Abstract
Objective. Five loci—the shared epitope (SE) of HLA-DRB1, the PTPN22 gene, a locus on 6q23, the STAT4 gene and a locus mapping to the TRAF1/C5 genetic region—have now been unequivocally confirmed as conferring susceptibility to RA. The largest single effect is conferred by SE. We hypothesized that combinations of susceptibility alleles may increase risk over and above that of any individual locus alone.
Methods. We analysed data from 4238 RA cases and 1811 controls, for which genotypes were available at all five loci.
Results. Statistical analysis identified eight high-risk combinations conferring an odds ratio >6 compared with carriage of no susceptibility variants and, interestingly, 10% population controls carried a combination conferring high risk. All high-risk combinations included SE, and all but one contained PTPN22. Statistical modelling showed that a model containing only these two loci could achieve comparable sensitivity and specificity to a model including all five. Furthermore, replacing SE (which requires full subtyping at the HLA-DRB1 gene) with DRB1*1/4/10 carriage resulted in little further loss of information (correlation coefficient between models = 0.93).
Conclusions. This represents the first exploration of the viability of population screening for RA and identifies several high-risk genetic combinations. However, given the population incidence of RA, genetic screening based on these loci alone is neither sufficiently sensitive nor specific at the current time.
Keywords: Rheumatoid arthritis, Genetics
Introduction
RA (MIM 180 300) is a complex autoimmune disease characterized by chronic inflammation and destruction of synovial joints, leading to disability and joint damage. It affects an estimated 0.8% of the UK population, and imposes a significant economic burden on healthcare systems [1]. Both genetic and environmental factors contribute to the aetiology. The heritability of RA has been estimated to be between 50 and 60% [2] suggesting that the genetic component of the disease has a significant impact on disease susceptibility.
The strongest genetic association with RA susceptibility has been established since 1970s and lies within the HLA region [3], in particular HLA-DRB1 (MIM 142 857). Alleles associated with RA share a conserved amino acid sequence in the third hyper-variable region of the DRβ1 chain and are referred to as the shared epitope (SE) [4]. The SE has reproducibly been shown to be associated with RA susceptibility and severity in many different populations.
More recently, other RA susceptibility loci have been identified and confirmed. A non-synonymous single nucleotide polymorphism (SNP) in the gene encoding protein tyrosine phosphatase non-receptor 22 (PTPN22) (MIM 600 716)—R620W—confers the second largest risk of susceptibility to RA [5]. A locus lying between OLIG3 (MIM 609 323) and TNFAIP3 (MIM 191 163) on chromosome 6q was identified in a genome-wide association study (GWAS) of seven common diseases, including RA, carried out by the WTCCC [6]. Association with 6q23 has been replicated in populations from the UK and USA [7, 8]. A GWAS in US and Swedish populations identified a novel locus mapping between TRAF1 (MIM 601 711) and C5 (MIM 120 900) associated with RA [9]. This association has been replicated in samples from UK, Greek, Dutch and North American populations [9–12]. Finally, the STAT4 (MIM 600 558) locus has been identified as a confirmed RA susceptibility locus in UK, Korean, Swedish, US, Greek, Colombian, Spanish and US populations [12–17].
The identified loci are neither necessary nor sufficient to cause RA. The largest single effect comes from the SE [odds ratio (OR) ranging from 2 to 3] with effect sizes for the other susceptibility genes ranging from 1.1 to 1.8. It is hypothesized that combinations of susceptibility alleles may further increase the risk of RA. Indeed, several commercial companies offer genetic screening tests to the general public quantifying the level of risk of developing RA over a lifetime. The loci tested vary and not all include the confirmed loci listed above. In particular, the SE is not included in any of the tests, presumably because the cost of subtyping at the HLA-DRB1 locus to define SE alleles is both time consuming and expensive. As SE confers the highest single genetic risk of RA, calculations failing to incorporate this factor may lead to inaccurate risk predictions.
The aim of the current work was, first, to investigate whether combinations of five confirmed RA susceptibility loci were associated with higher risk of developing RA than SE alone; secondly, to explore the extent of information loss by replacing SE subtyping with DRB*01, *04, and *10 broad typing as this could influence screening costs dramatically and, thirdly, to assess whether genotyping at these loci alone would be useful for population screening to identify ‘at risk’ individuals.
Materials and methods
Subjects
A total of 4238 RA cases and 1811 controls were selected, for whom individual genotype data were available at all five susceptibility loci. These data were generated for previously published studies [6, 8, 10]. All RA cases were >18 years old, and satisfied the ACR criteria for RA [18]. All cases and controls were Caucasians from the UK, and all provided informed consent. The study was approved by the North West Ethics Committee (MREC 99/8/84).
Genotyping
Genotyping of the PTPN22, 6q23, STAT4 and TRAF1/C5 loci was undertaken using the Sequenom MassArray platform as described and published previously [8, 10, 19]. For HLA genotyping, genomic DNA was amplified using the Dynal RELI SSO HLA-DRB1 kits as described previously [20]. PCR amplicons were identified by a reverse line assay using sequence-specific oligonucleotide (SSO) probes with the Dynal RELI SSO strip detection reagent kit (http://www.dynalbiotech.com/). Assay results were interpreted using the Pattern Matching Program provided by Dynal (Invitrogen, Paisley, UK). Broad HLA genotyping and subtyping were performed to identify the presence of the SE in the HLA-DRB1 locus.
Susceptibility loci tested
For each of the five susceptibility loci selected for investigation, the most significantly associated SNP identified to date in the UK population was tested, except in the case of the SE where full subtyping was available. Susceptibility loci were defined as: PTPN22, carriage of the minor risk allele T at rs2 476 601; 6q23, carriage of the minor risk allele A at rs6 920 220; STAT4, carriage of the minor risk allele T at rs7 574 865; TRAF1/C5, carriage of the major risk allele A at rs10 760 130; SE, defined by the presence of any of the following alleles: HLA-DRB1*0101, HLA-DRB1*0102, HLA-DRB1*0104, HLA-DRB1*0401, HLA-DRB1*0404, HLA-DRB1*0405, HLA-DRB1*0408 and HLA-DRB1*1001; and DRB1*01/04/10 status, defined as carriage of either *01, *04 or *10 allele/s.
Statistical analysis
Statistical analysis of the data was carried out using STATA version 9.2. Analysis was conducted by carriage of the risk allele for each locus: carriage of the risk allele at each locus was defined as 1, and not carrying the risk allele was defined as 0. Therefore, for the five loci, 32 (25) possible gene combinations were identified. Logistic regression was performed and genotypic ORs and CIs for each gene combination were generated. High-risk combinations were arbitrarily defined as those conferring an OR >6 and with 95% CIs that did not encompass unity. ORs were compared with base odds of the population, who did not carry risk alleles at any of the susceptibility loci to create comparable OR. If carriage of a particular combination was compared with non-carriage, different individuals would be included in the denominator resulting in non-comparable OR. Each individual could only be included once in the table.
ORs were calculated as:
where B0 denotes controls with no risk alleles; A0 denotes cases with no risk alleles; Di denotes controls with the i-th combination of risk alleles; and Ci denotes cases with i-th combination of risk alleles. Numbers of cases and controls for each allele combination class were tabulated. As each individual can only appear once, some genotype combinations were poorly represented; therefore, results from any class with less than five individuals in the case or control group were regarded as unreliable.
Statistical modelling was performed to determine whether the five-locus model could be simplified, using maximum likelihood tests implemented in STATA. First, combinations of SE carriage with different numbers of the other four loci were compared with the full model. Secondly, SE status was replaced by broad DRB*1, *4 or *10 status and compared with the full model. Log likelihood estimates showing the goodness of fit of the simplified model to the full model were calculated. The sensitivity and specificity of each model were calculated, setting probability thresholds at 80%. For population-based screening, it is recommended that a model should have >80% power of discrimination, in order to be useful in the identification of high-risk individuals [21]. Receiver operator curve (ROC) curves were generated, defining the sensitivity and specificity of each model, and the area under the curves was compared. Likelihood ratios were calculated and, from these, chi-square tests highlighted any significant differences between the models. Correlation coefficients of the models were compared to the full model. Sensitivity and specificity were compared using the ‘diagt’ command in STATA, which calculates summary statistics of a diagnostic test using a 2 × 2 table. Numbers of true positives, false negatives, false positives and true negatives defined by the test are compared with the true disease status of each individual to calculate the sensitivity and specificity.
Gender analysis
RA is three times more prevalent in women; therefore, the analysis was also performed with stratification by gender to investigate whether different genetic risk combinations were present in males and females.
Stratification by anti-citrullinated peptide antibody status
There is increasing evidence of differences in risk conferred by the loci identified in anti-citrullinated peptide (anti-CCP)-positive and -negative individuals. Analyses were therefore repeated in the anti-CCP antibody-positive subgroup.
Results
Clinical characteristics
The clinical and demographic features of the cases included in the analysis are shown in Table 1, and are typical of a hospital-based cohort of RA subjects.
Table 1.
Clinical and demographic data
| Characteristics | Cases, n (%) | Controls, n (%) |
|---|---|---|
| Male | 1493 (35.3) | 803 (44.6) |
| Female | 2738 (64.7) | 999 (55.4) |
| Caucasians | 4238 (100) | 1811 (100) |
| Erosions | 697 (68.7) | NA |
| Nodules | 750 (35) | NA |
| RF positive | 1781 (71) | NA |
| Anti-CCP positive | 1100 (66.5) | NA |
Clinical and demographic data were not available for all individuals. Percentages are given as a percentage of the data available.
Identification of high-risk combinations including SE
Statistical analysis of the whole dataset (6049 individuals) is shown in Table 2. Approximately 14% of the controls carried a combination of genotypes that conferred a relative risk of >5 and ∼10% carried >6 compared with carriage of no susceptibility variants. In total, eight combinations resulted in a relative risk of RA >6 compared with carriage of no susceptibility variants. All contained SE and all but one contained PTPN22.
Table 2.
Results for all 32 possible allele combinations
| Allele combination class | Combination | OR (95% CI) | Controls | Cases | Total |
|---|---|---|---|---|---|
| 1 | Negative for all | 54 | 42 | 96 | |
| 2 | TRAF only | 1.03 (0.66, 1.62) | 230 | 185 | 415 |
| 3 | STAT only | 1.31 (0.74, 2.34) | 45 | 46 | 91 |
| 4 | STAT+TRAF | 1.30 (0.82, 2.07) | 147 | 149 | 296 |
| 5 | 6q23 only | 2.01 (1.10, 3.66) | 32 | 50 | 82 |
| 6 | 6q23+TRAF | 1.25 (0.78, 1.99) | 136 | 132 | 268 |
| 7 | 6q23+STAT | 1.96 (0.99, 3.88) | 21 | 32 | 53 |
| 8 | 6q23+STAT+TRAF | 1.62 (1, 2.62) | 97 | 122 | 219 |
| 9 | PTPN22 only | 1.45 (0.66, 3.17) | 16 | 18 | 34 |
| 10 | PTPN22+TRAF | 1.29 (0.76, 2.19) | 64 | 64 | 128 |
| 11 | PTPN22+STAT | 3.64 (1.32, 10.04) | 6 | 17 | 23 |
| 12 | PTPN22+STAT+TRAF | 2 (1.11, 3.62) | 34 | 53 | 87 |
| 13 | PTPN22+6q23 | 1.93 (0.72, 5.15) | 8 | 12 | 20 |
| 14 | PTPN22+6q23+TRAF | 2.62 (1.43, 4.80) | 28 | 57 | 85 |
| 15a | PTPN22+6q23+STAT | 0.57 (0.16, 1.98) | 9 | 4 | 13 |
| 16 | PTPN22+6q23+STAT+TRAF | 3.51 (1.72, 7.19) | 15 | 41 | 56 |
| 17 | SE only | 3.75 (2.27, 6.18) | 59 | 172 | 231 |
| 18 | SE+TRAF1 | 3.92 (2.54, 6.05) | 200 | 610 | 810 |
| 19 | SE+STAT4 | 4.56 (2.59, 8.04) | 31 | 110 | 141 |
| 20 | SE+STAT+TRAF | 3.94 (2.52, 6.15) | 140 | 429 | 569 |
| 21 | SE+6q23 | 4.36 (2.49, 7.64) | 33 | 112 | 145 |
| 22 | SE+6q23+TRAF | 4.21 (2.70, 6.58) | 138 | 452 | 590 |
| 23 | SE+6q23+STAT | 5.98 (3.19, 11.22) | 20 | 93 | 113 |
| 24 | SE+6q23+STAT+TRAF | 5.23 (3.28, 8.33) | 89 | 362 | 451 |
| 25 | SE+PTPN22 | 9 (4.02, 20.16) | 9 | 63 | 72 |
| 26 | SE+PTPN22+TRAF | 6.91 (4.08, 11.69) | 40 | 215 | 255 |
| 27 | SE+PTPN22+STAT | 8.57 (3.32, 22.12) | 6 | 40 | 46 |
| 28 | SE+PTPN22+STAT+TRAF | 9.94 (5.45, 18.10) | 22 | 170 | 192 |
| 29 | SE+PTPN22+6q23 | 4.82 (2.27, 10.24) | 12 | 45 | 57 |
| 30 | SE+PTPN22+6q23+TRAF | 5.84 (3.41, 10) | 37 | 168 | 205 |
| 31 | SE+PTPN22+6q23+STAT | 6.06 (2.44, 15.06) | 7 | 33 | 40 |
| 32 | SE+PTPN22+6q23+STAT4+TRAF | 6.92 (3.87, 12.38) | 26 | 140 | 166 |
| Total | 1811 | 4238 | 6049 |
ORs are compared with base odds of no susceptibility loci. High-risk combinations are highlighted in bold. aAllele combination Class 15 has an OR of 0.57, suggesting a protective effect; however, this is due to the low sample numbers making the result unreliable (nine controls and four cases).
The highest risk was conferred by SE+PTPN22 +STAT4 + TRAF1/C5 (OR = 9.94; 95% CI 5.45, 18.10) followed by SE+PTPN22 (OR = 9, 95% CI 4.02, 20.16) alone. In order to calculate the risk of carriage of SE and PTPN22 compared with the rest of the population as opposed to just those subjects with carriage of no susceptibility alleles at any of the five loci, a further analysis was carried out comparing a group carrying SE+PTPN22 regardless of other susceptibility loci (1033 individuals), against a group negative for both SE and PTPN22 (1520 individuals), resulting in an OR of 5.53 (95% CI 4.54, 6.73).
Simplifying the model
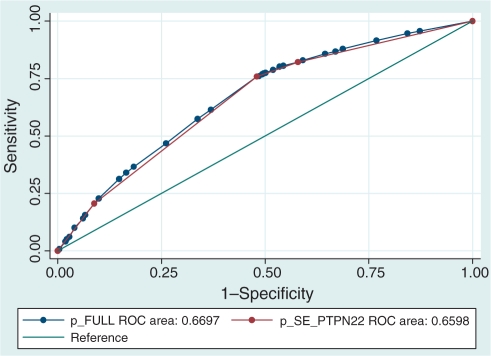
As both SE and PTPN22 were included in all but one of the high-risk combinations in Table 2, we first investigated whether these two loci alone would give as much information as including all five loci in the model. ROC curve analysis showed that the areas under the ROC curves were similar (0.67 full model, 0.66 SE+PTPN22 model) (Fig. 1), and the correlation coefficient between the two models was 0.98. However, the difference between the models was statistically significant (P = 0.0004); this arises due to the size of the study, meaning that trivial differences of no practical importance achieve statistical significance.
Fig. 1.
ROC curve analysis comparing the full model containing all five susceptibility loci (blue line) against the simpler model containing only SE and PTPN22 loci (red line).
Replacing DR*01/*04/*10 status for SE status
The analysis was repeated including broad HLA-DRB1 typing in place of full subtyping. For that analysis, carriage of any DR*01, DR*04 or DR*10 allele denoted an individual as SE positive. The results are shown in Table 3 and are very similar to the results including SE subtypes. ROC analysis showed that the area under the curve was similar to the full model (0.65), and the correlation coefficient was 0.93. Again, however, statistical comparison showed a significant difference between the areas under the two curves (P = 2 × 10−6).
Table 3.
Results for all 32 allele combinations generated by replacing SE with DRB1*1\4\10
| Allele combination class | Combination | OR (95% CI) | Controls | Cases | Total |
|---|---|---|---|---|---|
| 1 | Negative for all | 48 | 37 | 85 | |
| 2 | TRAF only | 1.09 (0.68, 1.75) | 206 | 173 | 379 |
| 3 | STAT only | 1.33 (0.71, 2.50) | 36 | 37 | 73 |
| 4 | STAT+TRAF | 1.27 (0.78, 2.07) | 133 | 130 | 263 |
| 5 | 6q23 only | 2.08 (1.11, 3.88) | 30 | 48 | 78 |
| 6 | 6q23+TRAF | 1.29 (0.78, 2.12) | 120 | 119 | 239 |
| 7 | 6q23+STAT | 2.05 (1, 4.20) | 19 | 30 | 49 |
| 8 | 6q23+STAT+TRAF | 1.72 (1.03, 2.87) | 86 | 114 | 200 |
| 9 | PTPN22 only | 1.60 (0.68, 3.73) | 13 | 16 | 29 |
| 10 | PTPN22+TRAF | 1.32 (0.75, 2.32) | 57 | 58 | 115 |
| 11 | PTPN22+STAT | 3.46 (1.23, 9.71) | 6 | 16 | 22 |
| 12 | PTPN22+STAT+TRAF | 2.24 (1.19, 4.19) | 29 | 50 | 79 |
| 13 | PTPN22+6q23 | 1.78 (0.65, 4.88) | 8 | 11 | 19 |
| 14 | PTPN22+6q23+TRAF | 2.65 (1.38, 5.07) | 24 | 49 | 73 |
| 15a | PTPN22+6q23+STAT | 0.58 (0.16, 2.02) | 9 | 4 | 13 |
| 16 | PTPN22+6q23+STAT+TRAF | 3.34 (1.57, 7.07) | 14 | 36 | 50 |
| 17 | DRB1*1\4\10 only | 3.53 (2.11, 5.91) | 65 | 177 | 242 |
| 18 | DRB1*1\4\10+TRAF1 | 3.60 (2.29, 5.68) | 224 | 622 | 846 |
| 19 | DRB1*1\4\10+STAT4 | 3.86 (2.21, 6.75) | 40 | 119 | 159 |
| 20 | DRB1*1\4\10+STAT+TRAF | 3.77 (2.37, 6.02) | 154 | 448 | 602 |
| 21 | DRB1*1\4\10+6q23 | 4.23 (2.38, 7.49) | 35 | 114 | 149 |
| 22 | DRB1*1\4\10+6q23+TRAF | 3.92 (2.46, 6.24) | 154 | 465 | 619 |
| 23 | DRB1*1\4\10+6q23+STAT | 5.60 (2.98, 10.54) | 22 | 95 | 117 |
| 24 | DRB1*1\4\10+6q23+STAT+TRAF | 4.80 (2.96, 7.78) | 100 | 370 | 470 |
| 25 | DRB1*1\4\10+PTPN22 | 7.03 (3.32, 14.88) | 12 | 65 | 77 |
| 26 | DRB1*1\4\10+PTPN22+TRAF | 6.10 (3.58, 10.38) | 47 | 221 | 268 |
| 27 | DRB1*1\4\10+PTPN22+STAT | 8.86 (3.40, 23.11) | 6 | 41 | 47 |
| 28 | DRB1*1\4\10+PTPN22+STAT+TRAF | 8.31 (4.61, 15) | 27 | 173 | 200 |
| 29 | DRB1*1\4\10+PTPN22+6q23 | 4.97 (2.31, 10.70) | 12 | 46 | 58 |
| 30 | DRB1*1\4\10+PTPN22+6q23+TRAF | 5.57 (3.22, 9.62) | 41 | 176 | 217 |
| 31 | DRB1*1\4\10+PTPN22+6q23+STAT | 6.12 (2.43, 15.37) | 7 | 33 | 40 |
| 32 | DRB1*1\4\10+PTPN22+6q23+STAT4+TRAF | 6.97 (3.85, 12.62) | 27 | 145 | 172 |
| Total | 1811 | 4238 | 6049 |
ORs are compared with base odds of no susceptibility loci. High-risk combinations are highlighted in bold. aAllele combination Class 15 shows a protective OR; however, this is due to the low sample numbers making the result unreliable.
An additional model containing DR*01/*04/*10 and PTPN22 was compared with the previous SE+PTPN22 model. ROC analysis showed that the area under the curve was similar to the SE+PTPN22 model (0.64), although it was slightly reduced by the exclusion of the other four loci. The correlation coefficient between the two models was 0.93 (data not shown).
Choice of cost-efficient screening model
In terms of general population screening test, the genetic factors included should be amenable to genotyping on medium-throughput platforms in a time- and cost-efficient manner. For this reason, we investigated a model containing DRB1*01/*04/*10 with the four other loci. DR*01/*04/*10 can be typed using PCR sizing-based methods; whereas, for many multiplex platforms, there would be little time or cost savings in genotyping fewer SNPs (i.e. there would be no benefit in genotyping just the PTPN22 locus rather than including the STAT, TRAF1/C5 and 6q23 loci as well). The combination of DR*01/*04/*10 + PTPN22 + STAT + 6q23 + TRAF has a sensitivity and specificity similar to the full model (Table 4), and the correlation coefficient for the predicted probabilities of the two models was 0.93. For population-based screening models, it is recommended that both sensitivity and specificity should exceed 80% [21]. Whereas all the models are specific (i.e. having a low risk of falsely identifying a subject as being at high risk), even the best model including SE and the remaining four risk variants lacks sensitivity and, consequently, individuals at risk of developing disease will be missclassified.
Table 4.
Sensitivity and specificity of screening models
| Model | Sensitivity, % | Specificity, % |
|---|---|---|
| Full | 31.4 | 85.2 |
| SE+PTPN22 | 20.6 | 91.2 |
| DRB1*1/4/10+four loci | 21.2 | 90.1 |
Probability thresholds were set at >0.8 for all models. The full model is based on carriage of: SE, plus PTPN22, STAT4, TRAF1 and 6q23 risk alleles; SE+PTPN22 model is based on carriage of: SE and PTPN22 risk allele; DRB*1\4\10 + four loci model is based on carriage of: DRB1*1\4\10, plus PTPN22, STAT4, TRAF1 and 6q23 risk alleles. See ‘Materials and Methods’ section for classification of carriage.
Stratification by gender
Analysis was repeated separately in male cases compared with male controls and in female RA cases and female controls. There was no statistically significant difference observed in the high-risk combinations identified (Supplementary Tables 1 and 2, available as supplementary data at Rheumatology Online).
Stratification by CCP status
Statistical analysis was repeated in individuals who were known to be anti-CCP positive (1100) and all controls (1811) (Supplementary Table 3, available as supplementary data at Rheumatology Online). Seventeen combinations resulted in a relative risk of >6 compared with carriage of no susceptibility variants. All but one high-risk combinations contained SE. The highest risk was conferred by SE+PTPN22 + STAT4 + TRAF1/C5 (OR = 21.39; 95% CI 8.47, 54) followed by SE+PTPN22 (OR = 18.86; 95% CI 6.24, 56.94).
Discussion
We have shown that carriage of combinations of confirmed RA susceptibility alleles can increase the risk of RA over and above any risk allele alone. In particular, genotyping only the SE and PTPN22 risk alleles was able to identify most individuals with an OR >6 of developing RA compared with carriage of no risk alleles. This is not surprising as these two loci confer the highest genetic risk for susceptibility to RA individually.
Identification of a number of high-risk combinations, present at an appreciable frequency in the control population, opens the debate about population screening to identify a group at high risk of developing RA. However, population screening using SE is not commercially viable due to the time required and the expense involved in subtyping for these susceptibility alleles. Indeed, companies offering a screening service for the risk of developing RA do not currently include SE among the loci tested. Excluding this locus significantly reduces the sensitivity and specificity of the model (ROC area 0.56) (Supplementary Figure 1, available as supplementary data at Rheumatology Online). For example, removal of SE information reduced the number of possible gene combinations from 32 to 16, and of those 16 gene combinations, the greatest risk was conferred by PTPN22 + STAT4 (OR 2.5). Hence, inclusion of the HLA-DRB1 status appears essential. However, our data suggest that DRB*01/*04/*10 broad typing can replace full HLA-DRB1 subtyping for SE with little loss in sensitivity and a gain in specificity; and a direct comparison of the OR shows little difference (SE only: OR 3.75; Table 2) (DRB*01/*04/*10 only: OR 3.53; Table 3).
There is increasing evidence that RA may be split into two genetically distinct subsets based on the presence of antibodies to CCPs. For example, although the SE confers the largest genetic association with RA, this association is restricted to anti-CCP-positive RA cases [22]. Therefore, subgroup analysis was carried out to see if risk prediction could be improved in this subset of individuals. Analysis of anti-CCP-positive individuals increased the number of high-risk combinations (OR >6) from 8 to 17; the highest relative risk was conferred by SE+PTPN22 + STAT4 + TRAF1/C5 (OR 21.39) compared with 9.94 in the original analysis. The area under the ROC curve was also increased from 0.67 to 0.71, showing an increase in sensitivity and specificity when all five susceptibility variants are genotyped in anti-CCP-positive individuals. Stratification by anti-CCP has shown that risk prediction is improved in this subset of individuals; however, some individuals will still be misclassified.
Ideally, it would also be important to test whether the models can accurately predict the risk of developing RA in a particular year. As the background risk of RA is at best 1 : 1000 per annum [1], genetic screening alone is unlikely to identify a group at very high risk. For example, the high-risk combinations identified in the current study increase risk compared with subjects not carrying any of the risk alleles and even then, an OR 6 still equates to a small absolute risk of developing RA. Although the model discriminates reasonably well between low-risk (OR <2) and high-risk (OR >5) groups (OR 5.12; 95% CI 4.32, 6.06), the absolute risk, calculated by combining all individuals at high risk (OR >5) and comparing them to the rest of the cohort results in an OR of 2.64 (95% CI 2.27, 3.07), suggesting a model based on these genetic loci is not sufficient to predict the risk. Furthermore, the sensitivity and specificity of the models using only genetic information are not sufficiently accurate to support population screening. An alternative approach would be to use genetic screening in a population that is already identified as being at higher risk of disease due to the presence of other predisposing factors; this is termed as genetic testing. Several environmental risk factors have been identified and confirmed in different populations. These include age, gender, family history, smoking history, obesity, a history of an adverse pregnancy outcome as well as other factors (reviewed in [23]). It will require further investigation to determine whether a two-stage screening process in which genetic testing was undertaken in individuals with environmental RA susceptibility factors could identify a group at high enough risk to recommend intervention. Furthermore, previous studies have found that specific SE allele combinations confer a higher risk than others [24]. It may be that these high-risk SE genotypes, combined with the other established RA risk loci, could be used to identify a group of individuals at very high risk of RA. Such individuals could be offered lifestyle advice, or be followed closely such that early intervention could be offered as soon as symptoms or signs of inflammatory arthritis became apparent. This is particularly important as there is mounting evidence that a ‘window of opportunity’ exists during which early diagnosis and treatment of RA can reduce the extent of joint damage, and could also help limit the involvement of extra articular tissues [25, 26].
Unfortunately, it was not possible to include environmental risk factors in the current analysis. As further susceptibility factors, both genetic and environmental, emerge from ongoing studies, the number of possible combinations increases exponentially. For example, the inclusion of another risk factor would increase the number of possible combinations to 64. Despite testing large numbers of cases and controls with information at the five susceptibility loci in the current study, there remained some genetic combinations for which results were unreliable, because of the insufficient numbers of subjects carried those combinations. Hence, to incorporate additional risk factors, it will be necessary to investigate combinations in even larger sample sizes so that accurate predictive models can be developed [27]. A limitation of this study is that the analysis has been confined to Caucasian individuals, meaning that the model could not be extended to a more ethnically diverse population. However, depending on the origins of the population tested, different susceptibility loci may need to be included within the model; for example, the PADI4 gene has reproducibly been associated with RA in a Japanese population but not in a Caucasian population [28].
Our analysis presents a thorough exploration of the viability of population screening for RA incorporating the five most widely confirmed susceptibility loci identified, to date, in large sample sizes. We conclude that population screening based on these genetic loci alone cannot currently be recommended. This is in line with several recent studies which have concluded that, even in diseases such as prostate cancer and breast cancer, with a higher prevalence in the population than RA, whole population screening using genetics alone does not provide the sensitivity and specificity required to accurately classify individuals at high risk [21, 29–31]. Further work to test whether genetic testing in patients with established risk factors including family history can be used to identify a group at very high risk of disease is required.
Although we have shown that genetic screening for RA is not currently viable, it may become so in the future; and this type of analysis represents a step en route to using genetics in a fully translational approach to inform clinical practice.

Supplementary data
Supplementary data are available at Rheumatology Online.
Acknowledgements
We thank the Arthritis Research Campaign for their support, ARC grant reference no. 17 552, and also acknowledge the use of data from the Wellcome Trust Case Control Consortium. We are also grateful to the NIHR Manchester Biomedical Research Centre for support.
Biologics in RA Control (BIRAC) consortium members are: Derbyshire Royal Infirmary, Derby (Dr L. J. Badcock, Dr C. M. Deighton, Dr S. C. O’Reilly, Dr M. R. Regan, Dr Snaith, Dr G. D. Summers and Dr R. A. Williams); Russells Hall Hospital, Dudley (Dr J. Delamere, Dr K. Douglas, Dr N. Erb, Prof. G. D. Kitas, Dr R. Klocke, Dr A. Pace and Dr A. Whallett); Glasgow Royal Infirmary and Gartnavel Hospital, Glasgow (Dr D. Porter, Dr J. Hunter, Dr M. M. Gordon, Dr M. Gupta, Prof. H. Capell, Prof. R. Sturrock, Prof. I. McInnes, Dr R. Madhok and Dr M. Field); Hope Hospital, Salford (Dr R. Cooper, Dr A. Herrick, Dr T. O’Neil, Prof. A. Jones and Dr R. Benitha); East Cheshire NHS Trust, Macclesfield (Dr A. Barton, Dr S. Knight and Prof. D. Symmons); Manchester Royal Infirmary, Manchester (Dr R. M. Bernstein, Dr I. N. Bruce, Dr K. Hyrich and Prof. A. Silman); Norfolk & Norwich University Hospital, Norfolk (Dr K. Gaffney, Prof. A. J. Macgregor, Dr. T. Marshall, Dr P. Merry and Prof. D. G. I. Scott); Poole General Hospital, Poole (Dr P. W. Thompson and Dr S. C. Richards); Queen Alexandra Hospital, Portsmouth (Dr R. G. Hull, Dr J. M. Ledingham, Dr F. Mccrae, Dr M. R. Shaban, Dr A. L. Thomas and Dr S. Young Min); St Helens Hospital, St Helens (Dr V. E. Abernethy, Dr J. K. Dawson and Dr M. Lynch); and Haywood Hospital, Stoke-On-Trent (Dr E. H. Carpenter, Dr P. T. Dawes, Dr C. Dowson, Dr A. Hassell, Prof. E. M. Hay, Dr S. Kamath, Dr J. Packham, Dr E. Roddy and Dr M. F. Shadforth).
Funding: The study was funded by an ARC grant (reference no. 17 552). Funding to pay the Open Access publication charges for this article was provided by the ARC.
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1.Symmons D, Turner G, Webb R, et al. The prevalence of rheumatoid arthritis in the United Kingdom: new estimates for a new century. Rheumatology. 2002;41:793–800. doi: 10.1093/rheumatology/41.7.793. [DOI] [PubMed] [Google Scholar]
- 2.Macgregor AJ, Snieder H, Rigby AS, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–7. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Stastny P. Mixed lymphocyte cultures in rheumatoid arthritis. J Clin Invest. 1976;57:1148–57. doi: 10.1172/JCI108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 5.Begovich AB, Carlton VEH, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–3. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis – a genome wide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton A, Thomson W, Ke X, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17:2274–9. doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurreeman FAS, Padyukov L, Marques RB, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4:e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zervou MI, Sidiropoulos P, Petraki E, et al. Association of a TRAF1 and a STAT4 gene polymorphism with increased risk for rheumatoid arthritis in a genetically homogeneous population. Hum Immunol. 2008;69:567–71. doi: 10.1016/j.humimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Barton A, Thomson W, Ke X, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17:2274–9. doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HS, Remmers EF, Le JM, Kastner DL, Bae S-C, Gregersen PK. Association of STAT4 with rheumatoid arthritis in a Korean population. Mol Med. 2007;13:455–60. doi: 10.2119/2007-00072.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomino-Morales RJ, Rojas-Villarraga A, Gonzalez CI, Ramirez G, Anaya JM, Martin J. STAT4 but not TRAF1/C5 variants influence the risk of developing rheumatoid arthritis and systemic lupus erythematosus in Colombians. Genes Immun. 2008;9:379–82. doi: 10.1038/gene.2008.30. [DOI] [PubMed] [Google Scholar]
- 16.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gisela O, Behrooz Z, Alizadeh, Angélica M, Delgado-Vega, et al. Association of STAT4 with rheumatoid arthritis: a replication study in three European populations. Arthritis Rheum. 2008;58:1974–80. doi: 10.1002/art.23549. [DOI] [PubMed] [Google Scholar]
- 18.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;13:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 19.Hinks A, Eyre S, Barton A, Thomson W, Worthington J. Investigation of genetic variation across the protein tyrosine phosphatase gene in patients with rheumatoid arthritis in the UK. Ann Rheum Dis. 2007;66:683–6. doi: 10.1136/ard.2006.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho PYPC, Barton A, Worthington J, et al. Investigating the role of the HLA-Cw*06 and HLA-DRB1 genes in susceptibility to psoriatic arthritis: comparison with psoriasis and undifferentiated inflammatory arthritis. Ann Rheum Dis. 2008;67:677–82. doi: 10.1136/ard.2007.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Annette HMvdHM, Tom WJ, Huizinga, de Vries Rene RP, Toes Rene EM. Emerging patterns of risk factor make-up enable subclassification of rheumatoid arthritis. Arthritis Rheum. 2007;56:1728–35. doi: 10.1002/art.22716. [DOI] [PubMed] [Google Scholar]
- 23.Oliver JE, Silman AJ. Risk factors for the development of rheumatoid arthritis. Scand J Rheumatol. 2006;35:169–74. doi: 10.1080/03009740600718080. [DOI] [PubMed] [Google Scholar]
- 24.Wordsworth P, Pile KD, Buckely JD, et al. HLA heterozygosity contributes to susceptibility to rheumatoid arthritis. Am J Hum Genet. 1992;51:585–91. [PMC free article] [PubMed] [Google Scholar]
- 25.Bukhari MAS, Wiles NJ, Lunt M, et al. Influence of disease modifying therapy on radiographic outcome in inflammatory polyarthritis at five years. Arthritis Rheum. 2003;48:46–53. doi: 10.1002/art.10727. [DOI] [PubMed] [Google Scholar]
- 26.Lard LR, Visser H, Speyer I, et al. Early versus delayed treatment in patients with recent-onset rheumatoid arthritis: comparison of two cohorts who received different treatment strategies. Am J Med. 2001;111:446–51. doi: 10.1016/s0002-9343(01)00872-5. [DOI] [PubMed] [Google Scholar]
- 27.Burton PR, Hansell AL, Fortier I, et al. Size matters: just how big is BIG?: quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2008;38:263–73. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burr ML, Naseem H, Hinks A, et al. PADI4 genotype is not associated with rheumatoid arthritis in a large UK Caucasian population. Ann Rheum Dis. 2008 doi: 10.1136/ard.2009.111294. Advance Access published Advanced May 25, 2009, doi:10.1136/ard.2009.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 30.Pharoah PDP, Antoniou AC, Easton DF, Ponder BAJ. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 31.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–19. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.