Abstract
Fusarium head blight is a devastating disease of cereal crops whose worldwide incidence is increasing and at present there is no satisfactory way of combating this pathogen or its associated toxins. There is a wide variety of trichothecene mycotoxins and they all contain a 12,13-epoxytrichothecene skeleton but differ in their substitutions. Indeed, there is considerable variation in the toxin profile across the numerous Fusarium species that has been ascribed to differences in the presence or absence of biosynthetic enzymes and their relative activity. This article addresses the source of differences in acetylation at the C15 position of the trichothecene molecule. Here, we present the in vitro structural and biochemical characterization of TRI3, a 15-O-trichothecene acetyltransferase isolated from F. sporotrichioides and the “in vivo” characterization of Δtri3 mutants of deoxynivalenol (DON) producing F. graminearum strains. A kinetic analysis shows that TRI3 is an efficient enzyme with the native substrate, 15-decalonectrin, but is inactive with either DON or nivalenol. The structure of TRI3 complexed with 15-decalonectrin provides an explanation for this specificity and shows that Tri3 and Tri101 (3-O-trichothecene acetyltransferase) are evolutionarily related. The active site residues are conserved across all sequences for TRI3 orthologs, suggesting that differences in acetylation at C15 are not due to differences in Tri3. The tri3 deletion mutant shows that acetylation at C15 is required for DON biosynthesis even though DON lacks a C15 acetyl group. The enzyme(s) responsible for deacetylation at the 15 position of the trichothecene mycotoxins have not been identified.
Keywords: Fusarium head blight, trichothecene mycotoxin, deoxynivalenol, T-2 toxin, Fusarium graminearum, Fusarium sporotrichioides, acetyltransferase, coenzyme A, BAHD superfamily
Introduction
Fusarium head blight (FHB) is a serious disease of cereal crops whose worldwide incidence is increasing and is a major factor limiting wheat production in many parts of the world.1 The disease is caused by several species of the fungus Fusarium which pose a dual threat: first by reducing the yield and quality of grain and secondly by contaminating the food grains with trichothecene mycotoxins. In 1998–2000, economic losses in the United States alone were estimated to exceed $ 2.7 billion2 which had devastating effects on farm communities.3 At present there is no satisfactory way of combating this pathogen or the associated toxins. This problem is accentuated by an incomplete understanding of the pathway and enzymes responsible for the biosynthesis of the trichothecene mycotoxins.
Trichothecene mycotoxins are sesquiterpene epoxide secondary metabolites that inhibit protein translation in eukaryotes and have several acute adverse effects in animals, including food refusal, diarrhea, and alimentary hemorrhaging.4 There is a wide variety of trichothecene mycotoxins, and they all contain a 12,13-epoxytrichothecene skeleton, but differ in their substitutions (see Fig. 1). Indeed, the substitution pattern on the core ring structure differs markedly between phylogenetically closely related Fusarium species where these alternative substitution patterns can have drastic effects on cytotoxicity. As much as 5 × 103 fold difference in LC50 has been reported between trichothecene variants.5 Studies have shown that acetylation at the C3 position of trichothecenes can decrease the phytotoxicity6,7 indicating the importance of acetylation. This study focuses on the structure and function of the acetyltransferase that is responsible for acetylation of the hydroxyl group at C15.
Figure 1.
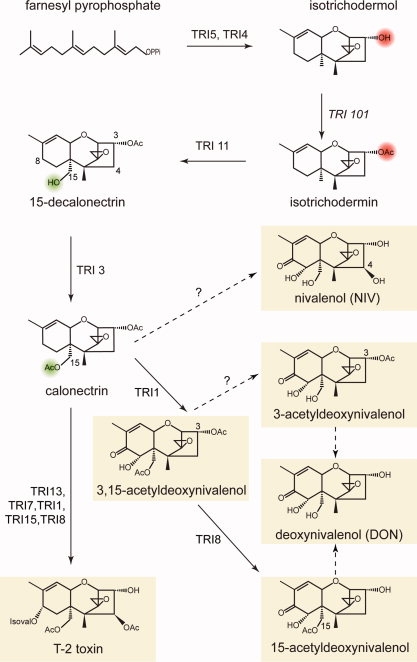
Abbreviated biosynthetic pathway for 15-acetyl deoxynivalenol and the structures of the common trichothecene myocotoxins. T-2 toxin is an A-type trichothecene with an ester-linked isovaleryl group at C8, whereas DON and nivalenol (NIV) are both B-type trichothecenes with a ketone moiety at C8 and are further differentiated by their substitution patterns at C4. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The biosynthetic pathway has been largely determined in the T-2 toxin producing strain, F. sporotrichioides through the use of genetic mutants and precursor feeding studies.8–11 The core trichothecene ring structure is formed from the cyclization of farnesyl pyrophosphate by trichodiene synthase encoded by Tri512 and subsequent multiple oxygenations by TRI4, yielding the toxic intermediate, isotrichodermol.13 The fungus then protects itself by acetylating the C3 hydroxyl group through the action of TRI101, which reduces the toxicity of trichothecene mycotoxins ∼ 100 fold.14 Thereafter, a complex series of coupled reactions lead to a final ensemble of toxins (see Fig. 1).
Modifications to the C4 and C15 positions are performed by the P450 monoxygenase/acetyltransferase pairs TRI13/TRI7 and TRI11/TRI3, respectively.9,15–18 C8 oxygenation is performed by TRI1,19–21 and this position is further modified by TRI16 to an isovaleryl group in T-2 toxin producers.22 The last step in the biosynthetic pathway involves removal of the protecting acetyl group at C3 by the action of TRI8.23
The toxins are often classified according to their substitution at C8 (see Fig. 1).24 Trichothecenes that carry an ester side chain at C8, or no side chain at all are classified as Type A, whereas a ketone functional group at this position defines the Type B toxins. T2-toxin and deoxynivalenol (DON) are examples of A-Type and B-Type toxins, respectively. Fusarium strains have, by extension, been classified into chemotypes on the basis of the toxins that they produce.25 The description and assignment of a Fusarium to a trichothecene chemotype in the literature has been complicated by the use of different growth substrates, such as sterile rice grains, sterile wheat grains, and liquid culture media, and different extraction methods.26–30 Furthermore, single Fusarium isolates often produce mixtures of acetylated and deacetylated trichothecenes, making their chemotype classification difficult. Given the importance of the toxin profile in the management of FHB, care should be taken when correlating a DNA sequence or genotype of the fungus with a chemical phenotype.31
It is generally assumed that the alternative substitution patterns on the core ring are determined by the presence or absence of functional copies of the biosynthetic genes in a Fusarium strain.18 For example, the basis for variations at C4 arise from differences in the coding sequence of Tri13. Nivalenol (NIV) types have a functional copy, whereas this gene is inactivated in DON producing strains. A switch from DON to NIV can be accomplished by heterologous expression of a functional TRI13 in a DON producing strain. NIV producers often produce mixtures of 4-acetyl-NIV and NIV and the inactivation of Tri7 will shift the chemotype to NIV only.18
The genetic and biochemical basis for differences of acetylation at C15 are less clear than for differences at C4. The predicted amino acid sequences for Tri3 from strains that produce 3-acetyl-deoxynivalenol (3ADON) and strains that produce 15-acetyl-deoxynivalenol (15ADON) are highly homologous, sharing greater than 90% sequence identity.27 Furthermore, there are no obvious mutations which would render these orthologs nonfunctional. Transcription of Tri3 was reported in a 3ADON producing strain (Fusarium graminearum F15), however, recombinant protein expressed in E. coli exhibited poor activity with DON.32 Several hypotheses for the reduced levels of C15 acetylation in DON and 3ADON chemotypes can be envisaged. First, the C15 position is deacetylated by an as yet unidentified enzyme analogous to the TRI101/TRI8 acetylation and deacetylation of C3, second, the Tri3 gene is transcribed but not translated, or third, the enzyme is produced but is nonfunctional. The last hypothesis of a transcribed but nonfunctional TRI3 in chemotypes that produce 3ADON would be similar to the control of modification at the C4 position.
The apparent dichotomy between the conserved sequence of TRI3 and its inactivity in similar strains of Fusarium is in contrast to the promiscuous behavior of TRI101 which is another acetyltransferase from the trichothecene biosynthetic pathway. In our previous work with TRI101, we demonstrated that this self protection enzyme from F. graminearum was equally effective at performing the 3-O-trichothecene acetyltransferase reaction with T-2, DON, or NIV as substrates. In contrast, the ortholog of TRI101 in F. sporotrichioides, which had been used in transgenic resistance strategies with wheat and barley, had a 70-fold reduced catalytic efficiency, kcat/Km, with DON compared to T-2 toxin.33 Differences in activity of TRI101 between T-2 toxin and DON chemotype producers correlate directly with variations in residues lining the active site. This raises the question of whether the differences in C15 chemotypes can be similarly ascribed to differences in the active site of TRI3. To address these questions a structural and functional analysis of TRI3 from F sporotrichioides was initiated.
Here, we present the in vitro structural and biochemical characterization of TRI3, a 15-O-trichothecene acetyltransferase isolated from F. sporotrichioides and the “in vivo” characterization of Δtri3 mutants of DON producing F. graminearum strains. The kinetic results indicate that TRI3 is an efficient enzyme with the native substrate, 15-decalonectrin, and is inactive with either DON or NIV. The structural studies provide an explanation for the specificity and reveal that Tri3 and Tri101 are likely evolutionarily related. Furthermore, the structural studies show that the residues that line the active site of Tri3 are strictly conserved, even in strains that do not appear to have C15 acetylation. Finally, biochemical characterization of the Tri3 deletion mutants reveal its gene product is essential for the biosynthesis of B type trichothecene products.
Results and Discussion
Tertiary and quaternary structure of TRI3
TRI3 was crystallized in space group P212121 with unit cell dimensions a = 64.28 Å, b = 81.56 Å, and c = 95.88 Å. The unit cell dimensions remained unchanged during heavy atom derivative soaks and freezing, thus allowing the apo-TRI3 structure to be determined by multiple isomorphous replacement (MIR) phasing with two heavy atom derivatives. One monomer is present in the asymmetric unit. Gel filtration analysis confirms that TRI3 is a monomer in solution (data not shown). Comparison of the apo structure with the binary complex of 15-decalonectrin bound to TRI3 reveals no major structural differences in the presence or absence of substrate (root mean square difference (rmsd) 0.26 Å for 463 Cα).
The structure of TRI3 is best described as a two domain protein whose N- and C-terminal domains associate to form a doughnut-shaped protein [Fig. 2(A)]. The active site lies in the doughnut hole formed at the interface between the two domains. The N-terminal domain consists of two mixed β-sheets with three and five strands each. The five-stranded β-sheet is built from four N-terminal domain strands (β-2,5,6,7) and one domain swapped strand (β12). Packed on both faces of the five stranded β-sheet are nine α-helices, eight are from the N-terminal domain (α-1,2,3,4,5,6,7,8), and the ninth helix is from the C terminal domain swapped loop (α17 + β12). The three-stranded β-sheet (β-1,3,4) is on the surface of the protein distal from the base of the active site. This sheet does not participate in the domain interface or ligand binding. The proposed catalytic histidine is located on the loop between β7 and α6 at the interface of the two domains.
Figure 2.
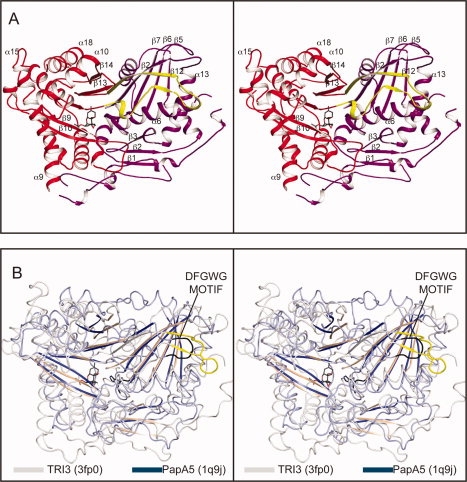
Structural representations of TRI3. (A) Stereoview of TRI3 complexed with 15-decalonectrin (PDB accession number 3fp0). The N- and C-terminal domains are colored magenta and red, respectively, and the domain swapped β-strand 12 is colored yellow. Bound ligand 15-decalonectrin is colored dark gray. (B) Stereo overlay of TRI3 (colored white) and vinorine synthase, PDB ID 2bgh, (colored light blue). The loop between residues Asp362 and Gly366 of vinorine synthase are colored black and the corresponding loop residues, Glu449-Ser461, of TRI3 are colored yellow.
The C-terminal domain contains a six-stranded mixed β-sheet (β-8,9,10,11,13,14) located at the interface. Six α-helices (α-10,11,13,14,15,18) are packed against the exterior face of this β-sheet. Three α-helices (α-9,12,16) form part of the domain interface and are packed between the C-terminal mixed β-sheet (β-9,10) and the N-terminal domain. A topology drawing is provided in Supporting Information Figure 1.
Comparison with BAHD family members
A search for structurally homologous proteins with the SSM server34 shows that the fold observed in TRI3 belongs to the BAHD superfamily. The closest structural relatives are: phthiocerol dimycocerosyl transferase from Mycobacterium tuberculosis (PapA5),35,36 vinorine synthase (VS),37 and TRI101 from F. graminearum33 which exhibit Q values of 0.29, 0.20, and 0.19, respectively. All of these proteins show very limited sequence similarity to TRI3. For example, VS shares only 10% sequence identity even though 306 structurally equivalent α-carbons superimpose with an rmsd of 3.1 Å. If only the secondary structure elements are considered, these structures align remarkably well with an rmsd of 2.6 Å for 145 stereochemically equivalent α-carbons. A superposition of TRI3 with VS is shown in Figure 2(B).
All enzymes of the BAHD family catalyze acyl transfer reactions and have two conserved sequence motifs: a catalytically important HX3D and a structurally important DFGWG motif.38 The HX3D motif is conserved in TRI3, where residue His186 superimposes closely with other BAHD family member catalytic histidine residues. In both the apo and 15-decalonectrin-bound TRI3 structures, the Asp190 residue of the HX3D motif is directed away from the substrate, forming a salt bridge with Arg342 and participating in helix capping. This structural role for the aspartate residue of the HX3D motif is consistent with previous structures of other BAHD enzymes.37,39
The DFGWG motif, which is believed to have a structural role in the BAHD family members,33,37 is not present in TRI3. In all other BAHD members this motif forms a turn between the domain swapped α17 helix and β12 strand. The recent structures for two trichothecene 3-O-acetyltransferases, FsTRI101 and FgTRI101, suggested that the fourth position of this DFGWG motif is variable and may only require a large hydrophobic residue for anchoring the turn. In TRI3, the residues which take the place of the DFGWG motif form a much larger loop extending from residues Glu449-Ser461. The structure of TRI3 demonstrates that this structural motif is not strictly required for membership within the BAHD family of enzymes.
Trichothecene binding site
The position of the active site was unequivocally identified by the location of the electron density for 15-decalonectrin. A molecule of 15-decalonectrin was unambiguously modeled into the Fo − Fc difference density [Fig. 3(A)]. The trichothecene binding site is located in the tunnel formed at the interface of the N- and C-terminal domains. Most of the interactions with the substrate are hydrophobic, including the binding of the core ring epoxide moiety into a cavity formed by residues of the N-terminal domain's α-1 helix, Ala36, and Val40 [Fig. 3(B)]. Carbon C8 is bound 4.5 Å from the Cγ of Val469.
Figure 3.
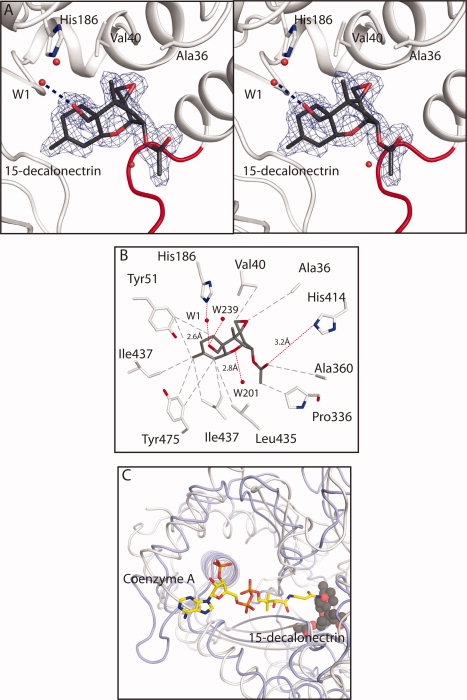
Details of the interaction of trichothecene intermediate 15-decalonectrin with TRI3. (A) Stereoview of the electron density corresponding to 15-decalonectrin (dark gray) bound to TRI3 (white). The map, contoured at 3σ was calculated from coefficients of the form Fo − Fc where the ligand was omitted from the phase calculation and refinement. Residues that interact with the epoxide moiety are depicted as sticks. Loop residues Lys424-Pro434, proposed to allow active site access are colored red. (B) Schematic representation of the interactions made by TRI3 with 15-decalonectrin. Polar and apolar interactions and distances in Å are indicated as red and gray dashes, respectively. Distance between the catalytic His186 and C15 Oxygen is not ideal for hydrogen bonding. (C) Coenzyme A binding site model. The proposed CoA binding site of TRI3 based on a structural superposition of the structure of FgTRI101 complexed with CoA and DON onto the structure of the TRI3·15-decalonectin complex is shown. TRI3 is colored white, FgTRI101 is colored light blue, CoA from the FgTRI101 structure is shown in stick representation, and 15-decalonectrin is depicted with spheres.
The C15 hydroxyl group which is acetylated by TRI3 is directed toward the proposed catalytic histidine's (His186) Nɛ, at a nonideal hydrogen bonding distance of 4.4 Å. However, there is a water molecule positioned between the catalytic histidine and the C15 hydroxyl group which forms a hydrogen bond with Nɛ of His186 but not with the toxin substrate. There are only two hydrogen bonds to 15-decalonectrin, one is between His414 and the C3 OAc carbonyl oxygen, and the other is between the core ring ether, O1, and a water molecule trapped in the active site. Of note, the C3 O-acetyl group is directed into a small depression within a larger pocket that has four water molecules trapped within it. This large cavity may be able to accommodate a large C3 moiety, or may provide the route of substrate access into the active site.
The toxin binding site is occluded from solvent in both the apo and 15-decalonectrin-bound structures, although the apo structure does not contain substrate, it appears to be maintained in the closed state because of a glycerol molecule in the active site. Indeed, in the present structures there is no obvious way that 15-decalonectrin can enter the active site without a conformational change. Both structures were determined with molecules in their active sites and in a closed conformation, where the toxin binding site is occluded from solvent. In the apo structure, a glycerol molecule from the cryoprotectant was observed bound in the active site. It was not possible to obtain substrate bound crystals with glycerol as the cryoprotectant. The substrate bound crystals were obtained by soaking substrate into apo crystals, requiring a structural rearrangement to either open the active site for binding or to close the active site following binding. The structural homology with other BAHD members predicts that loop Lys424-Pro434 is the loop that undergoes this rearrangement to provide access to the active site [Fig. 3(A)].
Attempts to soak acetyl-CoA or CoA into the active site were unsuccessful, however based on structural similarity with other BADH family members, it is expected that CoA will bind to TRI3 in a similar manner as other BAHD family members. In these structures acetyl-CoA binds in the active site tunnel on the opposite side of the catalytic histidine. A model of the complex between CoA and TRI3 and 15-decalonectrin was created based on a structural superposition of the tertiary complex of F. graminearum TRI101 with Coenzyme A and DON structure (PDB ID: 3b2s) as shown in Figure 3(C).
Kinetic studies
TRI3 has previously been shown to catalyze the acetylation of 15-decalonectrin at the trichothecene C15 hydroxyl position.9 However, the substrate specificity or catalytic efficiency of this enzyme with the native substrate, 15-decalonectrin, or other trichothecene intermediates has not been assessed. This kinetic analysis reveals that TRI3 is an efficient enzyme with catalytic parameters similar to TRI101 in the same pathway.33 Acetylation of C15 by this TRI3 ortholog with the final mycotoxins DON or NIV was not detectable in this assay. The kcat, Km, and kcat/Km kinetic constants determined for FsTRI3 with 15-decalonectrin are presented in Table I.
Table I.
Kinetic Constants, Km and kcat, for Two Acetyltransferases of the Trichothecene Biosynthetic Pathway
| Toxin | Km (μM) | kcat (s−1) | kcat/Km (M-1 s−1) | |
|---|---|---|---|---|
| FsTRI3 | 15-decalonectrin | 3.8 ± 0.1 | 7.2 ± 0.3 | 1.9 × 106 |
| FgTRI101a | DON | 11.7 ± 3.5 | 13.5 ± 2.1 | 1.2 × 106 |
| FgTRI101a | Isotrichodermol | 10.2 ± 3.5 | 411 ± 93 | 4.0 × 107 |
From Ref. 33.
Trichothecene binding comparisons between TRI3 and TRI101
TRI3 and TRI101 both are BAHD family members whose secondary structures and catalytic motifs align closely. Additionally, both enzymes catalyze the transfer of an acetyl group from acetyl coenzyme A to a trichothecene mycotoxin. The structure of FgTRI101 can be superimposed with TRI3 with an rmsd of 2.6 Å for 145 α-carbons of secondary structure. That both enzymes are in the same pathway and have strong structural and functional homology suggests an evolutionary relationship where the catalytic mechanism has been retained following a gene duplication event. Maintaining the catalytic mechanism freed the redundant gene to evolve new substrate specificity.
It is not possible to determine which gene, Tri3 or Tri101, was the primordial gene and which evolved the new specificity for binding the different trichothecenes in very different orientations. The hydroxyl moiety at different positions of the core ring structure must be positioned proximally to the conserved catalytic histidine. In TRI3 the C15 hydroxyl group is not positioned within hydrogen bonding distance to the catalytic histidine. A water molecule observed in the high resolution 15-decalonectrin-bound structure is positioned such that it could be activated by His186 for proton abstraction of the C15 hydroxyl [Fig. 3(A)]. This structure suggests that the TRI3 mechanism may be distinct from other BAHD family members, using an activated water for the proton abstraction.
An additional requirement for substrate specificity in this pathway is to keep the epoxide moiety in a nonreactive environment. In TRI3 and TRI101 the reactive epoxide moiety is bound into a hydrophobic pocket. However, the pocket is formed by residues from the N- and C-terminal domains in TRI3 and TRI101, respectively [Fig. 4(A,B)]. In TRI101 the epoxide is bound into a hydrophobic cavity formed by the residues of the C terminal domain's central βsheet (β9 + β11). The 15-decalonectrin bound TRI3 structure reveals the epoxide inserted into a shallow pocket formed by residues of the N-terminal domain's α1 helix. Given the reactive nature of epoxides, it is surprising that during divergent evolution the substrate was able to rotate 180° in the active site and a new epoxide binding pocket was formed.
Figure 4.
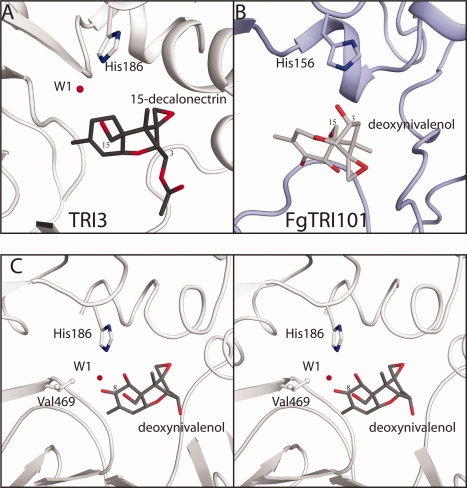
Comparison of trichothecene binding orientations in the structurally homologous TRI3 and FgTRI101. (A) TRI3 complexed with 15-decalonectrin. The 15-decalonectrin epoxide moiety is oriented into a hydrophobic depression formed by residues of the N-terminal domain, this requires a substantial reorientation of the substrate as compared to the TRI101 structures. In this structure the catalytic His186 does not form a hydrogen bond with the C15 hydroxyl moiety, but does form a hydrogen bond with a trapped water molecule. (B) FgTRI101 complexed with CoA and DON (3B2S). The DON substrate's epoxide moiety is oriented toward the C-terminal domain and the catalytic His156 is within ideal hydrogen bonding distance. (C) Proposed model of TRI3 complexed with DON. DON has been superimposed onto the core trichothecene ring structure of the TRI3·15-decalonectrin complex. This binding position of DON in the active site would position the C8 ketone moiety in close proximity to the Val469 side chain, thus creating a poor interaction that accounts for the lack of catalytic activity observed with DON as a substrate.
Role of TRI3 in determining chemotype
A hypothesis that Tri3 is transcribed but nonfunctional in 3ADON chemotypes was suggested by the poor C15-O-acetyltransferase activity of recombinantly expressed TRI3 from a 3ADON producer with the final mycotoxin, DON.32 The crystal structure of TRI3 with 15-decalonectrin bound provides a structural basis for interpreting these earlier results. The structure of TRI3 indicates that binding of trichothecene mycotoxins differentiated to A or B-type by the activity of TRI1 would form an unfavorable interaction with the C8 moiety. Modeling of DON into the 15-decalonectrin position shows that the C8 isovaleryl or ketone moieties of A or B type trichothecenes, respectively, would be bound into a hydrophobic environment with close proximity to Val469 [Fig. 4(C)]. This unfavorable interaction is proposed to be the basis for the lack of C15 acetylation activity that we and others have observed with DON and NIV.
While Tri3 genomic polymorphisms correlate with the B type trichothecene mixtures, these variations do not lie in the substrate binding pockets. A mapping of the sequence conservation of a ClustalW alignment from available TRI3 amino acid sequences onto the TRI3 structure is presented in Figure 5. This figure reveals that the trichothecene and predicted acetyl-coenzyme A binding site residues are highly conserved while the variable residues are located on surface exposed loops. The strict conservation of active site architecture suggests that both and A and B type trichothecene producing strains would be expected to possess a functional TRI3, in contrast to the earlier hypothesis based upon a lack of activity with a final mycotoxin. To examine whether TRI3 activity is essential in the biosynthesis of B-type trichothecenes, Tri3 was deleted in a B-type trichothecene producer and the toxin profiles analyzed.
Figure 5.

Sequence conservation mapped onto a stereo representation of TRI3. TRI3 (white ribbon) with bound 15-decalonectrin (colored black) and CoA (colored gray) modeled in as described above. Residues with a sequence conservation score derived from the T-coffee ClustalW alignment of TRI3 homologs from different chemotype Fusarium strains are shown as spheres colored purple = 1–4 (low), teal = 5 (mid), light blue = 6–8 (high).40
Effects of Δtri3 in F. graminearum
The structural analysis of TRI3 suggested that this enzyme would be functional in DON producing Fusarium strains, unfortunately attempts to obtain recombinant TRI3 from F. graminearum PH-1 were unsuccessful (the protein was expressed in an unfolded form). To confirm the role of the Tri3 gene product, “in vivo” deletion mutants of F. graminearum strain GZ3639 were generated and their trichothecene mycotoxin profiles assayed (see Fig. 6). In 2-stage liquid cultures, the wild-type strain produces primarily 15-ADON and a small amount of 3,15-diADON. Tri3 gene deletion mutants produced 15-decalonectrin and 3,15-didecalonectrin. The presence of both 15-decalonectrin and 3,15-didecalonectrin reflects the activity of TRI8 which removes the C3 acetyl moiety. Tri8 is the last step in the formation of 15ADON from 3,15-diADON but it acts on a variety of substrates.23 While a small amount of 15-ADON was detected in several of the Tri3 transformants, this was less than 5% of wild-type production and no 3-ADON or DON was detected in the liquid cultures.
Figure 6.
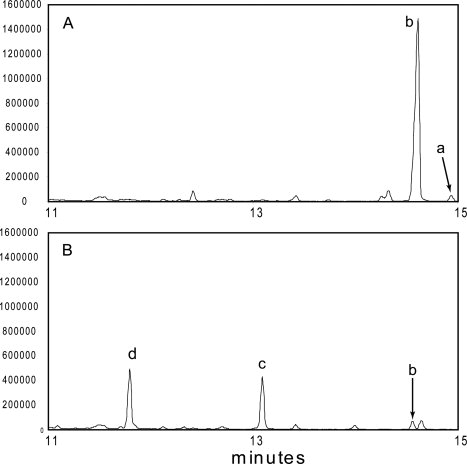
GC-MS metabolic profiles of (A) Fusarium graminearum wild-type strain GZ3639 and (B) FgTri3#228, a transformant with a disrupted Tri3. Y-axis indicates relative abundance. Retention times are (a) 3,15-diacetyldeoxynivalenol (14.9 min), (b) 15-acetyldeoxynivalenol (14.6 min), (c) 15-decalonectrin (13.1 min), and (d) 3,15-didecalonectrin (11.8 min).
The lack of TRI3 in both A and B type trichothecene producers results in the accumulation of 15-decalonectrin and 3,15-didecalonectrin demonstrating that the enzymes downstream of TRI3 in the biosynthetic pathway are reduced in their ability to modify the core ring structure without C15 acetylation (this study and Ref. 9). The genetic, structural, and kinetic data presented here suggest that the biosynthetic pathways to produce B type trichothecene mycotoxins require the C15 acetylation activity of TRI3 and this acetyl group is therefore removed later in the pathway. This implies that an unidentified deacetylase from the producing Fusarium strain or from the host organism is responsible for the lack of acetylation at C15 in the final mycotoxins.
The trichothecene mycotoxins have been identified as an important virulence factor in Fusarium infections of wheat and barley. Currently only Tri5 disruption mutants, blocked at the start of the biosynthetic pathway, have been shown to be severely impaired in their ability to spread between florets and induce disease. Although the analysis of virulence of Tri3 disruption mutants is incomplete, these strains are reduced in their virulence (unpublished data). The Δtri3 mutants produce trichothecenes with a 12,13-epoxy group but the specific compounds are not very phytotoxic.6,7 The reduced virulence of Δtri3 strains and essential role in biosynthesis of trichothecene mycotoxins establishes TRI3 as an additional target for combating FHB.
Materials and Methods
Plasmid and strain construction
All cloning was done in E. coli strain Top10. Primers were designed to amplify the F. sporotrichioides TRI3 gene (accession # AAK33072) using TRI3_forward primer 5′ GCTAGCATGAGCGCTT CATCCTCCT CCGCTTTACC3′ and TRI3_reverse primer 5′ gCTgAgCCTAAAGTCGGAAGGCAAGCA TGAACTCGACTATACTCTGC3′ from genomic DNA of F. sporotrichioides strain NRRL 3299 (a gift from Dr. Jaechuk Yu, UW Madison). Product bands were gel extracted using QIAquick® gel extraction kit (Qiagen), ligated into Zero Blunt Topo (Invitrogen) and then sequenced using M13F and M13R(−27) primers. Sequencing reactions were prepared using nonradioactive BigDye® protocols (ABI PRISM) and resolved at the UW-Madison Biotechnology Center. A single mutation from the deposited sequence of AAK33072 was observed, A291D. Introns were removed by overlap PCR extension.41 The exons were amplified with the following primer pairs: TRI3_forward + Intron_1R 5′GGTTGAAATTTCATACCCCGGCTTTTATAATTGGCTACAAGAGC3′, Intron_1F 5′GC TCTTGTAGCCAATTATAAAAGCCGGGGTATGAAATTTCAACC3′ + Intron_2R 5′CCC TTCGAACGGAATTGGTTTTGCGTGCAGATAGCTCGCC3′, Intron_2F 5′GGCGAGCTAT CTGCACGCAAAACCAATTCCGTTCGAAGGG3′ + Intron_3R 5′GCCATCACTGATGAA CAATGGGTTTGCTTCCCCTTCG3′, Intron_3F 5′CGAAGGGGAAGCAAACCCA TTGTTCATCAGTGATGGC3′ + Intron_4R 5′CCAGCTGTCCAGTCGGATTGCCAGATATGGCAAAGACTGG3′, Intron_4F 5′CCAG TCTTTGCCATATCTGGCAATCCGACTGGACAGCTGG3′ + TRI3_Reverse. The amplified exons were gel purified as described above and then joined by fusion PCR using TRI3_forward and TRI3_reverse primers. The validity of the gene was verified by sequencing.
Plasmid TRI3/pKLD37
The “intron-less” TRI3 allele was then excised from the Topo vector with restriction enzymes NheI and BlpI (New England Biolabs), and ligated with T4 DNA ligase (Fermentas) into a modified pET31b (Novagen) vector cut with NheI and BlpI. The pET31b vector was previously modified to contain an N terminal His6 tag followed by a TEV protease cut site two amino acids upstream of the gene start. The N-terminal amino acid sequence of this construct is MSYYHHHHHHDYDIPTSELYFQGASM1S2.… where the location of the TEV protease cleavage site is underlined.42 The validity of the final expression construct was verified by sequencing.
Protein expression and purification
Native TRI3 protein was overproduced utilizing plasmid TRI3/pKLD37 transformed into E. coli strain HMS174(λDE3). A starter culture from a single colony was grown overnight at 37°C in M9 minimal medium supplemented with ampicillin (0.2 μg/mL). The following day, 10 mL of the starter culture was used to inoculate 650 mL M9 minimal media + ampicillin in a 2-L shaker flask. Cultures were grown at 37°C until they reached an optical density (OD600) of ∼ 1.2. Cultures were transferred to a 16°C incubator and allowed to equilibrate for 30 min. After equilibration, CoA (Sigma) was added to 15 μM, TRI3 expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM. After a 20-h incubation period with IPTG under aerobic conditions, cells were harvested by centrifugation at 5000g, flash frozen in liquid nitrogen, and stored at −80°C until used.
TRI3 protein was purified from 30 g of cells thawed and resuspended in 210 mL of lysis buffer, which contained 0.5 mg/mL Lysozyme (Sigma), 50 mM NaH2PO4/K2HPO4, 300 mM NaCl, 20 mM Imidazole, 5 mM 2-mercaptoethanol, 1 tablet/50 mL of Complete Inhibitor (Roche), at pH 8.0. This mixture was incubated with stirring at 4°C for 1 h and then subjected to three rounds of sonication, each round of sonication continued until the temperature reached 10°C and then was left on ice until the temperature returned to 4°C. Cellular debris was removed by centrifugation at 40,000g for 30 min. The supernatant was loaded onto a 10-mL column of nickel-nitrilotriacetic acid-agarose (Qiagen) previously equilibrated with lysis buffer. The column was washed with lysis buffer until the A280 of the outflow reached background level. TRI3 protein was eluted with a linear gradient of 20–300 mM imidazole in lysis buffer. Fractions containing TRI3 protein were identified with SDS-PAGE43 and Coomassie Blue stain 3 and were pooled and dialyzed against 10 mMN-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), 250 mM NaCl, 0.5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 2 mM ethylenediaminetetraacetic acid, pH 7.5 buffer 4°C. The N-terminal hexahistidine (H6) tag was removed by treatment with recombinant tobacco etch virus (rTEV) protease.44 The remaining uncleaved TRI3 protein enzyme and rTEV protease were removed from the mixture with nickel-nitrilotriacetic acid-agarose affinity chromatography. Tagless TRI3 protein was concentrated to ∼ 20 mg/mL in a centriprep YM30 (Millipore) concentrator. Concentrated protein was dialyzed against a buffer containing 20 mM HEPES, 20 mM NaCl, 0.5 mM TCEP (pH 7.5 at 4°C).
Crystallization and structural determination of TRI3
A search for crystallization conditions was conducted at 4°C via the hanging drop method of vapor diffusion utilizing an “in-house” designed sparse matrix screen composed of 144 conditions. The crystals for 13 mg/mL enzyme were observed from hanging drop experiments with precipitant solutions of 2M ammonium sulfate buffered with HEPES buffer (100 mM, pH 7.5 at 25°C). Single crystals of dimensions 0.1 × 0.1 × 0.01 μm, were transferred stepwise to 1M ammonium sulfate, 0.1M HEPES, pH 7.5 at 25°C solution until the crystal edges began to visibly degrade. Each crystal was then transferred to a hanging drop equilibrated for 48 h prepared by mixing equal volumes of 13 mg/mL TRI3 and precipitant solution of 2M ammonium sulfate, 0.1M HEPES, pH 7.5 at 25°C. Wild type apo crystals of dimensions 0.6 × 0.25 × 0.05 μm were transferred stepwise into a cryoprotectant solution of 20% glycerol, 2M ammonium sulfate, 50 mM NaCl, 0.1M HEPES, pH 7.5 at 25°C for 24 h, then flash frozen directly into liquid nitrogen.
A mercury heavy atom derivative was prepared by soaking an apo crystal in 25 mM CH3HgCl2, 2M ammonium sulfate, 50 mM NaCl, 0.1M HEPES, pH 7.5 at 25°C for 75 min. The crystal was then transferred stepwise every minute into a 20% Glycerol, 1.6M ammonium sulfate, 50 mM NaCl, 0.1M HEPES, pH 7.5 at 25°C, then flash frozen directly into liquid nitrogen.
A platinum heavy atom derivative was prepared by soaking an apo crystal in 20 mM K2PtCl4, 2M ammonium sulfate, 50 mM NaCl, 0.1M HEPES, pH 7.5 at 25°C for 75 min. The crystal was then transferred stepwise every minute into a 20% Glycerol, 1.6M ammonium sulfate, 50 mM NaCl, 0.1M HEPES, pH 7.5 at 25°C, then flash frozen directly into liquid nitrogen.
X-ray data were collected from crystals of native and heavy atom derivatives on a CCD detector at SBC Beamline 19-BM (Advanced Photon Source, Argonne National Laboratory, Argonne, IL) The X-ray data were processed and scaled with HKL2000.45 X-ray data collection statistics are presented in Table II.
Table II.
Crystallographic Statistics
| Data collection | TRI3 apo | TRI3·15-decalonectrin |
|---|---|---|
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å, °) | a = 64.1 | a = 64.3 |
| b = 81.1 | b = 82.0 | |
| c = 95.3 | c = 95.4 | |
| α = β = γ = 90 | α = β = γ = 90 | |
| Wavelength | 0.97907 | 1.54 |
| Resolution range (Å) | 62–1.75 | 62–1.9 (2) |
| Reflections: measured | 272,276 | 202,367 |
| Reflections: uniquea | 44,076 (3,935) | 40,098 (5,384) |
| Redundancy | 3.3 (2) | 4.96 (2.35) |
| Completeness (%) | 98.2 (86.3) | 98.3 (90) |
| Average I/σ | 20.8 (3.16) | 10.7 (1.9) |
| Rsym (%)b | 4.8 (16.8) | 9.2 (37.3) |
| Rwork (%)c | 17.7 (21.3) | 18.0 (24.1) |
| Refinement | ||
| Rfree (%) | 22.2 (25.7) | 23.5 (30) |
| No. of protein atoms | 3,976 | 3,986 |
| No. of water molecules | 415 | 379 |
| Wilson B-value (Å2) | 16.9 | 15.0 |
| Average B factors (Å2) | ||
| Protein | 15.8 for 3,976 atoms | 14.2 for 3,986 atoms |
| Ligand | 20.5 for 52 atoms | |
| Solvent | 25.5 for 415 atoms | 21.1 for 379 atoms |
| Ramachandran (%) | ||
| Most favored | 94.1 | 92.6 |
| Additionally allowed | 5.5 | 6.7 |
| Generously allowed | 0.4 | 0.7 |
| Disallowed | 0 | 0 |
| rms deviations | ||
| Bond lengths (Å) | 0.012 | 0.016 |
| Bond angles (°) | 1.45 | 1.654 |
| Chiral | 0.095 | 0.116 |
| Disordered residues in loops | Asp150, Ser212, Lys323-Asp326 | Ser212, Pro324-Asp326 |
Data in parentheses represent highest resolution shell.
Rsym = ∑|I(hkl) – I|/ ∑ |I(hkl)|, where the average intensity I is taken over all symmetry equivalent measurements and I(hkl) is the measured intensity for a given reflection.
Rfactor = ∑|F(obs) – F(calc)|/∑ |F(obs)|, where Rwork refers to the Rfactor for the data utilized in the refinement and Rfree refers to the Rfactor for 5% of the data that were excluded from the refinement.
The structure of F. sporotrichioides TRI3 was solved via MIR phasing with crystals of the mercury and platinum derivatives. The software package SOLVE was utilized to determine the positions of three Hg and two Pt atoms in the asymmetric unit and to generate initial protein phases (figure of merit = 0.46).46 Solvent flattening with RESOLVE (figure of merit = 0.73) resulted in an interpretable electron density map calculated to 2.2 Å resolution.47 The initial map allowed for 295 out of 519 amino acid residues in the asymmetric unit to be modeled. This incomplete model was then used as the starting model for ARP/wARP48 which traced 491 residues into the electron density map for the native apo data set.
Alternate cycles of manual model building and least squares refinement with the programs COOT49 and Refmac50 reduced the R-factor to 17.5% for all X-ray data from 50–1.75 Å resolution. Refinement statistics are presented in Table II. In this model there are three breaks in the polypeptide chain between Asn149 and Asn151, Arg211 and Asp213, Leu322 and Leu327, the N terminus is disordered to residue Leu9. The Ramachandran plot as calculated by PROCHECK51 has no residues in the disallowed regions, 94.1% in the most favored, 5.5% in the additionally allowed, and 0.4% in the generously allowed region.
Structural determination of the TRI3·15-decalonectrin complex
Crystals of the complex of TRI3 with 15-decalonectrin mycotoxin were prepared by soaking apo crystals in 500 μM 15-decalonectirn, 2M ammonium sulfate, 25% sucrose, 0.1M HEPES, pH 7.5 at 25°C for 3 h. The crystal was then flash frozen directly into liquid nitrogen. X-ray data were collected with a Bruker AXS Platinum 135 CCD detector controlled with the PROTEUM software suite (Bruker AXS, Madison, WI). The X-ray source was CuKα radiation from a Rigaku RU200 X-ray generator equipped with Montel optics, operated at 50 kV and 90 mA. The X-ray data were processed with SAINT version 7.06 A (Bruker AXS) and internally scaled with SADABS version 2005/1 (Bruker AXS). X-ray data collection statistics are presented in Table II.
The structure of TRI3 complexed to 15-decalonectrin was solved by molecular replacement with the program MOLREP52 starting from the apo native TRI3 model. Alternate cycles of manual model building and least squares refinement with the programs COOT49 and Refmac50 reduced the Rfactor to 18% for all X-ray data from 50–1.9 Å resolution. Refinement statistics are presented in Table II. In this model there are three breaks in the polypeptide chain between Arg211 and Asp213, Lys323 and Leu327, the N terminus is disordered to residue Ser5. The Ramachandran plot as calculated by PROCHECK51 has no residues in the disallowed regions, 92.6% in the most favored, 6.7% in the additionally allowed, and 0.7% in the generously allowed region.
Acetyltransferase enzymatic assay
The trichothecene 15-O-acetyltransferase reaction catalyzed by TRI3 was monitored by following the production of CoA in a 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) coupled continuous assay (λ = 14,150 cm−1 M−1).53 Reaction mixtures were prepared at 25°C by combining 50 μL of 1.5 mM acetyl CoA (Sigma), 0.6 mM DTNB (Sigma), 0.1M potassium phosphate buffer, pH 8, with 50 μL of trichothecene toxin, 4.5% DMSO, 0.1M potassium phosphate buffer, pH 8.0. The reaction was initiated by the addition of 50 μL of 332 ng/mL enzyme, 200 μg/mL BSA, 0.1M potassium phosphate buffer, pH 8. After mixing, 100 μL of the reaction mixture was transferred to a 1 cm pathlength cuvette and the change in absorbance at 412 nm was followed. The rates of reaction were determined at various concentrations of 15-decalonectrin, NIV, and DON and then fit by nonlinear regression to the Michaelis-Menten equation.
Construction of ΔTri3 deletion strain
FgTri3 disruption was made using a gene replacement strategy by amplifying genomic GZ3639 (available as FGSC 8630 from the Fungal Genetics Stock Center, Department of Microbiology, University of Kansas Medical Center, Kansas City) DNA using primers specific to sequences prior to the 5′ translation start site (5′-CTATGGCGCCGCATCACC-3′ paired with 5′-CGCGTTAACGCTCAGAAGATACTTGGC-3′) and primers specific to sequences beyond the 3′ translation stop site (5′-CGCGTTAACGGCAACACTTTGGGTGG-3′ paired with 5′-GCTGAGATGTACCTCGCC-3′). The amplified fragments had a HpaI site incorporated (underlined sequence). Each segment of DNA was cloned separately into pCR2.1 (Invitrogen, Carlsbad, CA) and then cut with HpaI. The plasmid containing the 3′ fragment was further digested with NotI and this fragment was inserted into the HpaI-cut 5′-segment vector. The resulting vector was then cut with HpaI and the hygB (hygromycin) selectable marker was inserted. This final vector contained ∼ 975 bp of sequence upstream of the FgTri3 start site, followed by 2.5 bp of hygB, followed by 1070 bp of sequence downstream of the translational stop site of FgTri3.
Fungal transformations of GZ3639 protoplasts were carried out as described previously.54 All transformants were single-spored to ensure purity, tested by PCR to confirm disruption of FgTri3, and tested by Southern analysis55 to further confirm gene disruption.
Trichothecene toxin analysis
Trichothecenes were produced in liquid culture with a two-stage media protocol, (modified from Ref. 56). Strains were maintained in glycerol stocks. Cultures were grown initially on V-8 juice agar plates under alternating 12 h, 25°C-12 h 22°C dark cycle. Mycelia were washed from V8 plates with 3.5 mL water and used to inoculate 50 mL of 1st stage media (3 g NH4Cl, 2 g MgSO4-7H20, 0.2 g FeSO4-7H2O, 2 g KH2PO4, 2 g peptone, 2 g yeast extract, 2 g malt extract, 20 g glucose in 1 L distilled water) in 250 mL flasks. The cultures were grown at 28°C on a rotary shaker at 200 rpm in the dark for 3 days. The culture was then transferred to a 250 mL beaker and dispersed with a Toastmaster stick blender. The macerated culture was transferred to a 50-mL conical tube and centrifuged 5 min at 1600 rpm. Half of the supernatant was removed and the remaining fungal mass and medium was mixed well. Second stage cultures were initiated by adding 1.5 mL of the concentrated culture to 20 mL of 2nd stage medium (1 g (NH4)2HPO4, 3 g KH2PO4, 0.2 g MgSO4-7H2O, 5 g NaCl, 50 g sucrose, 10 g glycerol in 1 L distilled water) in a 50 mL flask. After 4 days, 5 mL aliquots were removed from each culture and extracted with 2 mL ethyl acetate. The extract was analyzed by GC-MS.
GC-MS measurements were made with a Hewlett-Packard 5890 Gas Chromatograph fitted with a 30 m fused silica capillary column (HP-5MS; 0.25 μm; J&W Scientific, Palo Alto, CA). The column was held at 120°C at injection; then heated to 210°C at 15°C/min and held for 1 min; then heated to 260°C at 5°C/min and held for 3 min. Trichothecenes were identified by comparison of retention times and mass spectral fragmentation patterns with authentic standards.
Conclusions
The ensemble of toxins synthesized by a particular Fusarium species is an important biological characteristic that is generally assumed to be controlled by the genetic make up of the fungus responsible for the toxin biosynthesis. The study here reveals that the DNA polymorphisms of TRI3 which are predictive of C15 chemotype in trichothecene producers do not map to residues lining the enzyme's active site, which suggests that, if expressed, all TRI3 orthologs should be capable of acetylating 15-calonectrin. Furthermore, deletion mutants of Tri3 in F. graminearum are blocked in their trichothecene biosynthesis and accumulate the pathway intermediates 15-decalonectrin and 3,15-didecalonectrin. This implies that acetylation at the 15 position is an obligate step in mycotoxin biosynthesis. Since the majority of the final mycotoxins generated by F. graminearum grown on cereal substrates are not acetylated at the 15 position there must be another activity responsible for the removal of the 15-acetyl moiety. The enzyme(s) responsible for removal of the acetyl group from the 15 position of the trichothecene mycotoxins have yet to be identified.
The structure of TRI3 with bound native substrate, 15-decalonectrin, reveals a high structural homology with another acetyltransferase in the same biosynthetic pathway, TRI101. The bound structures of TRI3 and TRI101 reveal an evolutionary relationship between enzymes within the trichothecene mycotoxin biosynthetic pathway where the catalytic mechanism has been maintained and substrate specificity has evolved. This has led to a restructuring of the active sites to bind the core trichothecene ring in two different orientations, binding the reactive epoxide moiety into the N-terminal domain in TRI3 and into the C-terminal domain in TRI101. In TRI3 this new binding orientation places the C8 ring position in close proximity to a hydrophobic patch of the active site, thereby preventing substrate promiscuity with the later products of the biosynthetic pathway that are modified at C8. The biochemical, structural, and kinetic data presented in this study identify TRI3 as a potential target in combating FHB.
Coordinates
The atomic coordinates and structure factors for the apo TRI3 and TRI3·15-decalonectrin complex have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/) with accession numbers 3FOT and 3FP0, respectively.
Acknowledgments
The authors thank Kirsten Dennison for creating the modified pET vector utilized in construction of the over expression plasmid for TRI3 and Dr. Martin St. Maurice for input to the kinetic assay design. This is a cooperative project with the U.S. Wheat and Barley Scab Initiative. SPM and NJA are supported by the U.S. Department of Agriculture NP 108 Food Safety. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the U.S. Department of Agriculture. Use of the Structural Biology BM19 beamline Argonne National Laboratory Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
Glossary
Abbreviations:
- 3ADON
3-acetyl-deoxynivalenol
- 15ADON
15-acetyl-deoxynivalenol
- DON
deoxynivalenol
- FHB
Fusarium head blight
- NIV
nivalenol.
References
- 1.Stack RW. 1999. http://www.apsnetorg/online/feature/FHB/Top.html Return of an old problem: Fusarium head blight of small grains. Available at: (accessed on February 2, 2008)
- 2.Nganje WE, Bangsund DA, Leistritz FL, Wilson WW, Tiapo NM. Regional economic impacts of Fusarium head blight in wheat and barley. Rev Agric Econ. 2004;26:332–347. [Google Scholar]
- 3.Windels CE. Economic and social impacts of Fusarium head blight: changing farms and rural communities in the Northern Great Plains. Phytopathology. 2000;90:17–21. doi: 10.1094/PHYTO.2000.90.1.17. [DOI] [PubMed] [Google Scholar]
- 4.Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson DW, Black RM, Lee CG, Pottage C, Rickard RL, Sandford MS, Webber TD, Williams NE. Structure-activity studies of trichothecenes: cytotoxicity of analogues and reaction products derived from T-2 toxin and neosolaniol. J Med Chem. 1989;32:555–562. doi: 10.1021/jm00123a008. [DOI] [PubMed] [Google Scholar]
- 6.Alexander NJ, McCormick SP, Ziegenhorn SL. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Natural Toxins. 1999;7:265–269. doi: 10.1002/1522-7189(199911/12)7:6<265::aid-nt65>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 7.Desjardins AE, McCormick SP, Appell M. Structure-activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J Agric Food Chem. 2007;55:6487–6492. doi: 10.1021/jf0709193. [DOI] [PubMed] [Google Scholar]
- 8.Zamir LO, Devor KA, Sauriol F. Biosynthesis of the trichothecene 3-acetyldeoxynivalenol. Identification of the oxygenation steps after isotrichodermin. J Biol Chem. 1991;266:14992–15000. [PubMed] [Google Scholar]
- 9.McCormick SP, Hohn TM, Desjardins AE. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl Environ Microbiol. 1996;62:353–359. doi: 10.1128/aem.62.2.353-359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem. 2007;71:2105–2123. doi: 10.1271/bbb.70183. [DOI] [PubMed] [Google Scholar]
- 11.Tokai T, Koshino H, Takahashi-Ando N, Sato M, Fujimura M, Kimura M. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem Biophys Res Commun. 2007;353:412–417. doi: 10.1016/j.bbrc.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 12.Hohn TM, Beremand PD. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene. 1989;79:131–138. doi: 10.1016/0378-1119(89)90098-x. [DOI] [PubMed] [Google Scholar]
- 13.McCormick SP, Alexander NJ, Proctor RH. Fusarium Tri4 encodes a multifunctional oxygenase required for trichothecene biosynthesis. Can J Microbiol. 2006;52:636–642. doi: 10.1139/w06-011. [DOI] [PubMed] [Google Scholar]
- 14.Kimura M, Kaneko I, Komiyama M, Takatsuki A, Koshino H, Yoneyama K, Yamaguchi I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J Biol Chem. 1998;273:1654–1661. doi: 10.1074/jbc.273.3.1654. [DOI] [PubMed] [Google Scholar]
- 15.Alexander NJ, Hohn TM, McCormick SP. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl Environ Microbiol. 1998;64:221–225. doi: 10.1128/aem.64.1.221-225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet Biol. 2001;32:121–133. doi: 10.1006/fgbi.2001.1256. [DOI] [PubMed] [Google Scholar]
- 17.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet Biol. 2002;36:224–233. doi: 10.1016/s1087-1845(02)00021-x. [DOI] [PubMed] [Google Scholar]
- 18.Lee T, Han YK, Kim KH, Yun SH, Lee YW. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl Environ Microbiol. 2002;68:2148–2154. doi: 10.1128/AEM.68.5.2148-2154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown DW, Proctor RH, Dyer RB, Plattner RD. Characterization of a fusarium 2-gene cluster involved in trichothecene C-8 modification. J Agric Food Chem. 2003;51:7936–7944. doi: 10.1021/jf030607+. [DOI] [PubMed] [Google Scholar]
- 20.Meek IB, Peplow AW, Ake C, Jr, Phillips TD, Beremand MN. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl Environ Microbiol. 2003;69:1607–1613. doi: 10.1128/AEM.69.3.1607-1613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick SP, Harris LJ, Alexander NJ, Ouellet T, Saparno A, Allard S, Desjardins AE. Tri1 in Fusarium graminearum encodes a P450 oxygenase. Appl Environ Microbiol. 2004;70:2044–2051. doi: 10.1128/AEM.70.4.2044-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peplow AW, Meek IB, Wiles MC, Phillips TD, Beremand MN. Tri16 is required for esterification of position C-8 during trichothecene mycotoxin production by Fusarium sporotrichioides. Appl Environ Microbiol. 2003;69:5935–5940. doi: 10.1128/AEM.69.10.5935-5940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormick SP, Alexander NJ. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl Environ Microbiol. 2002;68:2959–2964. doi: 10.1128/AEM.68.6.2959-2964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueno Y, Sawano M, Ishii K. Production of trichothecene mycotoxins by Fusarium species in shake culture. Appl Microbiol. 1975;30:4–9. doi: 10.1128/am.30.1.4-9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller JD, Greenhalgh R, Wang Y, Lu M. Trichothecene chemotypes of three Fusarium species. Mycologia. 1991;83:121–130. [Google Scholar]
- 26.O'Donnell K, Kistler HC, Tacke BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O'Donnell K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci USA. 2002;99:9278–9283. doi: 10.1073/pnas.142307199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goswami RS, Kistler HC. Pathogenicity and in planta mycotoxin accumulation among members of the Fusarium graminearum species complex on wheat and rice. Phytopathology. 2005;95:1397–1404. doi: 10.1094/PHYTO-95-1397. [DOI] [PubMed] [Google Scholar]
- 29.Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, Geiser DM, Suga H, Toth B, Varga J, O'Donnell K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet Biol. 2007;44:1191–1204. doi: 10.1016/j.fgb.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Ward TJ, Clear RM, Rooney AP, O'Donnell K, Gaba D, Patrick S, Starkey DE, Gilbert J, Geiser DM, Nowicki TW. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet Biol. 2008;45:473–484. doi: 10.1016/j.fgb.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Desjardins AE. Natural product chemistry meets genetics: when is a genotype a chemotype? J Agric Food Chem. 2008;56:7587–7592. doi: 10.1021/jf801239j. [DOI] [PubMed] [Google Scholar]
- 32.Kimura M, Tokai T, O'Donnell K, Ward TJ, Fujimura M, Hamamoto H, Shibata T, Yamaguchi I. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 2003;539:105–110. doi: 10.1016/s0014-5793(03)00208-4. [DOI] [PubMed] [Google Scholar]
- 33.Garvey GS, McCormick SP, Rayment I. Structural and functional characterization of the TRI101 trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: kinetic insights to combating Fusarium head blight. J Biol Chem. 2008;283:1660–1669. doi: 10.1074/jbc.M705752200. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr Sect D. 2004;60(12, Part 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 35.Buglino J, Onwueme KC, Ferreras JA, Quadri LE, Lima CD. Crystal structure of PapA5, a phthiocerol dimycocerosyl transferase from Mycobacterium tuberculosis. J Biol Chem. 2004;279:30634–30642. doi: 10.1074/jbc.M404011200. [DOI] [PubMed] [Google Scholar]
- 36.Onwueme KC, Ferreras JA, Buglino J, Lima CD, Quadri LE. Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proc Natl Acad Sci USA. 2004;101:4608–4613. doi: 10.1073/pnas.0306928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma X, Koepke J, Panjikar S, Fritzsch G, Stockigt J. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem. 2005;280:13576–13583. doi: 10.1074/jbc.M414508200. [DOI] [PubMed] [Google Scholar]
- 38.D'Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 39.Bayer A, Ma X, Stockigt J. Acetyltransfer in natural product biosynthesis—functional cloning and molecular analysis of vinorine synthase. Bioorg Med Chem. 2004;12:2787–2795. doi: 10.1016/j.bmc.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 40.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 42.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Shih YP, Wu HC, Hu SM, Wang TF, Wang AH. Self-cleavage of fusion protein in vivo using TEV protease to yield native protein. Protein Sci. 2005;14:936–941. doi: 10.1110/ps.041129605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55(Part 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Part 8):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57(Part 10):1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12, Part 1):2126–2132a. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Part 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 52.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 53.Ellman GL. Tissue sulphydryl groups. Arch Biochem Biophys. 1959;82:70. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 54.Proctor RH, Hohn TM, McCormick SP. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact. 1995;8:593–601. doi: 10.1094/mpmi-8-0593. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1989. [Google Scholar]
- 56.Miller JD, Blackwell BA. Biosynthesis of 3-Acetyldeoxynivalenol and other metabolites by Fusarium-culmorum Hlx 1503 in a stirred jar fermenter. Can J Bot Rev Can Bot. 1986;64:1–5. [Google Scholar]


