approach enables transfection of the islet core, wherein most beta cells reside. We have previously shown that combining UTMD with a rat insulin promoter (RIP) enhances beta-cell specificity (11). We now demonstrate differential efficiency of transgene expression by varying the length of the RIP promoter sequence. We attempted to expand insulin gene promoter to include non-coding regions in exon1, intron1 and part of exon2 of the rat insulin gene 1, driving a downstream reporter gene. We report that a modified RIP promoter (RIP3.1) enables beta-cell specific, glucose-regulatable gene expression in cell culture. In addition, in vivo delivery of reporter genes under control of RIP3.1 by UTMD provides islet specific, efficient and regulated transgene expression in rats.
RESULTS
RIP-luc transfection on pancreatic cell lines
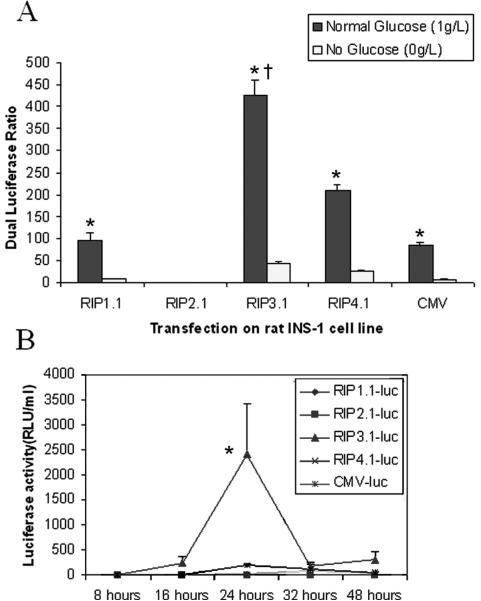
Figure 2 illustrates the dual luciferase ratio for the various RIP promoter lengths under with or without glucose after transfection on rat INS-1 cell line. Under glucose condition, the luciferase ratio of RIP3.1-luc is 7000-fold greater than RIP2.1-luc (truncated RIP promoter), 5-fold greater than RIP1.1-luc (full length RIP), and 5-fold greater than CMV-luc (p<0.001). Under absent glucose condition, the luciferase ratio is significantly blunted in all groups, but remains higher with RIP3.1 compared to other RIP lengths and CMV (p<0.001). For each different promoter, the ratio of glucose/no glucose luciferase activity was of the same order of magnitude, ranging from 8–15 fold. Surprisingly, luciferase activity could be detected from the culture media solution of RIP3.1 dish at 8, 16, 24, 32 and 48 hours after transfection under with glucose conditions (Fig. 2, bottom panel). No excretion of luciferase into culture media was seen under absent glucose (data not shown).
Figure 2.
Top Panel. Dual luciferase ratio of INS-1 cells lysates 48 hours after transfection with RIP-luc. Black bars indicate normal glucose concentration. White bars indicate no glucose. * p<0.001 vs no glucose; † p<0.001 vs all other promoters. Bottom panel. Luciferase activity of culture media solution at various times after transfection with RIP-luc with glucose. * p<0.0001 vs all other groups and timepoints.
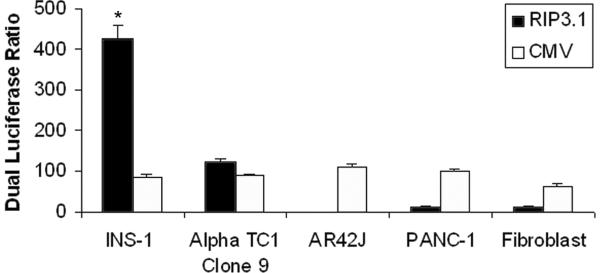
Figure 3 shows the results of RIP3.1-driven luciferase versus CMV-driven luciferase activity in various cell lines. The luciferase ratio is similar in all pancreatic cell lines when luciferase is under control of the CMV promoter. However, the RIP3.1 promoter results in a luciferase ratio that is 4-fold higher than CMV in INS-1 cells. Likewise, RIP3.1 results in 4-fold higher luciferase ratio in INS-1 cells than in alpha cells and 35-fold greater luciferase ratio compared to pancreatic exocrine cell lines (acinar, duct, and fibroblast cell lines) (p<0.001). RIP3.1 and CMV promoters exhibit similar activity in alpha cells, but is nearly absent in exocrine cell lines compared to CMV.
Figure 3.
RIP-luc and CMV-luc as a control transfected on pancreatic cell lines and dual luciferase ratio of fire-fly luc and renilla luc 48 hrs after transfection.
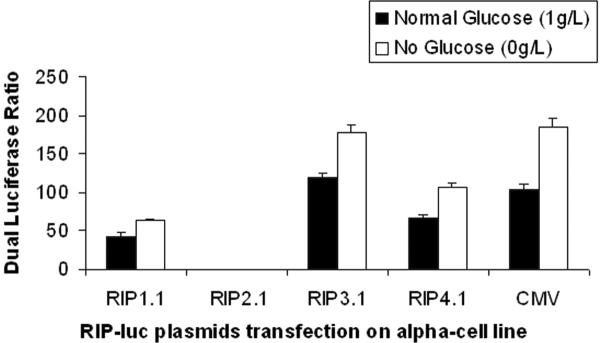
Figure 4 illustrates dual luciferase ratio for the various RIP promoter lengths under with or withou glucose after transfection on mouse alpha TC1 clone9 cell line. Under glucose condition, the luciferase ratio of RIP3.1-luc is 400-fold greater than RIP2.1-luc (truncated RIP promoter), and 3-fold greater than RIP1.1-luc (full length RIP) (p<0.001). However under absent glucose condition, the luciferase ratios are about 0.5 fold higher than under glucose condition (p<0.001). RIP3.1-luc and CMV-luc show nearly identical luciferase ratios regardless of glucose conditions.
Figure 4.
Dual luciferase ratio of Alpha TC1 Clone9 cells lysates 48 hours after transfection with RIP-luc with or without glucose.
Regulation of the RIP3.1 promoter by islet transcription factor genes
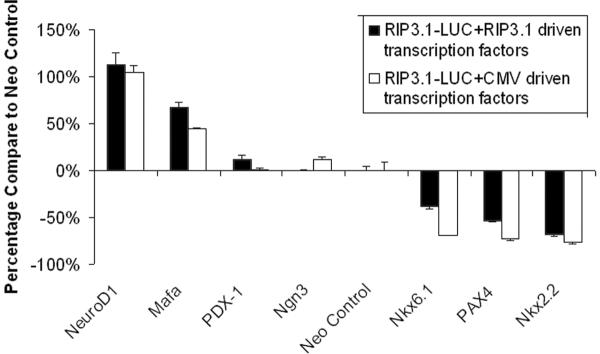
Figure 5 shows the regulation of the RIP3.1 promoter by various islet transcription factors. RIP3.1-NeuroD1 and RIP3.1-Mafa significantly up-regulate the RIP3.1-mediated luciferase activity (p<0.001), while the Nkx6.1, Nkx2.2, and PAX4 down-regulate it (p<0.001). PDX-1 and Ngn3 result in a minimal but statistically significant upregulation of RIP3.1-mediated luciferase activity (p=0.010274). Nearly identical results were obtained when the CMV was used to drive islet transcription factors, confirming that NeuroD1 and MafA significantly up-regulate the RIP3.1 promoter, whereas Pax4, Nkx2.2, and Nkx6.1 downregulate the RIP3.1 promoter.
Figure 5.
Co-transfection of RIP3.1 driving luciferase reporter gene and RIP3.1/CMV driving islet transcription factor genes to rat insulinoma cell line (INS-1).
RIP3.1-DsRed Plasmids Delivered to Pancreatic Islets in vivo with UTMD
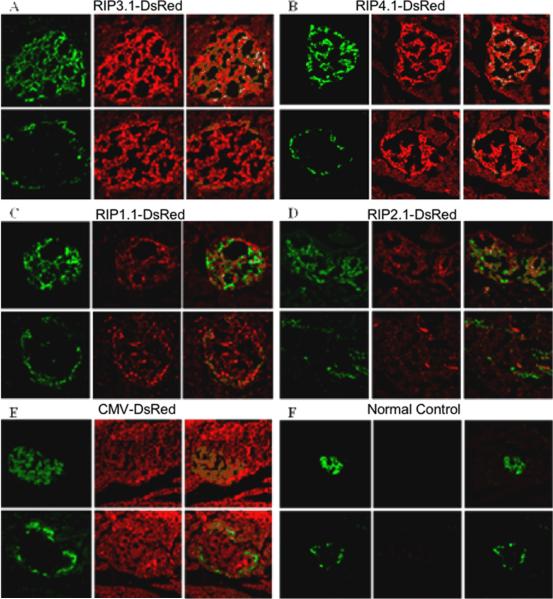
To better understand gene expression and regulation of these RIP promoters in islets of living animals, we further delivered these RIP-DsRed plasmids in vivo using UTMD. Rats were euthanized 4 days after UTMD. Figure 6 shows that DsRed protein under the regulation of RIP3.1 (panel A) and RIP4.1 (panel B) was detected in intact islets, but not in exocrine pancreas. Confocal images showed that DsRed protein was detected in both beta-cells and, to a lesser extent, in alpha-cells. DsRed protein signal was much lower with full length RIP1.1 (panel C) and nearly absent with truncated RIP2.1 (panel D). DsRed protein was seen in both the endocrine and exocrine pancreas in rats treated with CMV-DsRed plasmid (panel E). There was no detectable DsRed protein in the pancreas of normal control rats (panel F).
Figure 6.
In vivo immunohistology of rats treated with DsRed reporter gene using different RIP promoter lengths and controls. All images are magnified at 200X. For each group labeled A–F, a representative islet section is shown. Top panels are stained with green anti-insulin (left), red anti-DsRed (middle), and their confocal image (right). Bottom panels are adjacent sections of the same islet stained with green anti-glucagon (left), red anti-DsRed (middle), and their confocal image (right). A: RIP3.1-DsRed; B: RIP-4.1-DsRed; C: RIP-1.1-DsRed; D: RIP-2.1-DsRed slides, E: pCMV-DsRed; F: normal control.
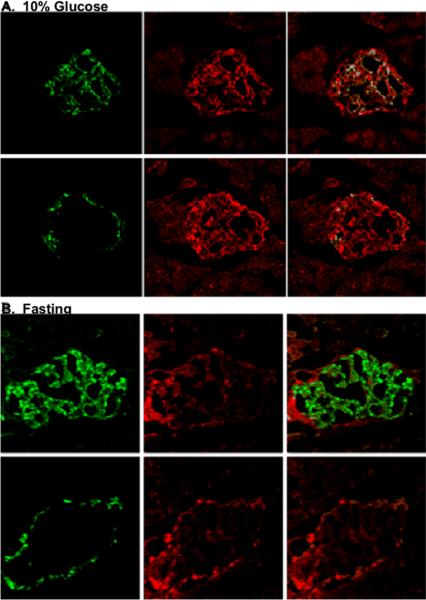
Next, we wanted to know if gene expression of delivered RIP plasmids was regulated by glucose as in the INS-1 cell line. We used RIP3.1-DsRed, because of its higher efficiency, and treated two groups of rats, fasting (12 hours) and feeding with 10% of glucose. Figure 7 (top panel) shows DsRed signal in both beta-cells and, to a lesser extent, alpha-cells of islets in rats fed 10% glucose. In contrast, fasting rats only exhibited DsRed signal in alpha-cells. This suggests that glucose feeding induced up-regulation of RIP3.1-DsRed gene expression in whole islets cells and that fasting (low glucose level) down-regulated RIP3.1-DsRed gene expression in beta-cells but not alpha-cells.
Figure 7.
Panel A. Sections from rat pancreas treated with pRIP3.1-DsRed with 10% glucose feeding. Top panels show an islet stained with anti-insulin (green) (left), anti-DsRed (red) (middle) and their confocal image (right). Bottom panels show an adjacent section stained with anti-glucagon (green)(left), anti-DsRed (red) (middle), and their confocal image (right); Panel B. Similar images from a rat treated with pRIP3.1-DsRed rats after fasting overnight.
Discussion
Most of the previous studies of the insulin promoter have been performed using the sequence before the transcriptional start site, usually ending after the TATA box (13–17). The RIP3.1 insulin promoter includes non-coding regions after the transcription start site including exon I, intron I, and part of exon II. Our study is the first report that the promoter RIP3.1 is much more efficient than the traditional insulin gene promoter, or the widely used CMV promoter. Thus, the non-coding exon and intron regions appear to be essential to generate robust beta-cell (and to a lesser extent, alpha-cell) specific transgene expression. Moreover, RIP3.1 showed secretion of the encoded protein under high glucose conditions in cell culture. Studies of the insulin promoter have revealed that the regulation of insulin gene is restricted to the pancreatic islets cells and regulated by glucose (18–21). The RIP3.1 promoter exhibited cell-specific gene expression in islets cell lines when compared to exocrine pancreatic cell lines; the luciferase activity of RIP3.1 plasmid in beta cell is 4-fold greater than in alpha cell, 35-fold greater than pancreatic duct cell and 300-fold greater than pancreatic acinar cells. The lower levels of gene expression of RIP3.1 in alpha cell lines are likely explained by homologous DNA sequence motifs like E1 or A boxes in regulation areas of the glucagon and insulin genes (22–23).
In addition, RIP3.1 promoter showed glucose responsiveness in both beta-cells and alpha-cells. In beta-cells, the promoter activity under with glucose condition (1g/L) is 10-fold greater than under absent glucose condition. It has already been demonstrated that the promoter elements upstream of −230 are involved in the glucose-mediated suppression of the insulin promoter activity (24). In alpha-cells, the promoter activity under absent glucose conditions is 1.5-fold greater than under with glucose condition. This might be because alpha-cells are activated under low glucose conditions, such that promoter activity is increased.
Our study is the first report the activity of 7 islets transcription factors (PDX-1, Ngn3, NeuroD1, Pax4, Nkx2.2, Nkx6.1, and Mafa) in regulating the insulin promoter in INS-1 cells (25–31). When RIP3.1-luc plasmids were co-transfected with these transcription factor genes under the CMV promoter, NeuroD1 and Mafa increased luciferase activity by 104% and 44%, respectively. Pdx-1 and Ngn3 increased luciferase activity only slightly, whereas Pax4, Nkx2.2 and Nkx6.1 significantly decreased it. When RIP3.1-luc was co-transfected with these transcription factors under the RIP3.1 promoter, the same effects were seen, confirming that NeuroD1 and Mafa upregulate the RIP3.1 promoter, while Pax4, Nkx2.2, and Nkx6.1 downregulate it. We designed a RIP3.1-neo control plasmid that could rule out the likely interference effects by the endogenous transcription factors when cotransfection in INS-1. Further studies needed to elucidate the mechanism(s) by which this occurs. However, it is likely that the effects of these transcription factors on the RIP3.1 promoter are explained by the presence of additional binding sites. For example, the insulin gene has four cAMP response elements (CRE), CRE1 at −210; CRE2 at −183; CRE3 at +18; and CRE4 at +61 (32). The RIP3.1 promoter contains two more of these CRE boxes than the standard RIP1.1 promoter. In addition, RIP3.1 contains additional cis-elements such as Nir-box (33), GAGA-box (34), Pl-box (35), and an AGGAT motif at +87 and +110 for the binding of the ets-family of transcription factors (36).
To confirm that the results of the in vitro experiments were physiologically relevant, we used UTMD to deliver the RIPs-dsRed plasmids to pancreas of the living animal in vivo. Immunohistological staining showed the RIP3.1 promoter is more effective than other promoter lengths for driving transgene expression in pancreatic islets. DsRed signal was only seen in the intact islets and not in exocrine pancreas. This high transgene expression by RIP3.1 might be related to two more CRE boxes and a C box in exon1 and intron1, which are not present in the truncated (RIP2.1) or the normal (RIP1.1) insulin promoters. We also found that RIP3.1 activity was regulated by glucose. This is the first report that we can manipulate transgene expression of pancreatic islets at the transcriptional level in living animals by fasting or feeding, which is not a feature of virus-mediated gene therapy. Careful attention to promoter biology may help target gene therapy for diabetes to the islets, and allow feedback regulation of the transgene in a physiological manner.
Materials and Methods
Rat insulin promoters and plasmids constructs
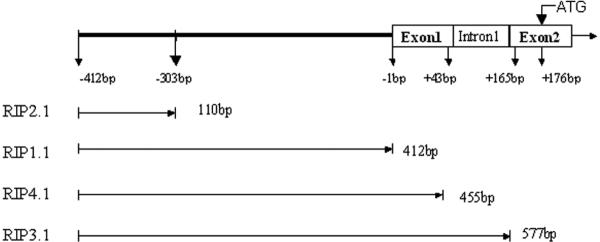
Rat genomic DNA was extracted from Sprague-Dawley rat peripheral blood with a QIAamp Blood kit (Qiagen Inc, Valencia, CA). Four different length rat insulin I gene promoters, named RIP2.1 (−412 to −303); RIP1.1 (−412 to −1); RIP4.1 (−412 to +43); RIP3.1 (−412 to +165), were designed (Fig. 1). The rat insulin I gene promoters were PCR amplified from rat genomic DNA by using forward and reverse primers containing either KpnI–HindIII or xhoI-EcoRI restriction site respectively. The sequences of these primers are listed in table I. The corresponding PCR products were verified by agarose gel electrophoresis and purified by QIAquick Gel Extraction kit (QIAGEN). To confirm the sequences, direct sequencing of PCR products was performed with dRhodamine Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster, CA) on an ABI 3100 Genomic Analyzer (Applied Biosystems, Foster, CA). Then PCR products were subcloned into the pGL3-basic firefly luciferase reporter plasmid (Promega, Madison, WI) and pDsRed1-1 reporter vector (Clonetech, Mountain View, CA). hMafa cDNA and hamster NKX6.1 cDNA were generously donated by the Olson laboratory at Michigan State University, and the Newgard laboratory at Duke University Medical Center, respectively. Rat Ngn3, NeuroD1, PAX4, and Nkx2.2 cDNAs were PCR products from Sprague-Dawley new-born rat pancreas cDNA pool that were reversed from their total RNA according to the manufacturer's instructions. The sequences of these primers are listed in table 1. Newborn rat pancreatic samples were flash frozen in liquid nitrogen and stored at −86°C. The frozen samples were thawed in 1 ml of RNA-STAT solution and immediately homogenized using a polytron 3000 homogenizer at 10,000 r.p.m. for 30s. Total RNA (30 ng) was reverse transcribed in 20 μl by using a Sensiscript RT Kit(Qiagen) with oligo(dT)16. The reaction mixture was incubated at 42°C for 50 min, followed by a further incubation at 70°C for 15 min. PCR was performed for all samples using a GeneAmp PCR System 9700 in 50 ul volume containing 2 ul cDNA, 25 ul of HotStarTaq Master Mix (Qiagen), and 20 pmol of each primer (Tab1). The corresponding PCR products were verified by agarose gel electrophoresis and purified by QIAquick Gel Extraction kit (QIAGEN). To confirm the sequences, direct sequencing of PCR products was performed with dRhodamine Terminator Cycle Sequencing Kit on an ABI 3100 Genomic Analyzer. All transcriptional factor gene cDNAs were subcloned to RIP3.1driving vector and pCI vector (Promega, Madison, WI). The plasmids digestion, ligation, subclone, isolation and purification were performed by standard procedures, and once again sequenced to confirm that no artifactual mutations were present.
Figure 1.
Schematic representation of the rat insulin promoter, including exon1, intron1 and exon2. RIPA2.1 contains only part of the promoter; RIPB1.1 is the full-length rat insulin promoter; RIPC4.1 includes promoter region plus exon 1; RIPD3.1 includes promoter, exon 1, intron 1 and the first three bases of exon 2.
Table I.
Primer sequences (Rat insulin gene promoters using the same forward primer).
| Primer | Sequences | GenBank accession no. |
|---|---|---|
| Rat insulin gene promoter forward | 5'- g CTg AgC TAA gAA TCC A | J00747 |
| Rat insulin gene promoter 2.1 reverse | 5'- CTg AgC ATT TTC CAC C | |
| Rat insulin gene promoter 1.1 reverse | 5'- ggg AgT TAC Tgg gTC TCC A | |
| Rat insulin gene promoter 4.1 reverse | 5'-CTg CTT gCT gAT ggT CTA | |
| Rat insulin gene promoter 3.1 reverse | 5'-gAC CTg gAA gAT Agg CAg ggT | |
| Rat Neurod1 cDNA forward | 5'-AAC Atg ACC AAA TCA TAC AgC | NM_019218 |
| Rat Neurod1 cDNA reverse | 5'-TgA AAC TgA AAC TgA CgT gCC | |
| Rat Neurogenin3 cDNA forward | 5'-ATg gCg CCT CAT CCC TTg gAT | NM_021700 |
| Rat Neurogenin3 cDNA reverse | 5'-ACA CAA gAA gTC TgA gAA CAC | |
| Rat PAX4 cDNA forward | 5'-AgC ATg CAg CAg gAC ggT CTCA | NM_031799 |
| Rat PAX4 cDNA reverse | 5'-TTA Tgg CCA gTg TAA gTA ATA | |
| Rat NKX2.2 cDNA forward | 5'-ATg TCg CTg ACC AAC ACA AAg AC | XM_345446 |
| Rat NKX2.2 cDNA reverse | 5'-TCA CCA AgT CCA CTg CTg ggC CT | |
| Rat PDX1 cDNA forward | 5'-gCC ACC Atg AAT AgT gAg gAg | NM_022852 |
| Rat PDX1 cDNA reverse | 5'-TCA gCC TgC ggT CCT CAC Cgg ggT | |
| Golden hamster NKX6.1 cDNA forward | 5'-CTg Tgg gAT gTT AgC TgT | X81409 |
| Golden hamster NKX6.1 cDNA reverse | 5'-TCA ggA CgA gCC gTg ggC CT | |
| Human MafA cDNA forward | 5'-ATg gCC gCg gAg CTg gCg AT | NM_201589 |
| Human MafA cDNA reverse | 5'-CTA CAg gAA gAA gTC ggC CgT |
Cell culture and transient transfection assays
The rat insulinoma cell line INS-1 (courtesy of the Newgard laboratory, Duke University Medical Center, Durham, NC), rat acinar cell line (AR42J), mouse alpha cell line (alpha TC1 Clone9), human pancreatic ductal cell line (PANC-1), and human fibroblast cell line were maintained in DMEM media. All cell lines were co-transfected with 1μg of luciferase reporter plasmid and 0.02μg of pTS-RL Renilla luciferase (internal control plasmid) in 100μl serum free DMEM with 3μl Lipofectamine 2000 in 100μl serum free DMEM each well. The cell harvest and firefly and Renilla luciferase activities were measured 48 hours after transfection using the dual luciferase assay system (Promega, Madison, WI) and a Turner TD 20/20 luminometer (Turner, Sunnyvale, CA).
Rat UTMD Protocol
Sprague-Dawley rats (250–350 g) were anesthetized with intraperitoneal ketamine (100mg/kg) and xylazine (5 mg/kg). A polyethylene tube (PE 50, Becton Dickinson, MD) was inserted into the right internal jugular vein by cutdown. The anterior abdomen was shaved and an S3 probe (Sonos 5500, Philips Ultrasound, Andover, MA) placed to image the left kidney and spleen, which are easily identified. The pancreas lies between them, so the probe was adjusted to target the pancreas and clamped in place. One ml of microbubble solution was infused at a constant rate of 3ml/h for 20 minutes using an infusion pump. Throughout the duration of the infusion, microbubble destruction was achieved using ultraharmonic mode (transmit 1.3 MHz / receive 3.6 MHz) with a mechanical index of 1.2–1.4 and a depth of 4 cm. The ultrasound pulses were ECG-triggered (at 80 ms after the peak of the R wave) to deliver a burst of 4 ultrasound pulses every 4 cardiac cycles. At the end of each experiment the jugular vein was tied off and the skin closed. All rats were monitored after the experiment for normal behavior. Rats were euthanized 4 days later and the pancreas was harvested.
Manufacture of Plasmid-Containing Lipid-Stabilized Microbubbles
Lipid-stabilized microbubbles were prepared as previously described in our laboratory (11–12). Briefly, a solution of DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine, Sigma, St. Louis, MO) 2.5 mg/ml; DPPE (1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine, Sigma, St. Louis, MO) 0.5 mg/ml; and 10% glycerol was mixed with 2 mg of plasmid solution in a 2:1 ratio. Aliquots of 0.5 ml of this phospholipid-plasmid solution were placed in 1.5 ml clear vials; the remaining headspace was filled with perfluoropropane gas (Air Products, Inc, Allentown, PA). Each vial was incubated at 40°C for 30 min and then mechanically shaken for 20 seconds by a dental amalgamator (VialmixTM, Bristol-Myers Squibb Medical Imaging, N. Billerica, MA). The lipid-stabilized microbubbles appear as a milky white suspension floating on the top of a layer of liquid containing unattached plasmid DNA. The subnatant was discarded and the microbubbles washed three times with PBS to removed unattached plasmid DNA. Mean diameter and concentration of the microbubbles were measured by a particle counter (Coulter Multisizer III, Beckman, Miami, FL).
Immunohistochemistry for Detection of DsRed protein, Insulin, and Glucagon
Cryostat sections 5–8 μm in thickness were fixed in 4% paraformaldehyde for 15 min at 4 °C and quenched for 5 min with10 mM glycine in PBS. Sections were then rinsed in PBS 3 times, and permeabilized with 0.5% Triton X-100 in PBS for 10 min. Sections were blocked with 10% goat serum at 37°C for 1hr and washed with PBS 3 times. The primary antibodies (mouse anti-insulin (sigma, 1:10000 dilution), rabbit anti-glucagon (chemicon 1: 500 dilution), rabbit anti-DsRed (1:500 dilution) were added and incubated at RT for 2 hours. After washing with PBS three times for 5 min, the secondary antibody (anti-mouse lgG conjugated with FITC) (1:500 dilution in block solution) and anti-rabbit IgG-Cy5 (1:250 dilution) were added and incubated for 1 hr at RT. Sections were rinsed with PBS for 10 min, 5 times, and then mounted.
Statistical Analysis
Differences in luciferase activity between experimental groups were compared by two-way ANOVA. A p value < 0.001 was considered statistically significant. Post-hoc Scheffe tests were performed only when the ANOVA F values were statistically significant.
Acknowledgements
This work is supported in part by NIH grant P02 DK 58398 (Newgard, PI, Grayburn, Director Islet Targeting Core Laboratory) and by the Mark Shepherd Fund of the Baylor Foundation.
References
- 1.Newgard CB. While tinkering with the ß-cell metabolic regulatory mechanisms and new therapeutic strategies: American Diabetes Association Lilly Lecture, 2001. Diabetes. 2002;51:3141–3150. doi: 10.2337/diabetes.51.11.3141. [DOI] [PubMed] [Google Scholar]
- 2.Bonner-Weir S, Weir GC. New sources of pancreatic ß-cells. Nat. Biotechnol. 2005;23:857–861. doi: 10.1038/nbt1115. [DOI] [PubMed] [Google Scholar]
- 3.Samson SL, Chan L. Gene therapy for diabetes: reinventing the islet. Trends in endocrinology and metabolism. 2006;17:92–100. doi: 10.1016/j.tem.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 4.McClane SJ, Chirmule N, Burke CV, Raper SE. Caracterization of the immune response after local delivery of recombinant adenovirus in murine pancreas and successful strategies for readministration. Human Gene Ther. 1997;8:2207–2216. doi: 10.1089/hum.1997.8.18-2207. [DOI] [PubMed] [Google Scholar]
- 5.Ayuso E, Chillon M, Agudo J, Haurigot V, Bosch A, Carretero A, et al. In vivo gene transfer to pancreatic beta cells by systemic delivery of adenoviral vectors. Human Gene Ther. 2004;15:805–812. doi: 10.1089/1043034041648426. [DOI] [PubMed] [Google Scholar]
- 6.Wang AY, Peng PD, Ehrhardt A, Storm TA, Kay MA. Comparison of adenoviral and adeno-associated viral vectors for pancreatic gene delivery in vivo. Human Gene Ther. 2004;15:405–413. doi: 10.1089/104303404322959551. [DOI] [PubMed] [Google Scholar]
- 7.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, et al. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 10.McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Ding JH, Bekeredjian R, Yang BZ, Shohet RV, Johnston SA, et al. Efficient gene delivery to pancreatic islets with ultrasonic microbubble destruction technology. Proc Natl Acad Sci U S A. 2006;103:8469–74. doi: 10.1073/pnas.0602921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Ding JH, Grayburn PA. Reversal of streptozotocin-induced diabetes in rats by gene therapy with betacellulin and pancreatic duodenal homeobox-1. Gene therapy. 2007;14:1102–10. doi: 10.1038/sj.gt.3302963. [DOI] [PubMed] [Google Scholar]
- 13.Hwung YP, Gu YZ, Tsai MJ. Cooperativity of sequence elementsmediates tissue specificity of the rat insulin II gene. Mol. Cell. Biol. 1994;4:1784–1788. doi: 10.1128/mcb.10.4.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nir U, Walker MD, Rutter WJ. Regulation of rat insulin 1 gene expression: evidence for negative regulation in nonpancreatic cells. Proc. Natl. Acad. Sci. U. S. A. 1986;10:3180–3184. doi: 10.1073/pnas.83.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen HV, Serup P, Leonard J, Michelsen BK, Madsen OD. Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc Natl Acad Sci U S A. 1994;91:10465–9. doi: 10.1073/pnas.91.22.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boam DS, Clark AR, Docherty K. Positive and negative regulation of the human insulin gene by multiple trans-acting factors. J. Biol. Chem. 1990;14:8285–8296. [PubMed] [Google Scholar]
- 17.Fukazawa T, Matsuoka J, Naomoto Y, Nakai T, Durbin ML, Kojima I, et al. Development of a novel beta-cell specific promoter system for the identification of insulin-producing cells in in vitro cell cultures. Exp Cell Res. 2006;312:3404–12. doi: 10.1016/j.yexcr.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Walker MD, Edlund T, Boulet AM, Rutter WJ. Cell-specifi expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature. 1983;306:557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- 19.Andrali SS, Sampley ML, Vanderford NL, Ozcan S. Glucose regulation of insulin gene expression in pancreatic beta-cells. Biochem J. 2008;415:1–10. doi: 10.1042/BJ20081029. [DOI] [PubMed] [Google Scholar]
- 20.Edlund T, Walker MD, Barr PJ, Rutter WJ. Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′ flanking elements. Science. 1985;230:912–916. doi: 10.1126/science.3904002. [DOI] [PubMed] [Google Scholar]
- 21.German MS, Moss LG, Rutter WJ. Regulation of insulin gene expression by glucose and calcium in transfected primary islet cultures. J Biol Chem. 1990;265:22063–22066. [PubMed] [Google Scholar]
- 22.Philippe J. Structure and pancreatic expression of the insulin and glucagon genes. Endocr Rev. 1991;12:252–71. doi: 10.1210/edrv-12-3-252. [DOI] [PubMed] [Google Scholar]
- 23.Cordier-Bussat M, Morel C, Philippe J. Homologous DNA sequences and cellular factors are implicated in the control of glucagon and insulin gene expression. Mol. Cell. Biol. 1995;15:3904–3916. doi: 10.1128/mcb.15.7.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson LK. Elevated glucose attenuates human insulin gene promoter activity in INS-1 pancreatic beta-cells via reduced nuclear factor binding to the A5/core and Z element. Mol Endocrinol. 2005;19:1343–60. doi: 10.1210/me.2003-0493. [DOI] [PubMed] [Google Scholar]
- 25.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cordle SR, Henderson E, Masuoka H, Weil PA, Stein R. Pancreatic beta-cell-type-specific transcription of the insulin gene is mediated by basic helix-loop-helix DNA-binding proteins. Mol. Cell. Biol. 1991;11:1734–1738. doi: 10.1128/mcb.11.3.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 28.Ohneda K, Mirmira R, Wang J, Johnson J, German M. The homeodomain of PDX-1 mediates multiple proteinprotein interactions in the formation of a transcriptional activation complex on the insulin gene promoter. Mol Cell Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;12:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 30.Mirmira RG, Watada H, German MS. Autoregulation and maturity onset diabetes of the young transcription factors control the human PAX4 promoter. J. Biol. Chem. 2000;19:14743–14751. doi: 10.1074/jbc.M005202200. [DOI] [PubMed] [Google Scholar]
- 31.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;13:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 32.German M, Ashcroft S, Docherty K, Edlund H, Edlund T, Goodison S, et al. The insulin gene promoter. A simplified nomenclature. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 33.Boam DSW, Docherty K. A tissue specific nuclear factor binds to multiple sites in the human insulin gene enhancer. Biochem J. 1989;264:233–239. doi: 10.1042/bj2640233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy GC, Rutter WJ. Pur-1, a zinc-finger protein that binds to purin-rich sequences, transactivates an insulin promoter in heterologous cells. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11498–11502. doi: 10.1073/pnas.89.23.11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohlsson H, Thor S, Edlund T. Novel insulin promoter-and enhancer-binding proteins that discriminate betwen pancreatic α- and β-cells. Mol. Endocrinol. 1991;5:897–904. doi: 10.1210/mend-5-7-897. [DOI] [PubMed] [Google Scholar]
- 36.Cordle SR, Henderson E, Masuoka H, Weil PA, Stein R. Pancreatic [3-cell-type-specific transcription of the insulin gene is mediated by basic helix-loop-helix DNAbinding proteins. Mol Cell Biol. 1991;11:1718–1734. doi: 10.1128/mcb.11.3.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]