Abstract
The molecular disassembly and DNA release dynamics of bioreducible poly(amido amine) polyplexes were studied in real time by atomic force microscopy (AFM). Molecular disassembly of polyplexes constitutes a critical step along the nonviral gene delivery pathway. DNA release is triggered by the depolymerization of high-molecular-weight polycations into low-molecular-weight oligocations utilizing the thiol and disulfide exchange reaction mechanism. The research is motivated by low cytotoxicity and higher transfection of the poly(amido amine)s containing the disulfide bond than those without the disulfide linkage. AFM images were captured in simulated physiological reducing environment by employing dithiothreitol (DTT). Distinctive stages of polyplex disassembly common to the polyplexes with different disulfide content, molecular weight, and polymer architecture are revealed though the DNA release rate is dependent on the disulfide content. In stage one, upon the depolymerization, polyplexes evolve from metastable structures into the thermodynamically favorable toroid structure. In the second stage, toroids aggregate and sometimes fuse into larger ones. In the last stage DNA gradually unravels from the polyplex resulting in highly decondensed wormlike chains and loops that are held by a central compact core. The results affirm the promising aspect of bioreducible poly(amido amine)s as controlled DNA delivery vectors. The study offers new physical insights into the DNA release pathway including intermediate structures with a high degree of structural heterogeneity and disassembly-induced particle growth. The study identifies disassembly-induced colloidal and morphological instability as an important issue to be addressed in the development of successful polyelectrolyte-based gene delivery systems.
Keywords: DNA condensation, DNA release, polyelectrolyte, colloidal stability, nonviral gene delivery, molecular disassembly
INTRODUCTION
The basic research on charged complexes, such as the Layer-by-Layer (LbL) assembly of bolaform amphiphiles and polyelectrolytes by electrostatic attraction of the hydrophilic headgroups and short-range van der Waals attraction among the nonpolar chains,1–4 has contributed to one of the most promising methods for localized gene delivery, i.e. the incorporation of plasmid DNA polyanion into the LbL assembly.5–7 An alternative method for gene delivery involving the DNA/polyelectrolyte complexes is the mixing of DNA and a polycation in aqueous solution, which results in a homogeneous solution containing nano-sized polyplexes. In light of the promising applications in gene delivery, our research is focused on the molecular disassembly process through which DNA is released from the LbL films and polyplexes. Molecular disassembly of polyplexes constitutes a critical step along the nonviral gene delivery pathway. In addition to the effect of polyplex particle size,8, 9 the timing and the degree of DNA release have been shown to affect transgene expression efficiency.10–13
In this study the DNA release is triggered by the depolymerization of high-molecular-weight polycations into low-molecular-weight oligocations utilizing the thiol and disulfide exchange reaction mechanism. Poly(amido amine)s with disulfide linkages were synthesized and their polyplexes were shown to display low cytotoxicity and significantly higher transfection than those without the disulfide linkage.14–16 The bioreducible polymers are less sensitive to hydrolysis than hydrolytically degradable polymers, such as poly(amino ester)s, and their polyplexes are expected to only release DNA rapidly inside the cell. The reduction of the disulfide-containing polymers to short α,ω-dithiol-oligocations proceeds via a thiol and disulfide exchange reaction mechanism. The degradation can be triggered by the redox potential gradient between extracellular environment and various subcellular organelles in states compatible with the physiological conditions.17–22 Bioreducible poly(amido amine)s in the linear and hyperbranched forms have been synthesized by Michael addition copolymerization reaction.23 With secondary and tertiary amines in the polymer backbone, poly(amido amine)s possess high buffering capacity, which enables endosomal escape of the gene delivery vectors.24 The LbL films of a hyperbranched poly(amido amine) and DNA showed higher and prolonged transfection than control polyethylenimine (PEI)-containing films in vitro and exhibited promising transfection activity in vivo.6 The interest in the use of hyperbranched polymers stems from the possibility to prepare polymers with higher molecular weights compared with similar linear polymers.6, 14, 16, 25 The higher molecular weight of hyperbranched polycations is expected to influence positively the stability and transfection activity of the DNA delivery vectors especially in vivo.
Recently, we applied atomic force microscopy (AFM) to visualize plasmid DNA in various decondensed states from reducible polypeptide polyplexes under simulated physiological reducing conditions.26 Prior to our work, AFM has largely being used to study DNA condensation and self-assembled nanostructure of polyplexes.27–37 Our study revealed new molecular disassembly dynamics including intermediate structures with high degree of structural heterogeneity, disassembly-induced aggregation, and dependence of DNA release rate on polymer structure and solution conditions. Striking differences between polyplexes based on polypeptides of histidine-rich peptide HRP (CKHHHKHHHKC) and nuclear localization signal NLS (CGAGPKKKRKVC) peptide were captured by AFM in real time and at the single polyplex level. The HRP and NLS polyplexes were similar in their initial morphology with a majority of them containing only one DNA plasmid. Upon reductive degradation by dithiothreitol (DTT), DNA was released from NLS abruptly regardless of the initial polyplex morphology, while DNA release from HRP polyplexes displayed a gradual decondensation that was dependent on the size of polyplexes. HRP polyplexes also displayed cooperative release behavior where smaller HRP polyplexes became unstable when they were in contact with neighboring decondensed chains.
This paper describes a new AFM investigation of polyplexes containing bioreducible poly(amido amine)s in linear and hyperbranched forms undergoing reductive molecular disassembly. The chemical composition of the polymer is varied by the reducible monomer to non-reducible monomer ratio during synthesis. Distinctive stages of polyplex disassembly are revealed that are common among the polyplexes though the DNA release rate is dependent on the disulfide content and perhaps also on the polymer architecture. The results affirm the promising aspect of bioreducible poly(amido amine)s as controlled DNA delivery vectors and offer new insights into DNA release dynamics relevant to in vitro and in vivo gene delivery.
EXPERIMENTAL METHODS
Materials
Plasmid DNA vector, gWizTM High-Expression Luciferase, containing luciferase reporter gene was purchased from Aldevron. The contour length of DNA with 6,732 base pairs is estimated to be 2.3 µm. Dithiothreitol (DTT, Sigma), 1-(2-aminoethyl)piperazine (AEPZ, Aldrich), 1-methylpiperazine (Aldrich), N,N’-methylenebisacrylamide (MBA, Aldrich), and N,N’-cystaminebisacrylamide (CBA, Polysciences) were purchased in the highest purity and used without further purification. All other chemicals were purchased from Sigma-Aldrich. Water was deionized to 18 MΩ×cm resistivity using Nanopure System from Barnstead. Grade V5 muscovite mica was purchased from Ted Pella and hand cleaved just before use.
Synthesis of bioreducible hyperbranched and linear poly(amido amine)s
The synthesis of hyperbranched and linear bioreducible poly(amido amine)s by Michael addition copolymerization was reported in an earlier paper.6 The distinct reactivity of the amines in AEPZ allows synthesis of either linear or hyperbranched polymers by simply changing the ratio of AEPZ-to-bisacrylamide monomers.23 Using 1:2 molar ratio of AEPZ to CBA+MBA produces hyperbranched polymers, while the use of 1:1 ratio leads to the formation of linear polymers (Scheme 1). The chemical composition of the hyperbranched polymers is further varied by the CBA to MBA ratio, i.e. the reducible disulfide chain density. Table 1 lists the chemical composition and molecular weight characteristics of all the poly(amido amine)s studied here. The chemical composition was characterized by 1H NMR and 13C NMR using a Varian spectrometer (400 MHz). Number-average (Mn) and weight-average (Mw) molecular weight and polydispersity index (Mn/Mw) were determined by size exclusion chromatography (SEC) in 0.03 M sodium acetate (pH 4.5) using Shimadzu LC-10ADVP liquid chromatography equipped with CTO-10ASVP Shimadzu column oven and Polymer Labs PL gel 5 mm mixed C column. SEC data were analyzed using Astra 5.3.1.4 software from Wyatt Technology. Refractive index increments ( dn/dC ) were determined by an interferometric refractometer and used in SEC analysis. All the bioreducible poly(amido amine)s displayed lower cytotoxicity than control branched polyethylenimine (PEI, 25 kDa) and their cytotoxicity decreased with increasing content of the reducible disulfide bonds (data not shown). Cytotoxicity of the synthesized polycations and control PEI were estimated by measuring cell viability at 50 µg/mL of the polymer. The low cytotoxicity of the bioreducible polymers is the primary motivation for this study.
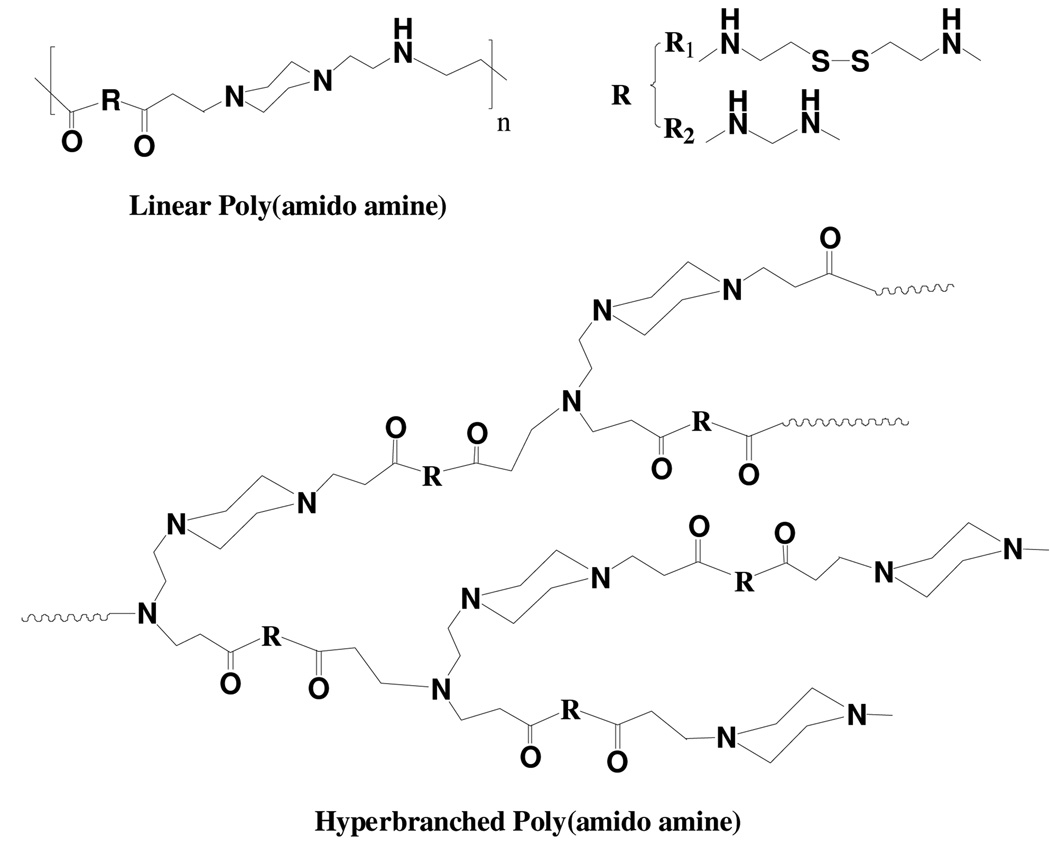
Scheme 1.
Molecular structures of linear and hyperbranched poly(amido amine)s. The disulfide bond content is varied by the feed molar ratio of reducible monomer CBA (R1) to non-reducible monomer MBA (R2).
Table 1.
Chemical composition and molecular weight characteristics of poly(amido amine)s.
| Polymer | CBA content (% mol) |
Mw | Mw/Mn |
|---|---|---|---|
| RHB 1 | 0 | 50,000 | 3.8 |
| RHB 2 | 15 | 115,000 | 6.4 |
| RHB 3 | 21 | 32,000 | 2.2 |
| RHB 4 | 32 | 66,000 | 1.7 |
| RHB 5 | 48 | 40,000 | 3.3 |
| RHB 6 | 100 | 127,000 | 2.2 |
| RL1 | 0 | 25,200 | 1.8 |
| RL2 | 15 | 54,500 | 3.8 |
| RL3 | 100 | 20,800 | 3.8 |
Polyplex preparation
The structure of polyplexes is sensitive to preparation conditions.38, 39 Here, 20 mg/L DNA solution in 0.03 M sodium acetate buffer (pH 4.5) was used to prepare polyplexes at the desired amine-to-DNA phosphate molar ratio (N/P). The poly(amido amine) was added to the DNA solution and mixed by vortexing at 3,200 rpm (Fisher Scientific Vortex Mixer) for 10 s and the solution was incubated at room temperature for 30 min. Twenty µL of polyplex solution was placed on approximately 1 cm2 freshly cleaved mica. Excess of solution was removed after 2 min, and surface was rinsed with deionized water.
AFM Characterization
AFM imaging was conducted using a Nanoscope III MultiMode atomic force microscope from Digital Instruments. Tapping Mode was performed in liquid using silicon nitride probes (NP, VEECO) with a nominal radius of curvature of 20 nm and cantilever spring constant of 0.38 N/m as provided by the manufacturer. A procedure to always image the adsorbed polyplexes in the same buffer (0.03 M sodium acetate buffer) and at the same ionic strength as the reducing solution prior to reduction was adopted in order to be certain of the polyplex stability and to minimize tip disturbance. Usually, the polyplexes were imaged in 40 µL of buffer solutions before 10 µL of DTT solution was injected. The final DTT concentration was maintained at 0.02 M in the liquid cell, which is at a similar level as the in vivo glutathione concentration in the nucleus.40 AFM imaging ensued immediately after the introduction of DTT. The surface was imaged continuously at an average rate of 1.5–2 Hz on a 2 × 2 µm2 until no significant changes occurring at the surface. The commonly used frequency, amplitude, integral, and proportional gains are approximately 8 kHz, 0.5–1 V, 0.5–2, and 0.75–3 respectively.
The AFM images were analyzed using Nanoscope Software Version 5.12b by Veeco. The bearing analysis command in Nanoscope Software was used to measure the particle volume and area in which the substrate was used as the threshold bearing plane. The particle volume above the threshold plane was calculated by integrating the depth histogram over the entire area above the threshold plane. The dynamic particle size results have a typical error of 30%. The apparent particle volume in our AFM analysis is higher than the actual volume due to the well-known tip convolution effect. Alternatively the particle volume can be determined by considering only the top half of the structure.48 In the case of cap-shaped objects likely adopted by the polyplexes after adsorption this method could lead to an underestimation of the particle size.
RESULTS AND DISCUSSION
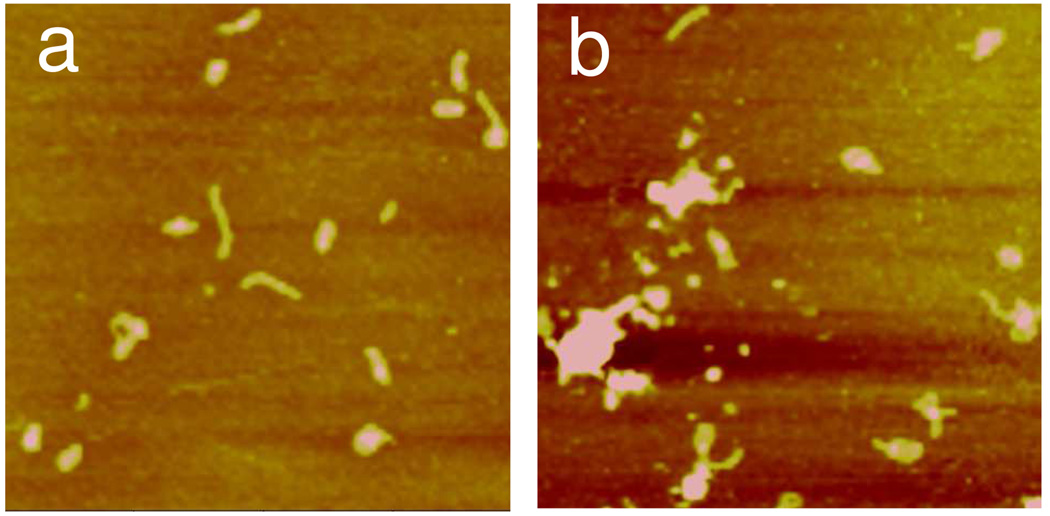
Firstly, the structure of polyplexes as a function of incubation time was probed. An incubation time of 30 min yielded the largest population of compact and non-aggregated polyplex structure. Incubation time of 30 min was therefore chosen for the preparation of all polyplexes described below. Shorter incubation time yielded significant amount of uncondensed and rod structure while significant aggregation occurred at longer than 30 min incubation time (Figure 1). Others have also reported larger percentage of rods near the start of incubation due to the more favorable formation kinetics of rods.41
Figure 1.
AFM height images of RL2/DNA polyplexes (N/P = 4) formed at incubation times of (a) 10 min and (b) 60 min. The images were captured in Tapping Mode in air. The scan size is 2 µm and the z range is 10 nm.
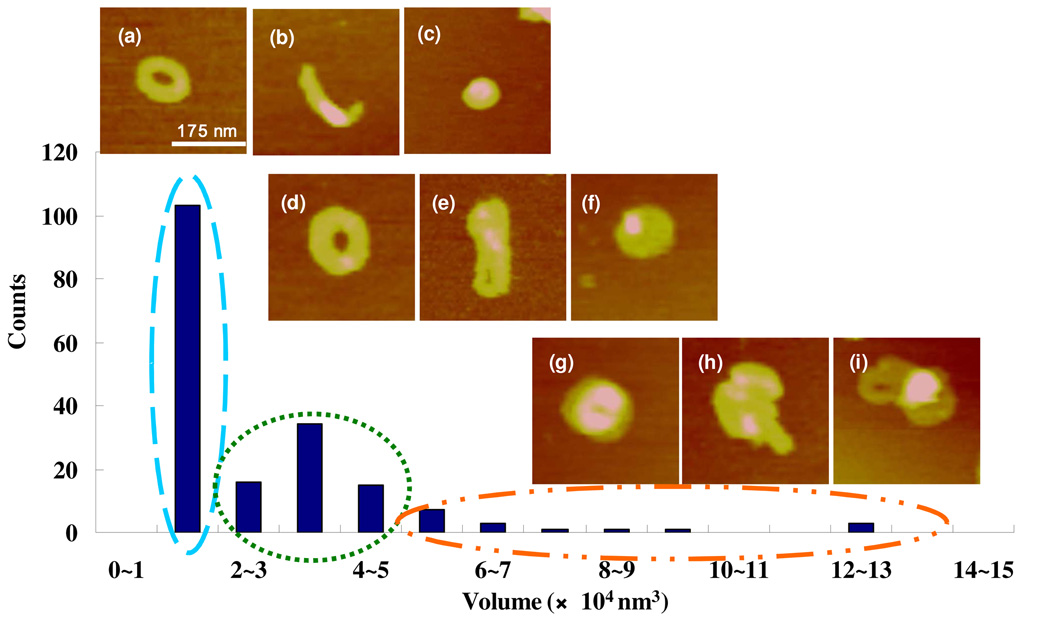
Secondly, the morphology and size distribution of as-formed polyplexes adsorbed on mica were studied by AFM tapping mode in deionized water. All the bioreducible poly(amido amine)s were capable of condensing plasmid DNA into nano-sized polyplexes at N/P > 2 at an incubation time of 30 min. No significant morphological difference was observed among polyplexes formed with the various poly(amido amine)s. The common morphology and size features are described here. Our results are similar to those reported previously using different multivalent condensing agents.30, 34, 37, 42 Figure 2 presents representative polyplex shapes and sizes together with their bearing volume histogram. The toroid structure is characterized by the distinctive hole in the middle. The spheroid is a globular structure without the central hole. The rod is an elongated object whose diameter and length are similar to the diameter and circumference of the toroid. The largest population of the polyplexes has a size range of 1– 2 × 104 nm3. This range matches the theoretical volume of a closely packed polyplex containing a single plasmid DNA with 6,732 bp, 1.4 × 104 nm3. The theoretical value was calculated by assuming an interhexagonal separation between neighboring polycation and DNA chain of 2.7 nm.43 Thus we can conclude that the largest population, regardless of morphology (toroid in Figure 2a, rod in Figure 2b, spheroid in Figure 2c), contains a single plasmid DNA, i.e. it belongs to the monomolecular condensate. The particles with a volume range of 2–5 × 104 nm3 contain 2–3 DNA plasmids and are also capable of forming toroids (represented by Figure 2d), rods or folded toroids (represented by Figure 2e), and spheroids (represented by Figure 2f). A minor population of polyplexes contains more than 3 DNA molecules (represented by Figure 2g – i). The shape of these larger particles is ill defined. The larger particles are generally the result of partial condensation or polyplex aggregation.
Figure 2.
AFM bearing volume histogram and representative AFM height images of the bioreducible polyplexes (N/P = 4). The images were captured in Tapping Mode in deionized water. The scan size is 350 nm. The z range is 15 nm for (g) and (i) and 8 nm for all others. Polyplexes whose volume is in the range of 1–2 × 104 nm3 are represented by (a)–(c). Polyplexes whose volume is in the range of 2–5 × 104 nm3 are represented by (d)–(f). Polyplexes whose particle volume is greater than 5 × 104 nm3 are represented by (g)–(i). (a), (e), and (i) are RHB3/DNA polyplexes and all others are RHB2/DNA polyplexes.
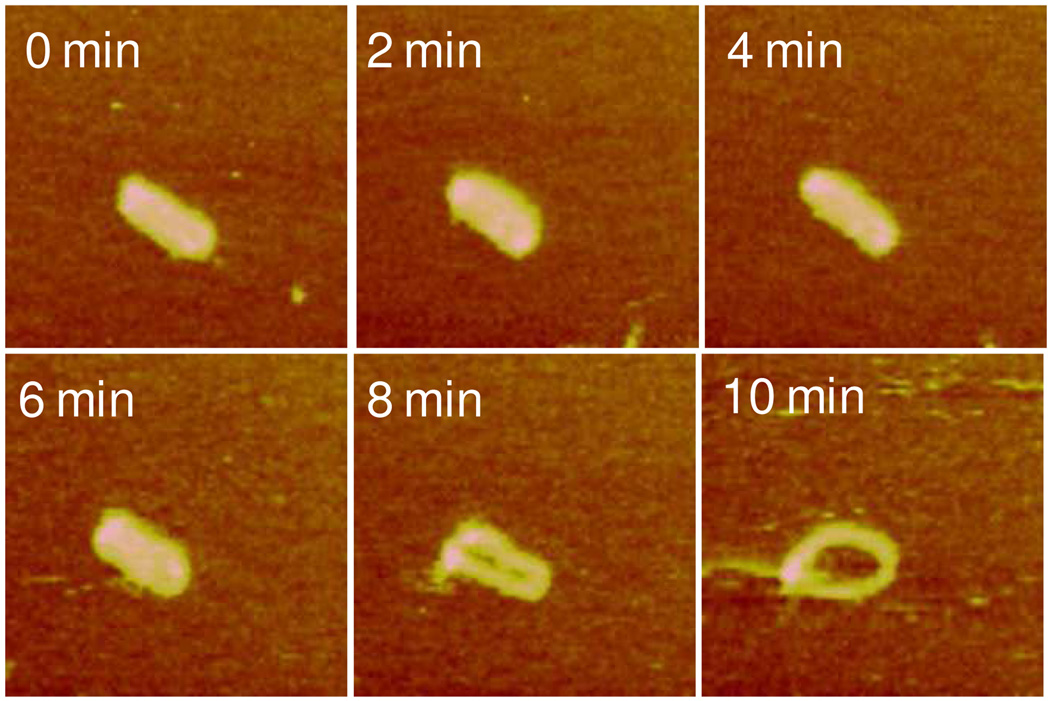
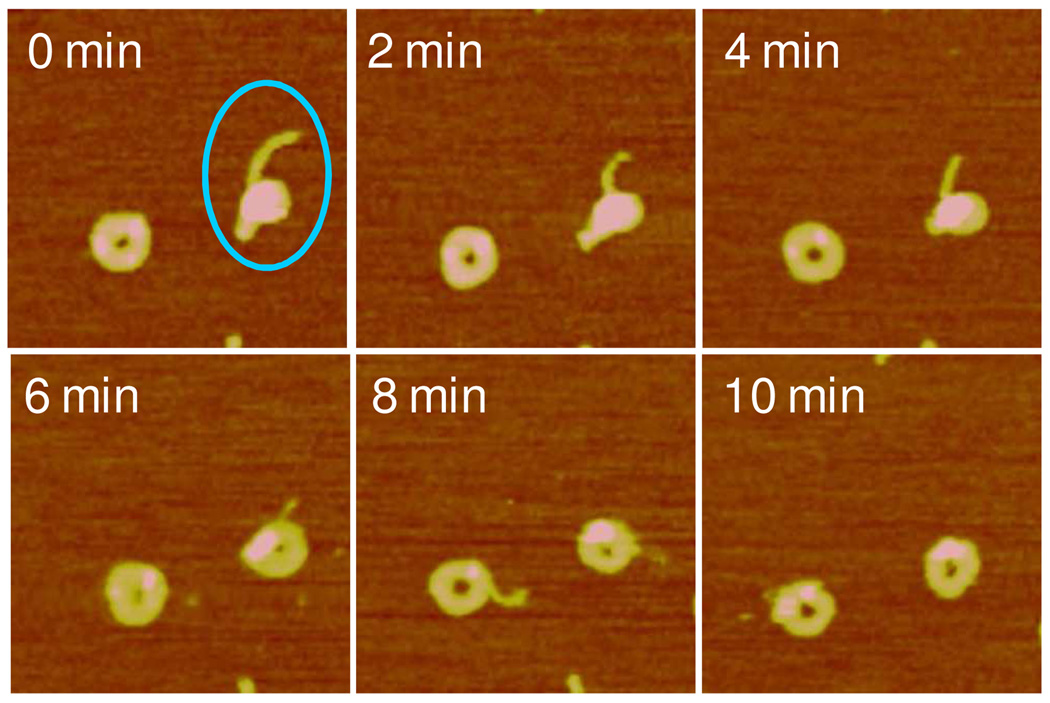
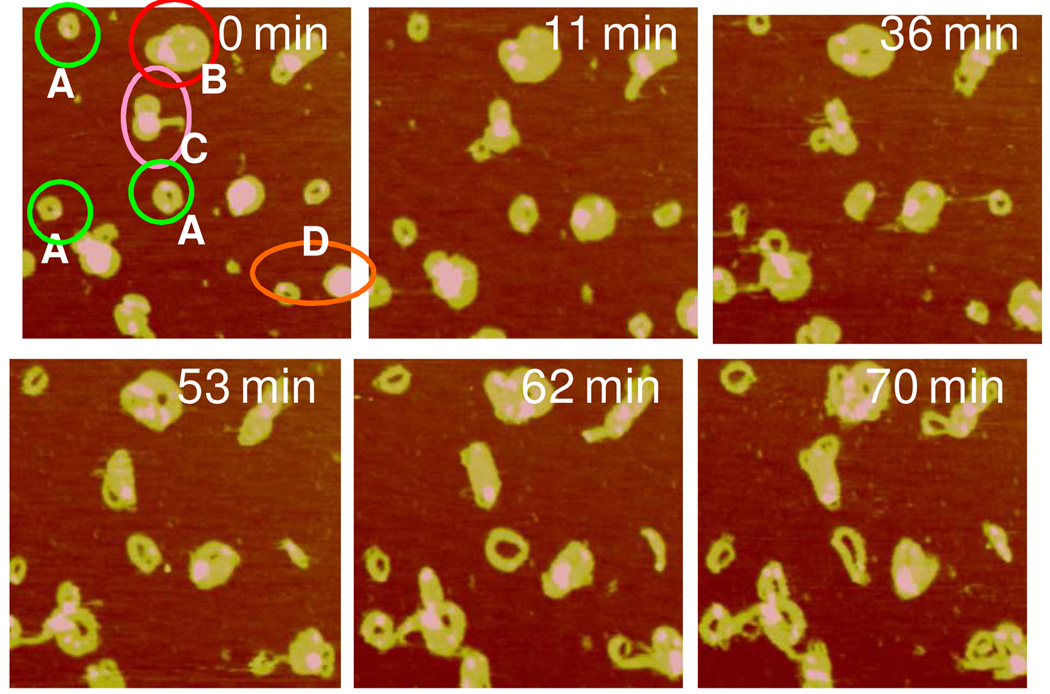
Next we studied DNA release from polyplexes in real time by contacting the adsorbed polyplexes with the reducing DTT solution. The images were captured every 2 min from the time of DTT addition. Upon DTT introduction, a trend of polyplexes of various shapes to converge into the toroid form was observed during the first 10 min prior to visible DNA release. Figure 3 shows such a transition of a rodlike object to toroid by gradually opening up in the center. The rod has a smaller length to width aspect ratio, 2.6, than that of a typical rod however. Figure 4 shows a similar transition to the toroid form by a spheroid with a small arm. With time the spheroid gradually developed a central hole and incorporated the outstretched arm into the single toroid. In contrast the toroid on the left remained unchanged in its overall morphology. Figure 5 presents images containing multiple polyplexes undergoing morphological transitions upon the addition of the DTT solution. The transition from compact structure to toroid with increasing diameter is evident (see particles marked by A). Larger particles containing more than one DNA plasmid also display the same trend toward the formation of toroid, e.g. particle marked by B. Particle B ultimately disentangled into several toroids (t = 70 min). The polyplexes also display mutual interactions during disassembly. The object marked by C was composed of aggregated spheroid-toroid-rod. It first developed into three distinct toroids (t = 36 min) and then the three toroids fused into one larger object (t = 53 min). Initially separated polyplexes, marked by D, were capable of forming connections with each other and eventually merged into each other during disassembly.
Figure 3.
Real-time AFM images showing a gradual transition of a single rodlike RHB3/DNA polyplex to a toroid with a well-defined central hole captured in the DTT solution. Time zero corresponds to the injection of the DTT solution. The scan size is 500 nm. The z range is 7 nm.
Figure 4.
Real-time AFM images showing a gradual transition of a single spheroid RHB4/DNA polyplex (circled) to a toroid captured in the DTT solution. The scan size is 600 nm. The z range is 8 nm.
Figure 5.
Real-time AFM images of RHB2/DNA polyplexes captured after the introduction of the DTT solution. Images were continuously captured every 2–5 min but only selective images are shown. The scan size is1.5 µm. The z range is 10 nm.
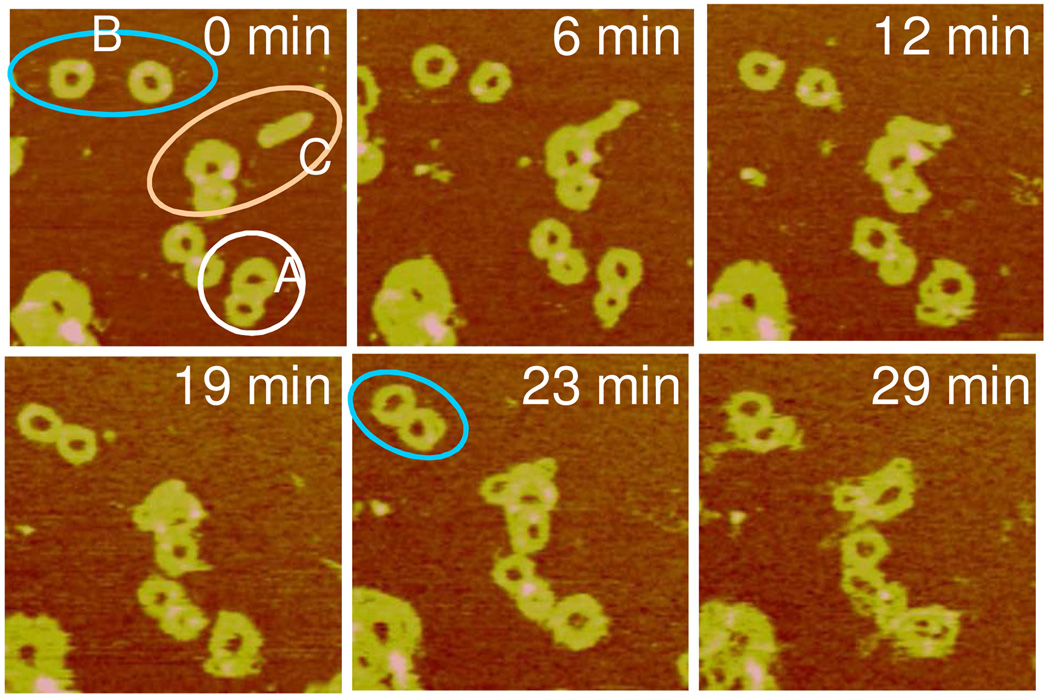
Figure 6 provides another example of disassembly-induced particle-particle interaction and aggregation. The two attached toroids prior to the addition of DTT (marked by A) merged into one 12 min later, and the merged toroid attached to another group 23 min later. The initially far apart toroids separated by ∼100 nm from each other (marked by B) were capable of attaching to each other during the observation period. The group of polyplexes (marked by C) demonstrates both mutual attraction and fusion. The rod polyplex first attached to the neighboring toroid (t = 6 min), and reoriented to the tangential direction of the toroid host (t = 12 min) before fusing into the host (t = 19 min). It should be pointed out that during particle fusion there is little loss in the overall particle volume. The overall volume changed from 4.8 × 104 nm3 to 4.4 × 104 nm3 and from 9.0 × 104 nm3 to 8.6 × 104 nm3 for group A and C respectively.
Figure 6.
Real-time AFM images of RL2/DNA polyplexes in the DTT solution showing particle-particle interaction and aggregation. Images were continuously captured every 2–5 min but only selective images are shown. The scan size is 1 µm and the z range is10 nm.
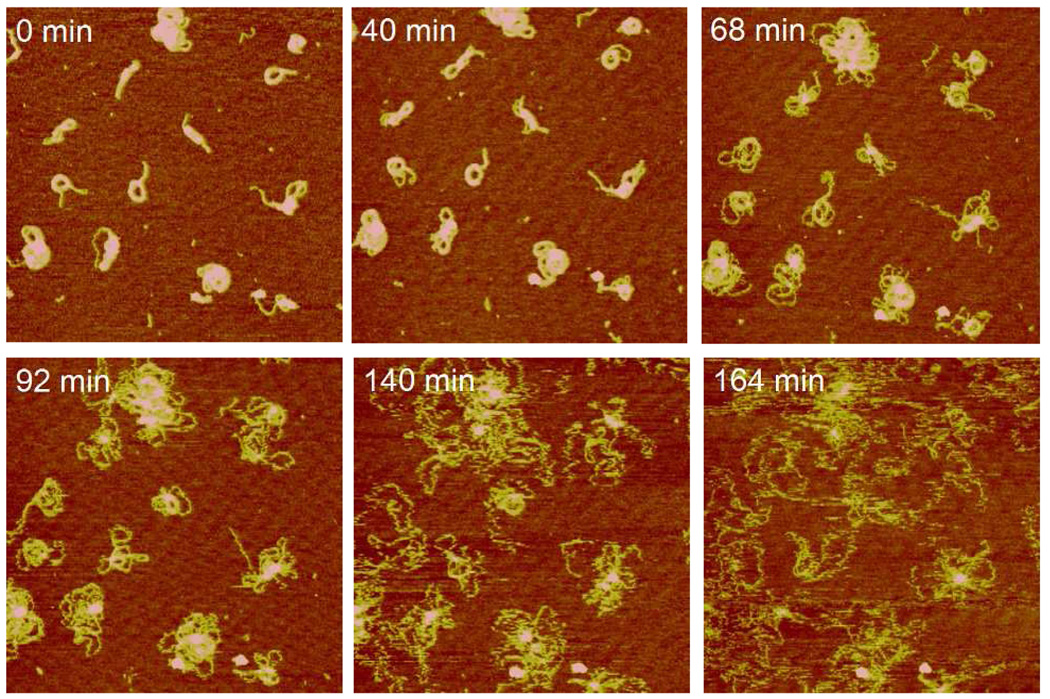
Following morphological changes and fusion as described above the bioreducible polyplexes underwent the final DNA decondensation and release transition. DNA decondensation is signified by thin wormlike chains emerging from the polyplexes. Figure 7 presents a typical DNA release sequence from the RHB2/DNA (N/P = 4) polyplex in the DTT solution. Significant amount of DNA chains was released from the polyplexes at 68 min. The decondensed DNA chains are thinner than the condensed portion of the polyplex. Loose DNA chains continued to emerge and expand up to the end of the observation period. The various shapes of condensed polyplexes tend to evolve into a common decondensed structure containing a central core surrounded by loose DNA loops and chains. This common structure during polyplex disassembly resembles closely the partially condensed DNA during polyplex assembly reported in literature. For example, Dunlap et al.29 reported several partially condensed DNA structures including folded loops of DNA surrounding central cores, loose coils with isolated nodes, and bundles. The less condensed site was believed to be the initiation sites for enzymatic attack.35
Figure 7.
Real-time AFM images of RHB2/DNA polyplexes in the DTT solution showing the DNA decondensation stage. Images were continuously captured every 2–5 min but only selective images are shown. The scan size is 2 µm and the z-range is 6 nm.
Similar dynamic AFM experiments were carried out on other poly(amido amine)s and the average DNA release times are listed in Table 2. The DNA release time was recorded as the time from DTT injection the time when remaining features exhibited height less than 3 nm, which corresponds to decondensed DNA chains adsorbed on substrate, or when the chain features disappeared from the image, which corresponds to complete desorption of DNA chains from the substrate. Only poly(amido amine)s containing the reducible disulfide bond were capable of releasing DNA, while the polyplexes formed with non-reducible poly(amido amines), i.e. RL1 and RHB1, were stable in the reducing solution. The DNA release rate from polyplexes can be tuned from almost instant release to sustained release by varying the CBA content. Complete DNA release from RHB6 and RL3 polyplexes took only a few minutes while 170 min elapsed before DNA was completely released from RHB2 polyplexes. The slow release kinetics of RHB2 polyplexes made it more feasible to study the intricate and multistep DNA release dynamics reported above. Presumably disulfide reaction with DTT is not the rate limiting step, the release rate is a function of oligocation chain length because the higher the CBA content the shorter the average chain length after the reduction reaction. DNA release from RHB2 polyplexes is slower than that form RL2 counterparts even though they have similar disulfide contents. There is insufficient evidence to determine whether the difference is due to polymer architecture, i.e. hyperbranched vs. linear, or molecular weight difference.
Table 2.
DNA release rate dependence on chemical composition and polymer chain architecture. The reducing solutions consists of 0.02 M DTT and 0.2 M NaCl, and 0.03 M sodium acetate buffer (pH = 4.5).
| CBA content (%) | 100 | 48 | 32 | 21 | 15 | 0 |
|---|---|---|---|---|---|---|
| Hyperbranched poly(amido amine) |
< 2 min |
∼ 5 min |
∼ 30 min |
∼ 50 min |
∼ 170 min |
No disassembly |
| Linear poly(amido amine) |
< 2 min |
∼ 50 min |
No disassembly |
DNA condensation remains an active topic for experimental and theoretical research44–49 because of its importance in cell biology, virology, polymer physics, and biotechnology. DNA condensation is driven by an entropy increase associated with the release of counterions upon the polyplex formation. The final condensate structure is determined by a variety of intermolecular forces including forces resisting condensation, such as bending, entropy loss upon demixing of polymer and solvent, and electrostatic repulsion among DNA chains, and favorable forces, such as correlated multivalent counterion fluctuation and cooperative hydration.48 Manning’s counterion condensation theory predicts that 90% of the DNA charges must be neutralized for condensation to occur.50 In the reverse process of decondensation, when a sufficient number of binding cations are lost, DNA is released from the polyplex. Our experimental results confirm the main principle behind controlled DNA release by bioreducible polycations, which is the binding affinity dependence on polycation chain length, though the decondensation process does not appear to be instant as predicted for stiff polymers such as DNA and the system exhibits a great deal of structural heterogeneity.
Various morphologies of DNA condensates have been reported including toroids, rods, spheroids, and less defined ones including rings and flower-like particles depending on condensing conditions, properties of DNA molecules, and condensing agents.51 The polyplex formation is described by the nucleation and growth process.45, 52, 53 It starts with the formation of a nucleation loop or rod followed by the intramolecular collapse of the entire DNA molecule to form a monomolecular toroid or rod. Monomolecular polyplexes grow into multimolecular toroids or rods by incorporating free DNA molecules.53 Molecular simulations of DNA condensation show that the monomolecular toroid is thermodynamically more stable than the rod morphology.54–56 Nucleation kinetics favors the rod form,32, 41 which is the reason why more rods were found at incubation times shorter than 30 min. Spheroids and flower-like polyplexes are often associated with high-molecular-weight polyelectrolytes57, 58 and are also considered to be kinetically trapped. Polyplexes are unstable relative to the aggregated phase and they are expected to grow in size with time controlled by both kinetic and thermodynamic factors.41, 42, 59 In low salt conditions, polyplex formation is dominated by kinetics and its structure is trapped in far from thermodynamic equilibrium state. The morphological instability of polyplexes represents one of the major obstacles for successful non-viral gene delivery systems.13, 60, 61
In our study, we found that the initial polyplex size and morphological distributions are rather insensitive to polymer structure. For example, we observed toroids with average outer diameter of 100 nm in a wide range of molecular weight (25,000–130,000 g/mol), and disulfide content (0–100% CBA) as well as different chain architecture (linear versus hyperbranched). Our experimental evidence points to polyplex formation dynamics dominated by kinetic factors. The relative stability among the different forms also indicates that the transition energy from one to another is prohibitively high. The polyplexes were formed in dilute solution during vigorous mixing, which ensured small polyplex size. In addition, N/P ratios greater than 2 plus the highly charged nature of the poly(amindo amine)s resulted in overall positive zeta potential values (data not shown). The positive surface charge further limits particle growth during polyplex formation.
DNA release from the bioreducible polyplexes is triggered by a depolymerization process to convert high-molecular-weight poly(amido amine)s into low-molecular-weight oligomers and monomers. The polyplex becomes unstable, i.e. the polyanion and polycation dissolve in solution, when the molecular weight of the polycation is greatly reduced. This chain length dependence originates from a loss of entropy as a result of a higher number of shorter chains bound to DNA. The depolymerization rate is much faster than the AFM time period. 62, 63 Therefore we interpret the polyplex morphologies after DTT injection as those exhibited by polyplexes containing low-molecule-weight cations. The experimental evidence points to the role of depolymerization in driving the transition from frozen states to thermodynamically stable state. When the high-molecular-weight polycation is converted to low-molecular-weight cations, the transition energy is lowered to allow different forms to converge into the lowest energy form, i.e. the toroid structure. Depolymerization weakens the electrostatic interaction and enables rearrangement by freeing counterion from kinetically constrained binding sites. The chain length reduction results in loss of excess cations thus allowing closer packing as well as loss of the electrostatically stabilizing shell of the excess polycation. The role of depolymerization in a sense is similar to that of salt. Our study shows that toroid is the thermodynamically favorable form in the case of low-molecular-weight counterions for both monomolecular and multimolecular polyplexes. Since all the in situ AFM experiments were conducted in DNA-free solutions, the structural transition can be accomplished without free DNA, thus providing a new mechanism for DNA morphological transition alternative to the DNA-assisted mechanism.41
Polyplex interaction during disassembly is another interesting aspect of this study. Two neighboring polyplexes are attracted to each other and fuse into one. The interparticle interaction is facilitated by a reduction in the positive charges or local charge reversal due to depolymerization. The same local charge reversal mechanism results in intersegment attraction in polyplex formation.62, 64 The less compact condensate starts to recover phosphate groups that make part of the chain negatively charged. The negative charged DNA interacts electrostatically with positively charged condensate surface. The hydrophobic interaction may also play a role if oligocations rearrange to expose the hydrophobic parts as in the ladder structure initially proposed by Kabanov. The interparticle interaction and particle growth in our case are not mediated by free DNA. Instead, the same correlated electrostatic fluctuation and hydrophobic interactions between neighboring particles and chains are the responsible factors. A previous study observed the commensurate reorganization of two DNA strands, which was considered as a precursor to the formation of a larger condensate.31 The commensurate association may also play a role in DNA release dynamics, e.g. the rod orientation from the radial to the tangential direction with respect to the nearby toroid before fusion (Figure 5 group C).
CONCLUSIONS
The bioreducible poly(amido amine) polyplexes provide an ideal system to study molecular disassembly and DNA decondensation dynamics by real-time AFM. The polyplexes are stable in the oxidizing environment representative of the non-reducing extracellular space. DNA release is triggered by mild DTT and salt concentration compatible with the physiological environment. In order to mimic conditions in cytosol, future experiments will be conducted in conditions more closely resembling cytosol, e.g. in glutathione instead of DTT. The results demonstrate DNA release dynamics from bioreducible polyplexes to consist of three stages that take place at different times. In stage one, upon the depolymerization; polyplexes evolve from metastable structures into the thermodynamically favorable toroid structure. In the second stage, toroids aggregate and sometimes fuse into larger ones. In the last stage DNA gradually unravels from the polyplex resulting in highly decondensed wormlike chains and loops held by a central compact core. Our data demonstrate that the DNA release rate can be precisely controlled by the disulfide bond content. The impact of polymer architecture and molecular weight needs further investigation. The polyplex colloidal stability, the intermediate structure, and their interactions impact its delivery efficiency and its effect on cell viability. The results reveal potentially rich DNA release dynamics relevant to in vitro and in vivo DNA delivery. The results point to the potential challenge in regulating the stability of gene delivery vectors during disassembly due to disassembly-induced colloidal and morphological instability. Higher level aggregation can be a disadvantage for gene delivery because of the difficulty in trafficking large particles and the introduction of too many DNA fragments into one cell.
ACKNOWLEDGEMENT AND DEDICATION
This work was supported partially by National Science Foundation Grants CBET-0553533 & CBET-0755654 (G.M.) and National Institute of Health Grant CA 109711 from the National Cancer Institute (D.O.). The manuscript is dedicated to Professor H. Ted Davis on his 70th birthday.
REFERENCES
- 1.Decher G, Hong JD. Buildup of Ultrathin Multilayer Films by a Self-Assembly Process .2. Consecutive Adsorption of Anionic and Cationic Bipolar Amphiphiles and Polyelectrolytes on Charged Surfaces. Berichte Der Bunsen-Gesellschaft-Physical Chemistry Chemical Physics. 1991;95(11):1430–1434. [Google Scholar]
- 2.Mao GZ, Tsao YH, Tirrell M, Davis HT, Hessel V, Ringsdorf H. Self-Assembly of Photopolymerizable Bolaform Amphiphile Monolayers and Multilayers. Langmuir. 1993;9(12):3461–3470. [Google Scholar]
- 3.Mao GZ, Tsao YH, Tirrell M, Davis HT, Hessel V, Ringsdorf H. Interactions Structure and Stability of Photoreactive Bolaform Amphiphile Multilayers. Langmuir. 1995;11(3):942–952. [Google Scholar]
- 4.Mao GZ, Tsao YH, Tirrell M, Davis HT, Hessel V, Vanesch J, Ringsdorf H. Monolayers of Bolaform Amphiphiles - Influence of Alkyl Chain-Length and Counterions. Langmuir. 1994;10(11):4174–4184. [Google Scholar]
- 5.Blacklock J, Handa H, Manickam DS, Mao GZ, Mukhopadhyay A, Oupicky D. Disassembly of layer-by-layer films of plasmid DNA and reducible TAT polypeptide. Biomaterials. 2007;28(1):117–124. doi: 10.1016/j.biomaterials.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 6.Blacklock J, You Y-Z, Zhou Q-H, Mao G, Oupick D. Gene delivery in vitro and in vivo from bioreducible multilayered polyelectrolyte films of plasmid DNA. Biomaterials. 2009;30(5):939–950. doi: 10.1016/j.biomaterials.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kakade S, Manickam DS, Handa H, Mao G, Oupicky D. Transfection activity of layer-by-layer plasmid DNA/poly(ethylenimine) films deposited on PLGA microparticles. International Journal of Pharmaceutics. 2009;365(1–2):44–52. doi: 10.1016/j.ijpharm.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen H, Kunath K, Martin AL, Stolnik S, Roberts CJ, Davies MC, Kissel T. Star-shaped poly(ethylene glycol)-block-polyethylenimine copolymers enhance DNA condensation of low molecular weight polyethylenimines. Biomacromolecules. 2002;3(5):926–936. doi: 10.1021/bm025539z. [DOI] [PubMed] [Google Scholar]
- 9.Vijayanathan V, Thomas T, Shirahata A, Thomas TJ. DNA Condensation by Polyamines: A Laser Light Scattering Study of Structural Effects. 2001;Vol. 40:13644–13651. doi: 10.1021/bi010993t. [DOI] [PubMed] [Google Scholar]
- 10.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnology and Bioengineering. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 11.Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Advanced Drug Delivery Reviews. 2001;53(3):341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- 12.Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-mediated RNA delivery: a novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Research. 2001;29(18):3882–3891. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. Journal of Pharmaceutical Sciences. 2003;92(2):203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- 14.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JFJ. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: Synthesis and in vitro gene transfer properties. Journal of Controlled Release. 2006;116(2):130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Hoon Jeong J, Christensen LV, Yockman JW, Zhong Z, Engbersen JFJ, Jong Kim W, Feijen J, Wan Kim S. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28(10):1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong ZY, Lin C, Engbersen JFJ, Feijen J. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjugate Chemistry. 2006;17(5):1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 17.Tang FX, Hughes JA. Use of dithiodiglycolic acid as a tether for cationic lipids decreases the cytotoxicity and increases transgene expression of plasmid DNA in vitro. Bioconjugate Chemistry. 1999;10(5):791–796. doi: 10.1021/bc990016i. [DOI] [PubMed] [Google Scholar]
- 18.Byk G, Wetzer B, Frederic M, Dubertret C, Pitard B, Jaslin G, Scherman D. Reduction-sensitive lipopolyamines as a novel nonviral gene delivery system for modulated release of DNA with improved transgene expression. Journal of Medicinal Chemistry. 2000;43(23):4377–4387. doi: 10.1021/jm000284y. [DOI] [PubMed] [Google Scholar]
- 19.Saito G, Swanson JA, Lee KD. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Advanced Drug Delivery Reviews. 2003;55(2):199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 20.Read ML, Bremner KH, Oupicky D, Green NK, Searle PF, Seymour LW. Vectors based on reducible polycations facilitate intracellular release of nucleic acids. Journal of Gene Medicine. 2003;5(3):232–245. doi: 10.1002/jgm.331. [DOI] [PubMed] [Google Scholar]
- 21.Chittimalla C, Zammut-Italiano L, Zuber G, Behr JP. Monomolecular DNA Nanoparticles for Intravenous Delivery of Genes. J. Am. Chem. Soc. 2005;127(32):11436–11441. doi: 10.1021/ja0522332. [DOI] [PubMed] [Google Scholar]
- 22.Oishi M, Hayama T, Akiyama Y, Takae S, Harada A, Yamasaki Y, Nagatsugi F, Sasaki S, Nagasaki Y, Kataoka K. Supramolecular Assemblies for the Cytoplasmic Delivery of Antisense Oligodeoxynucleotide: Polyion Complex (PIC) Micelles Based on Poly(ethylene glycol)-SS-Oligodeoxynucleotide Conjugate. Biomacromolecules. 2005;6(5):2449–2454. doi: 10.1021/bm050370l. [DOI] [PubMed] [Google Scholar]
- 23.Wu DC, Liu Y, Chen L, He CB, Chung TS, Goh SH. 2A(2)+BB ‘ B " approach to hyperbranched poly(amino ester)s. Macromolecules. 2005;38(13):5519–5525. [Google Scholar]
- 24.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong JH, Christensen LV, Yockman JW, Zhong ZY, Engbersen JFJ, Kim WJ, Feijen J, Kim SW. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28(10):1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Wan L, Manickam DS, Oupický D, Mao G. DNA Release Dynamics from Reducible Polyplexes by Atomic Force Microscopy. Langmuir. 2008;24(21):12474–12482. doi: 10.1021/la802088y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfert MA, Seymour LW. Atomic force microscopic analysis of the influence of the molecular weight of poly(L)lysine on the size of polyelectrolyte complexes formed with DNA. Gene Therapy. 1996;3(3):269–273. [PubMed] [Google Scholar]
- 28.Allen MJ, Bradbury EM, Balhorn R. AFM analysis of DNA-protamine complexes bound to mica. Nucl. Acids Res. 1997;25(11):2221–2226. doi: 10.1093/nar/25.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunlap DD, Maggi A, Soria MR, Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Research. 1997;25(15):3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansma HG, Golan R, Hsieh W, Lollo CP, Mullen-Ley P, Kwoh D. DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Research. 1998;26(10):2481–2487. doi: 10.1093/nar/26.10.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono MY, Spain EM. Dynamics of DNA condensates at the solid-liquid interface by atomic force microscopy. Journal of the American Chemical Society. 1999;121(32):7330–7334. [Google Scholar]
- 32.Martin AL, Davies MC, Rackstraw BJ, Roberts CJ, Stolnik S, Tendler SJB, Williams PM. Observation of DNA-polymer condensate formation in real time at a molecular level. Febs Letters. 2000;480(2–3):106–112. doi: 10.1016/s0014-5793(00)01894-9. [DOI] [PubMed] [Google Scholar]
- 33.Iwataki T, Kidoaki S, Sakaue T, Yoshikawa K, Abramchuk SS. Competition between compaction of single chains and bundling of multiple chains in giant DNA molecules. Journal of Chemical Physics. 2004;120(8):4004–4011. doi: 10.1063/1.1642610. [DOI] [PubMed] [Google Scholar]
- 34.Danielsen S, Maurstad G, Stokke BT. DNA–Polycation Complexation and Polyplex Stability in the Presence of Competing Polyanions. Biopolymers. 2004;77:86–97. doi: 10.1002/bip.20170. [DOI] [PubMed] [Google Scholar]
- 35.Chim YTA, Lam JKW, Ma Y, Armes SP, Lewis AL, Roberts CJ, Stolnik S, Tendler SJB, Davies MC. Structural study of DNA condensation induced by novel phosphorylcholine-based copolymers for gene delivery and relevance to DNA protection. Langmuir. 2005;21(8):3591–3598. doi: 10.1021/la047480i. [DOI] [PubMed] [Google Scholar]
- 36.Maurstad G, Danielsen S, Stokke BT. The Influence of Charge Density of Chitosan in the Compaction of the Polyanions DNA and Xanthan. Biomacromolecules. 2007;8(4):1124–1130. doi: 10.1021/bm0610119. [DOI] [PubMed] [Google Scholar]
- 37.Golan R, Pietrasanta LI, Hsieh W, Hansma HG. DNA Toroids: Stages in Condensation. Biochemistry. 1999;38(42):14069–14076. doi: 10.1021/bi990901o. [DOI] [PubMed] [Google Scholar]
- 38.Zelphati O, Nguyen C, Ferrari M, Felgner J, Tsai Y, Felgner PL. Stable and monodisperse lipoplex formulations for gene delivery. Gene Therapy. 1998;5(9):1272–1282. doi: 10.1038/sj.gt.3300707. [DOI] [PubMed] [Google Scholar]
- 39.Oupicky D, Konak C, Ulbrich K, Wolfert MA, Seymour LW. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. Journal of Controlled Release. 2000;65(1–2):149–171. doi: 10.1016/s0168-3659(99)00249-7. [DOI] [PubMed] [Google Scholar]
- 40.Oupicky D, Bisht HS, Manickam DS, Zhou Q-h. Stimulus-controlled delivery of drugs and genes. Expert Opin. Drug Deliv. 2005;2(4):653–665. doi: 10.1517/17425247.2.4.653. [DOI] [PubMed] [Google Scholar]
- 41.Vilfan ID, Conwell CC, Sarkar T, Hud NV. Time study of DNA condensate morphology: Implications regarding the nucleationgrowth and equilibrium populations of toroids and rods. Biochemistry. 2006;45(26):8174–8183. doi: 10.1021/bi060396c. [DOI] [PubMed] [Google Scholar]
- 42.Siqian He, Victor PGA, Bloomfield A. Condensation of DNA by multivalent cations: Experimental studies of condensation kinetics. Biopolymers. 2000;53(4):329–341. doi: 10.1002/(SICI)1097-0282(20000405)53:4<329::AID-BIP5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 43.Dauty E, Remy JS, Blessing T, Behr JP. Dimerizable Cationic Detergents with a Low cmc Condense Plasmid DNA into Nanometric Particles and Transfect Cells in Culture. J. Am. Chem. Soc. 2001;123(38):9227–9234. doi: 10.1021/ja015867r. [DOI] [PubMed] [Google Scholar]
- 44.Gosule; LC, Schellman JA. Nature. 1976;259:333–335. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 45.Bloomfield VA. DNA condensation. Current Opinion in Structural Biology. 1996;6(3):334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 46.Rolland A, Felgner PL. Non-viral gene delivery systems - Preface. Advanced Drug Delivery Reviews. 1998;30(1–3):1–3. doi: 10.1016/s0169-409x(97)00101-4. [DOI] [PubMed] [Google Scholar]
- 47.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nature Biotechnology. 2000;18(1):33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 48.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44(3):269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 49.Todd BA, Parsegian VA, Shirahata A, Thomas TJ, Rau DC. Attractive forces between cation condensed DNA double helices. Biophysical Journal. 2008;94(12):4775–4782. doi: 10.1529/biophysj.107.127332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson; RW; Bloomfield, A V. Counterion-induced condensation of deoxyribonucleic acid. A light-scattering study. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 51.Hansma HG. Surface biology of DNA by atomic force microscopy. Annual Review of Physical Chemistry. 2001;52:71–92. doi: 10.1146/annurev.physchem.52.1.71. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa K, Matsuzawa Y. Nucleation and growth in single DNA molecules. Journal of the American Chemical Society. 1996;118(4):929–930. [Google Scholar]
- 53.Conwell CC, Vilfan ID, Hud NV. Controlling the size of nanoscale toroidal DNA condensates with static curvature and ionic strength. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9296–9301. doi: 10.1073/pnas.1533135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noguchi H, Yoshikawa K. Folding path in a semiflexible homopolymer chain: A Brownian dynamics simulation. Journal of Chemical Physics. 2000;113(2):854–862. [Google Scholar]
- 55.Stevens MJ. Simple simulations of DNA condensation. Biophysical Journal. 2001;80(1):130–139. doi: 10.1016/S0006-3495(01)76000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ou ZY, Muthukumar M. Langevin dynamics of semiflexible polyelectrolytes: Rod-toroid-globule-coil structures and counterion distribution. Journal of Chemical Physics. 2005;123(7) doi: 10.1063/1.1940054. [DOI] [PubMed] [Google Scholar]
- 57.Vasilevskaya VV, Khokhlov AR, Kidoaki S, Yoshikawa K. Structure of collapsed persistent macromolecule: Toroid vs spherical globule. Biopolymers. 1997;41(1):51–60. [Google Scholar]
- 58.Akitaya T, Seno A, Nakai T, Hazemoto N, Murata S, Yoshikawa K. Weak interaction induces an ON/OFF switch whereas strong interaction causes gradual change: Folding transition of a long duplex DNA chain by poly-L-lysine. Biomacromolecules. 2007;8(1):273–278. doi: 10.1021/bm060634j. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen TT, Shklovskii BI. Kinetics of macroion coagulation induced by multivalent counterions. Physical Review E. 2002;65(3) doi: 10.1103/PhysRevE.65.031409. [DOI] [PubMed] [Google Scholar]
- 60.Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Advanced Drug Delivery Reviews. 2001;46(1–3):187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 61.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Advanced Drug Delivery Reviews. 2002;54(5):715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 62.Jensen KS, Hansen RE, Winther JR. Kinetic and Thermodynamic Aspects of Cellular Thiol-Disulfide Redox Regulation. Antioxidants & Redox Signaling 0. (ja) doi: 10.1089/ars.2008.2297. [DOI] [PubMed] [Google Scholar]
- 63.Whitesides GM, Lilburn JE, Szajewski RP. Rates of thiol-disulfide interchange reactions between mono- and dithiols and Ellman’s reagent. The Journal of Organic Chemistry. 1977;42(2):332–338. [Google Scholar]
- 64.Grosberg AY, Nguyen TT, Shklovskii BI. Colloquium: The physics of charge inversion in chemical and biological systems. Reviews of Modern Physics. 2002;74(2):329–345. [Google Scholar]