Abstract
The design of protein–peptide interactions has a wide array of practical applications and also reveals insight into the basis for molecular recognition. Here, we present the redesign of a tetratricopeptide repeat (TPR) protein scaffold, along with its corresponding peptide ligand. We show that the binding properties of these protein–peptide pairs can be understood, quantitatively, using straightforward chemical considerations. The recognition pairs we have developed are also practically useful for the specific identification of tagged proteins. We demonstrate the facile replacement of these proteins, which we have termed T-Mods (TPR-based recognition module), for antibodies in both detection and purification applications. The new protein–peptide pair has a dissociation constant that is weaker than typical antibody–antigen interactions, yet the recognition pair is highly specific and we have shown that this affinity is sufficient for both Western blotting and affinity purification. Moreover, we demonstrate that this more moderate affinity is actually advantageous for purification applications, because extremely harsh conditions are not required to dissociate the T-Mod-peptide interaction. The results we present are important, not only because they represent a successful application of protein design but also because they help define the properties that should be sought in other scaffolds that are being developed as antibody replacements.
Keywords: tetratricopeptide repeat protein (TPR), peptide binding, rational design, molecular recognition, antibody replacements, solvent accessible surface area, western blotting, hydrophobic interactions
Introduction
The design of proteins with novel binding specificities is a central goal of protein engineering with widespread applications. Here, we describe the rational redesign of a natural protein–peptide interaction to create an orthogonal recognition pair. This new class of protein-based detection agents is based on the tetratricopeptide repeat (TPR) motif which we name T-Mods (TPR-based recognition module). Not only does the redesign of protein-ligand interactions contribute to our fundamental understanding of molecular recognition but also the novel protein-peptide interaction pairs can be used in many practical applications. For example, in molecular and cellular biology, there are frequently instances in which it is essential to specifically identify, or purify, a protein of interest from the cellular milieu. A common strategy is to attach a peptide “tag” to the protein of interest, which can be detected by a specific antibody. Although this strategy has the advantage that similar detection and purification procedures can be used for different proteins with the same tag, in most of its current manifestations it has the disadvantage of requiring a peptide-specific antibody. Antibodies cannot be genetically encoded in an active form within the cell, they are large, sensitive to freeze-thaw cycles, and cannot easily be modified or engineered. The generation of antibodies by the injection of mammals is slow, difficult to control, and obviously involves the slaughter of many animals. The T-Mods that we have created are small, approximately one-tenth the size of an antibody (Fig. 1), stable, can be readily produced in large quantities in Escherichia coli, and can be effectively substituted for antibodies in both detection and purification applications.
Figure 1.
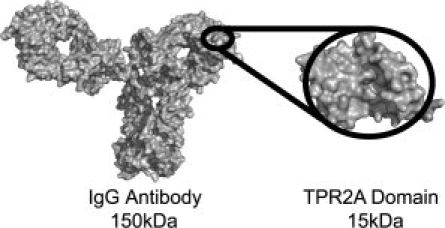
Relative size of an antibody and a T-Mod. Comparison of the TPR2A domain of HOP (PDB file 1ELR)3 and an IgG Antibody (PDB file 1HZH)4 to scale. The 3-TPR repeat is roughly one-tenth the size of an antibody. Images were rendered in PyMol (Delano Scientific).
Results
The scaffold
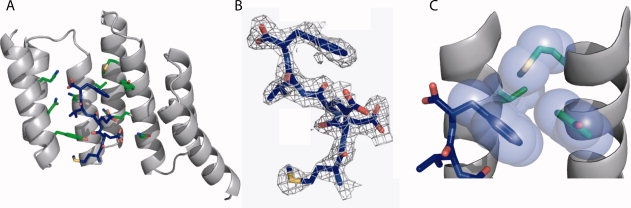
The tetratricopeptide repeat (TPR) is a 34 amino acid helix-turn-helix-turn motif.5 Here, we use the natural 3-TPR module, TPR2A from the heat shock organizing protein (HOP), as the framework on which to introduce novel binding activities. An attractive feature of TPR-based recognition modules as frameworks for protein design is that they undergo little or no structural change upon ligand binding [Fig. 2(A)].7 Figure 2(B) shows the crystal structure of the complex of TPR2A bound to its natural ligand, the C-terminal peptide of Hsp90, which has the sequence MEEVD. The peptide is bound in an extended conformation with its free C-terminus and most of its side-chains or backbone making specific interactions with side-chains of the TPR.3
Figure 2.
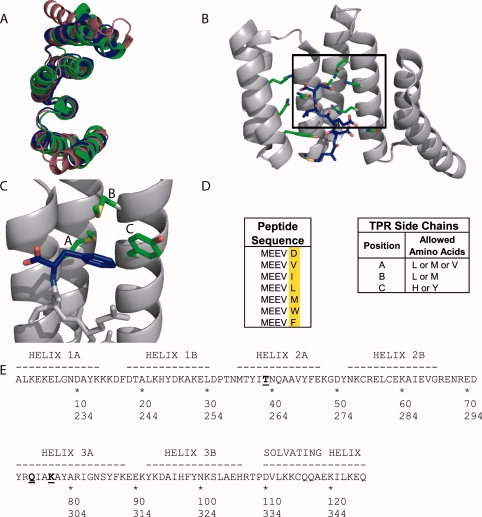
The TPR Scaffold and Library Design. (A) Overlay of the crystal structures of three natural 3-TPR domains: from PP5 (magenta, PDB file 1A17)6, TPR1 domain of HOP (blue, PDB file 1ELW)3, and TPR2A domain of HOP (green, PDB file 1ELR)3. The TPR1 and TPR2A structures are taken from the structures of the TPR in complex with its peptide ligand. The PP5 structure is of the TPR domain alone. (B) The cocrystal structure of TPR2A (gray ribbon) with its cognate ligand, the C-terminal peptide of Hsp90 (MEEVD) (blue sticks).3 Some of the side chains of TPR2A involved in interactions with the peptide, including all residues varied in this work, are shown as sticks. (C) Illustration of the hydrophobic cavity of a designed TPR in complex with the MEEVF peptide. This figure should not be interpreted as an optimal model for the T-Mod-peptide interaction, but instead a guide for the reader. The randomized TPR positions are shown in green and labeled A-C, and the C-terminal residue of the peptide is shown in blue with the carboxylate oxygen atoms in red. Images A-C were rendered in PyMol. (D) Sequences of peptides (left) and TPRs (right) used in this study. A library of 12 TPR proteins was constructed based on the sequence of TPR2A with randomizations introduced to the hydrophobic residues listed. (E) The sequence of TPRs used in this study compared to wild-type TPR2A.3 Wild-type TPR2A was used as the scaffold, with a hydrophobic cavity introduced by randomizing three key positions shown in bold and underlined. In addition, the mutation D110K (equivalent to position 334 in HOP) was introduced which gives a slight increase in the affinity of wt-TPR2A for Hsp90 (Tommi Kajander and Lynne Regan, manuscript in preparation). The first line gives the numbering scheme based on TPR2A alone, the second line shows the corresponding numbering in the full-length HOP protein. TPRs are labeled in the text based on the residues at the three randomized positions in the format (T-Mod(ABC)).
Our designs introduce minimal changes that alter the TPR2A-MEEVD binding specificity to create new TPR-peptide recognition pairs. The key feature of our design is to introduce a hydrophobic binding pocket onto the TPR to bind a hydrophobic residue at the C-terminal position of the peptide, rather than the negatively charged aspartic acid of the TPR2A cognate peptide [Fig. 2(C)]. We hypothesized that if all the other TPR-peptide binding interactions were retained in such a design, it was reasonable to expect that the position of the side-chain of the C-terminal residue of the peptide relative to the TPR would be maintained.
Library design and screening
We created several T-Mods with different combinations of hydrophobic residues to form a binding pocket for the side chain of the C-terminal amino acid of the peptide. We also made a small library of peptide sequences, which for convenience we expressed as GST-fusions with the sequence GST-MEEVX, where ‘X’ represents the hydrophobic amino acid to be varied [Fig. 2(D)]. Our aim was not to characterize all possible interacting pairs, but rather to identify the best T-Mod-peptide pairs for the desired applications.
The sequence of TPR2A with the three mutated positions indicated is shown in Figure 2(E). Each of these three positions was mutated to two or three different amino acids, resulting in 12 possible combinations [Fig. 2(D)].
As an initial screen for binding, we developed a “dot-blot” assay, in which the GST-peptide was immobilized on a membrane, probed with the T-Mods, and binding detected through the T-Mod's His tag. We envision using the T-Mod-peptide pairs in Western Blotting, so this screen allowed us to identify T-Mods with the desired functional properties. Several of the T-Mod-peptide pairs gave a positive signal in the dot blot assay (data not shown), and we selected three of these for further characterization.
We measured the thermal stability of several of these proteins, and all were very similar to the parent TPR2A, with a melting temperature of ∼54°C (Fig. 3). In future designs, the stability of the T-Mods could be increased by protein engineering if needed.8,9
Figure 3.
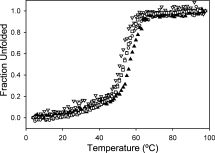
The Stability of Designed TPRs. The thermal denaturation curves of the designed TPRs (MMY open circles, MLY closed triangles, and VMY open triangles) overlay well with that of TPR2A (open squares), indicating there is no destabilization due to introduction of the hydrophobic pocket. T-Mods MLY and VMY have an additional K5H mutation that had no effect on melting temperature. The constructs used for all of the other experiments discussed in this work do not have this mutation.
We used surface-plasmon resonance (SPR) to assess the binding affinity and specificity of different T-Mod-peptide pairs. His-tagged T-Mods were immobilized on a Ni-NTA chip, the GST-peptide fusions were passed over the chip, and binding was monitored. We observed that a peptide with a particular hydrophobic C-terminal residue bound with similar affinity to any of the three T-Mods tested (T-Mod(MMY), T-Mod(MLY), T-Mod(VMY)). However, the identity of the C-terminal hydrophobic amino acid strongly influenced binding affinity, the larger the side-chain, the tighter the binding (Fig. 4). Neither the GST-only control nor the GST-MEEVD fusion showed any significant binding to any of the hydrophobic pocket T-Mods. An example of the SPR data for T-Mod(MMY) binding to GST-MEEVF, which has a dissociation constant of ∼1 μM, is shown in Figure 4.
Figure 4.
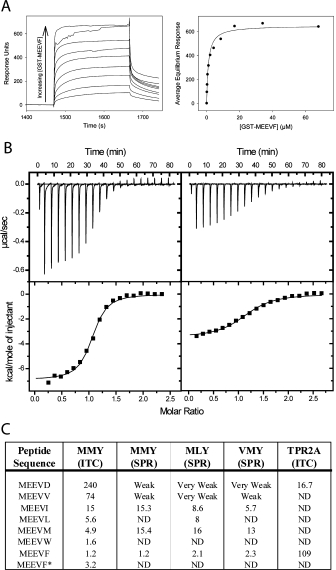
Quantifying T-Mod-Peptide Binding Interactions. (A) Surface Plasmon Resonance (SPR). Left: Example of a titration curve showing sensorgrams as a function of fusion protein (GST-MEEVF) concentration, with T-Mod(MMY) immobilized on the chip. The sensorgrams have been corrected for nonspecific binding by subtraction of a channel without TPR bound. Right: Equilibrium response plotted versus concentration of GST-MEEVF. The solid line is a fit of the data to a 1:1 binding model. This fit gives a dissociation constant (Kd) of 1.2 μM. (B) Isothermal titration calorimetry (ITC). T-Mod(MMY) was titrated into GST-MEEVF (left) and GST-MEEVM (right). The heats were integrated and fit to a 1:1 binding model. For GST-MEEVF, Kd = 1.2 μM and stoichiometry (N) = 1.03. For GST-MEEVM, Kd = 4.1 μM and N = 1.15. (C) Dissociation constants (μM) of different TPR-peptide interactions. All measurements were made with TPRs and GST-peptide fusions except for MEEVF*, where the 5-mer peptide was used. ND indicates measurement was not determined. When interactions were measured in duplicate, average values are shown, and for these measurements the reported values have an error of ±25% between complete duplicate experiments.
Isothermal titration calorimetry
We used isothermal titration calorimetry (ITC) to further characterize the interaction of T-Mod(MMY) with the different GST-peptide fusions. Under the conditions of our assay, we measured a dissociation constant (Kd) for the TPR2A-GST-MEEVD interaction of ∼17 μM, consistent with measurements reported by Scheufler et al.3
Representative binding data for the new T-Mod-peptide pairs are shown in Figure 4. The dissociation constants determined by ITC were in accord with those measured by SPR (Fig. 4). The ITC data also confirmed one to one binding stoichiometry for all the T-Mod-peptide pairs. To be sure that the binding we observed was a consequence of the interaction of only the C-terminal 5 amino acids with the T-Mod, we synthesized the 5-residue peptide MEEVF with a free C-terminus and acetylated N-terminus and measured its binding to T-Mod(MMY) using ITC. The Kd for the T-Mod(MMY)-MEEVF peptide interaction is ∼3 μM, consistent with the conclusion that the binding we are monitoring is dominated by the T-Mod-peptide interaction.
We also measured the dissociation constants for the noncognate pairs to determine if the new pair is indeed orthogonal to the natural pair. The Kd of the TPR2A-MEEVF peptide interaction is 110 μM, while that for the hydrophobic T-Mod with the MEEVD peptide is 240 μM. The observation of such weak binding for the noncognate pairs is consistent with the energetically unfavorable event of placing a negative charge into a hydrophobic environment.
Free energies correlate with structural predictions of packing of the hydrophobic cavity
We analyzed the interaction of T-Mod(MMY) with peptides having different hydrophobic C-terminal amino acids by plotting the ΔG of binding versus the solvent accessible surface area of the side-chain of the C-terminal residue. We noted a linear relationship between the surface area of the hydrophobic side-chain of the peptide10,11 and the free energy of binding to the T-Mod (Fig. 5). Two components presumably contribute to this increase in affinity with the increase in hydrophobic surface area of the C-terminal amino acid: removal of hydrophobic surface area from solvent when the peptide binds to the T-Mod and van der Waals interactions associated with “filling” the hydrophobic cavity of the T-Mod with the hydrophobic side-chain of the peptide. Several studies have sought to quantify the contribution of hydrophobic burial to protein stability, in the context of mutations in the hydrophobic core. Interestingly, the slope of our data gives a value of 40 calories per Å2 of buried hydrophobic surface area, which is consistent with the values obtained in those studies.12–16
Figure 5.
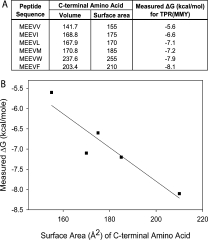
Correlation Between C-terminal Amino Acid Identity and Free Energy. (A) Free energy of binding (ΔG) measured using ITC for the series of GST-peptide fusions with T-Mod(MMY). These values are directly correlated with the surface area11 and volume10 of the C-terminal amino acid. (B) Plot of the measured free energy of binding versus the surface area of the C-terminal amino acid of the peptide. The linear regression gives a slope of about 40 calories per Å2 and a correlation coefficient of 0.93.
Because binding affinity increased with the increasing size of the C-terminal hydrophobic residue of the peptide, it was of interest to test if MEEVW bound even more tightly. We therefore made the GST-MEEVW construct and measured its binding to T-Mod(MMY) by ITC. If the linear relationship between surface area of the C-terminal amino acid side chain and the free energy of binding extended to tryptophan, its free energy of binding would be expected to be ∼−10 kcal/mole. The measured free energy of binding for GST-MEEVW to T-Mod(MMY) is −7.9 kcal/mole. This result suggests that the C-terminal phenylalanine side-chain of the peptide best fills the hydrophobic cavity on the TPR, and that the larger side-chain of tryptophan is too large to allow optimum binding. These results also emphasize that the strength of the T-Mod-peptide interaction can be fine-tuned as desired.
X-ray crystallography confirms the binding mode
To further characterize the T-Mod-peptide interactions, we attempted to cocrystallize several different T-Mod-peptide pairs. We were successful in crystallizing T-Mod(VMY) in complex with the MEEVF peptide. The crystal structure of this complex reveals that the peptide interacts with the T-Mod in the same conformation and location as that of the MEEVD peptide with TPR2A [Figs. 2(B) and 6].3 In the TPR2A-MEEVD crystal structure, a key interaction is that between the TPR and the “two-carboxylate clamp,” that is, the Asp side-chain and C-terminal carboxylate of the peptide. Mutagenesis studies have shown that this interaction contributes significantly to the stability of the complex.17 Our redesigns sought to maintain the interactions of the C-terminal carboxyl group of the peptide with the TPR, while changing the side chain from aspartic acid to a hydrophobic amino acid. The structure of the T-Mod(VMY)-MEEVF complex [Fig. 6(A)] clearly shows that the interaction between the C-terminal carboxylate and the TPR are maintained as in the natural pair. Moreover, the hydrophobic side-chain of the C-terminal residue, phenylalanine, fits nicely into the designed hydrophobic pocket of the T-Mod [Fig. 6(C)]. Looking in detail at the interaction of the C-terminal phenylalanine side chain with the T-Mod, we see that it inserts into the hydrophobic pocket, with only the δ1C retaining significant solvent accessibility. Using the program GETAREA,18 the solvent accessible surface area of the phenylalanine side chain in the complex is estimated to be about 26% relative to the accessible surface area of the Phe side chain in a Gly-Phe-Gly peptide. About half of this solvent accessibility can be attributed to the δ1C (19 Å2). Finally, as expected based on the hypothesis that the majority of the peptide-TPR interactions could be maintained while changing the identity of just one residue, the interactions of the remaining peptide residues with the TPR are unaltered. A structural alignment of the TPR2A-MEEVD complex with the T-Mod(VMY)-MEEVF complex overlays extremely well, with an RMSD of 0.41 Å. Much of this deviation comes from the loop regions between helices, which are likely the result of crystal packing contacts. These results confirm that the recognition pair forms a stable and specific interaction as envisioned, and that the structural data support our interpretation of the thermodynamic data.
Figure 6.
X-ray Crystallographic cocrystal structure of the T-Mod(VMY)-MEEVF Complex. (A) The crystal structure of T-Mod(VMY) (gray ribbon) is shown in complex with the MEEVF peptide (blue sticks). Some of the side chains of T-Mod(VMY) involved in interactions with the peptide, including all residues varied in this work, are shown as sticks. (B) A 2Fo − Fc electron density map in the region of the MEEVF peptide. (C) Close-up view showing the interaction of the C-terminal phenylalanine side chain of the peptide with the hydrophobic pocket of the T-Mod. The side chains involved in the packing are shown as spheres representing van der Waals radii. These images were rendered in PyMol.
We anticipate that appropriate T-Mod-peptide pairs can be developed for use in numerous applications in preference to antibodies. Here, we demonstrate a few such applications.
T-Mods can replace antibodies in Western blotting
A widely used technique for protein identification using antibodies is the Western Blot. We therefore sought to demonstrate that T-Mods can replace antibodies as the “primary antibody” in Western Blots. For proof of principle experiments, the T-Mod's N-terminal hexahistidine tag allows anti-His mouse monoclonal antibodies to be used as a secondary antibody, with alkaline phosphatase conjugated goat anti-mouse antibodies for detection [Fig. 7(A)]. One can envision, however, that when used more routinely, T-Mods can be conjugated directly to alkaline phosphatase, horseradish peroxidase, biotin, or a fluorescent protein to allow for efficient one or two-step detection, thus eliminating any requirement for antibodies.
Figure 7.
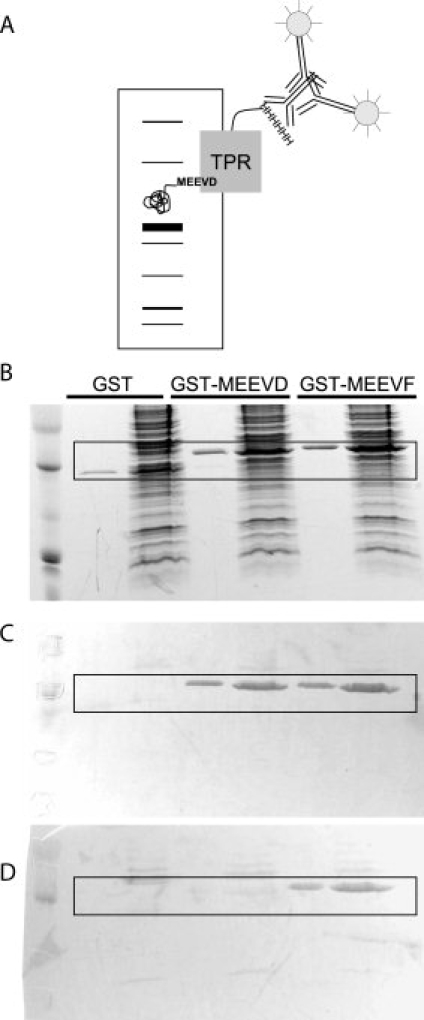
Western Blots using TPRs as Primary Antibodies. (A) Schematic illustrating Western blot strategy. Cellular extract containing a tagged target protein is separated by SDS-PAGE and then transferred to a membrane. The tagged protein is probed with His-TPRs, followed by anti-His antibodies and alkaline-phosphatase conjugated secondary antibodies. (B) A duplicate SDS-PAGE gel used for Western blotting is stained for total protein with Coomassie Brilliant Blue. 1.75 μg of purified tagged protein was diluted in water (lanes 2, 4, and 6) or cellular extract (lanes 3, 5, and 7). (C) Blot probed with His-TPR2A. Note that both the MEEVD and MEEVF tagged proteins are stained. (D) Blot probed with His-T-Mod(MMY). Note that only the MEEVF tagged protein is stained. The boxed areas in B–D highlight the presence or absence of GST-peptide staining.
To test if the T-Mods can be used to detect the peptide-tagged protein within a total cellular extract, we prepared samples of E. coli extract spiked with small amounts of GST-peptide. When these samples are separated by SDS-PAGE, staining with Coomassie Blue does not allow for direct identification of the protein of interest (GST-peptide) [Fig. 7(B)]. We performed Western Blots on extracts spiked with GST-MEEVF, GST-MEEVD, or untagged GST. The blots were probed with purified His-tagged TPRs, followed by anti-His antibodies and alkaline phosphatase-conjugated antibodies. Purified protein was run side-by-side for ease in determining the correct identification of the target protein. TPR2A detected both the MEEVD and MEEVF tagged GST [Fig. 7(C)]. Apparently, in this context even the weak affinity of TPR2A for MEEVF is sufficient. In contrast, the hydrophobic T-Mod(MMY) only detected the MEEVF tagged GST [Fig. 7(D)]. This clearly indicates that the new TPR-peptide pair has enhanced specificity when compared with the starting pair. In addition, it is important to note that for both the natural and designed T-Mods, there is minimal nonspecific staining of background bands, supporting the hypothesis that T-Mods can effectively function as primary antibody replacements.
T-Mod-GFP fusions can be used for one-step detection
To completely eliminate the need for antibodies in Western Blots using T-Mods, we created a T-Mod(MMY) fusion with monomeric-enhanced GFP (mEGFP) [Fig. 8(A)]. Others have reported the use of Protein A-GFP fusions in detection.19,20 In our procedure, in a single step, the membrane is incubated with the T-Mod-GFP and then directly visualized using a UV transilluminator typically used for imaging DNA gels [Fig. 8(B)]. This method completely eliminates the need for any antibodies or developing procedures, making Western Blotting far faster, simpler, and less costly. In addition, T-Mod(MMY)-mEGFP can be produced in large quantities in E. coli and purified in one step via its hexahistidine tag. The TPR-mEGFP fusion presented here can detect less than 25 ng of target protein using concentrations of T-Mod-mEGFP as low as 150 nM. To further enhance the detection limits and signal, the use of tandem TPRs and alternative fluorescent proteins can be explored.
Figure 8.
Antibodies can be eliminated from Western Blots by using T-Mod-GFP fusions. (A) Schematic of the detection reagent. Monomeric enhanced GFP (green) is fused to the C-terminus of T-Mod(MMY) (blue). (B) Duplicate gels were stained for total protein (left) or probed with T-Mod(MMY)-mEGFP (right). The lane on the right could be readily visualized using a 365 nm UV transilluminator typically used for DNA gels, and the image was captured with a digital camera. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
T-Mods can be used for affinity purification
To use T-Mods for purification of tagged proteins, for our proof of principle experiments, we immobilized the T-Mods on Ni-NTA resin via their hexahistidine tags. For more general use, however, it would be easy to conjugate the T-Mods directly to resin or beads to eliminate the need for a linker. We prepared total cellular extracts from E. coli expressing GST alone, GST-MEEVD, and GST-MEEVF and passed them over columns of immobilized T-Mod [Fig. 9(A)]. For the natural TPR2A with MEEVD-tagged GST, complete elution of reasonably pure target protein was accomplished with 1M NaCl in pH 8.0 Tris buffer [Fig. 9(B)]. For the designed hydrophobic T-Mod with MEEVF-tagged GST, only slight elution in 1M NaCl was observed, and salt-free or low pH buffers also did not elute the protein [Fig. 9(C)]. However, the addition of free MEEVF peptide effectively elutes the fusion protein from the resin [Fig. 9(D)]. This resistance to 1M salt allows for an extremely stringent washing step at high ionic strength before gentle elution.
Figure 9.
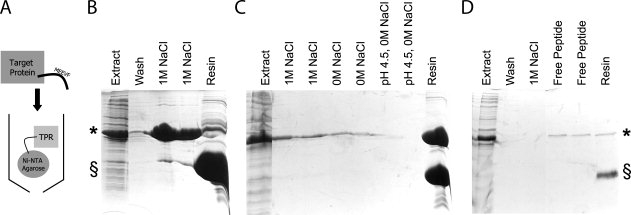
Affinity Purification of Tagged Proteins using TPRs. (A) Schematic illustrating affinity purification strategy. His-TPRs are immobilized on Ni-NTA resin and extracts containing MEEVF tagged target proteins are passed over the resin and purified. (B) Affinity purification of MEEVD tagged protein (*) using TPR2A (§), as illustrated by SDS-PAGE. Soluble lysate containing MEEVD tagged target protein (lane 1) is passed over the resin to bind. Following washes (with 50 mM Tris pH 8.0, 150 mM NaCl, 5 mM B-mercaptoethanol) (lane 2), the MEEVD tagged protein is eluted with wash buffer supplemented to 1M NaCl (lanes 3 and 4). Minimal target protein remains bound to the TPR (lane 5). (C) Affinity purification of MEEVF tagged protein (*) using T-Mod(MMY) (§). The same procedures were followed, except here, the interaction predominantly withstands 1M NaCl (lanes 2–3), 0M NaCl (lanes 4–5), and low pH conditions (lanes 6–7). Almost all of the tagged protein remains bound to the TPR (lane 8). The purification of MEEVF tagged protein using T-Mod(MMY) was repeated (D) and again the interaction withstands 1M NaCl (lane 3) before elution with 100 μg/mL free MEEVF peptide (lanes 4–5). Note that these bands are weaker because of the lower total amount of MEEVF tagged protein, but that far less MEEVF tagged protein remains bound to the TPR, indicating better elution (lane 6).
Discussion and Conclusions
A functional substitute for antibodies is desirable for a range of applications. There have been some prior attempts to design small and stable antibody replacements.1,2,21 In one such example, peptides were developed that were capable of binding other peptides.22 Although the affinities of these peptide–peptide interactions were of similar magnitude to those we report, the tag was significantly longer (13aa) and the interactions were not stable under typical conditions required for their use in Western Blots or in protein purification.
Here, we present the design, characterization, and application of a new T-Mod-peptide tag detection pair. Although the designs preserve some characteristics of the natural TPR2A-peptide interaction, new features are introduced to generate a completely novel TPR-peptide binding specificity. The designed T-Mod(MMY) binds the peptide tag MEEVF tightly and with high specificity. The correlation between the size of the peptide's C-terminal hydrophobic side-chain and the free energy of binding is consistent with burial of the C-terminal side-chain in the new hydrophobic pocket on the TPR, and this is confirmed by the crystal structure of the T-Mod-peptide complex.
We have shown that T-Mods have many desirable qualities, they are small, stable, contain no disulfide bonds, are rapidly produced from E. coli, and their affinity and specificity can be rationally engineered. But is the designed T-Mod-peptide pair useful? Is the interaction tight enough and specific enough for the desired applications? Yes. We have demonstrated that T-Mods can be readily substituted for antibodies in both detection and affinity purification applications. Thus, both their affinity and specificity are sufficient for practical applications.
When antibodies are used to purify target proteins, there are many drawbacks, the most prohibitive of which is the requirement of extremely low pH to elute the target protein. For instance, to purify FLAG tagged proteins using an anti-FLAG antibody, the manufacturer's protocol (Sigma) calls for an extremely low pH buffer along with the addition of free peptide competitor. The target protein must be transferred to a more neutral pH immediately, and the antibodies must be replaced often, because they are not tolerant of such extreme cycles of pH.
T-Mods have the distinct advantage of being able to be fine-tuned to select for properties desired for a particular application. For example, we show that TPR2A can be used to purify MEEVD-tagged proteins using 300 mM NaCl in the wash buffer and 1M NaCl as the elution buffer. For an even more stringent purification scheme, T-Mod(MMY) can be used to purify MEEVF-tagged proteins, washing with 1M NaCl and eluting with free peptide in pH 8.0 buffer.
Our results demonstrate that it is not necessarily extremely high affinity that is important for the application of T-Mods to protein purification and Western blotting. Rather, the T-Mod-peptide interaction must be specific enough and tight enough for the desired application. The T-Mods we have developed have high specificity, with minimal background binding to natural TPR2A or Hsp90. In future applications, T-Mods could potentially be used to track intracellular localization of tagged proteins, because they have been shown to be functional within live mammalian cells.9
Materials and Methods
Cloning
All enzymes for cloning were purchased from New England Biolabs (Beverly, MA), unless otherwise noted. All oligonucleotide synthesis and sequencing was performed by the Keck Facility, Yale University. When a representative sampling of the TPR and peptide libraries did not yield all the desired clones, in instances where the desired mutation was not represented by a single series of degenerate codons, or where specific mutants were required, a QuikChange Site-Directed mutagenesis kit (Stratagene) was used with the appropriate primers to incorporate the desired point mutations.
To assemble the TPR library, the TPR2A template was synthesized from six overlapping oligonucleotides: (1) 5′-atggctaagcaggcactgaaagaacacgagctggggaacgatgcctacaaga agaaagactttgacacagccttgaagcattacgacaaag-3′, (2) 5′-cttttcaaagtataccgctgctt gattcaBaatgtaagtcatgttagtggggtccagctccttggctttgtcgtaatgcttcaag-3′, (3) 5′-cagcggtat ctttgaaaagggcgactacaataagtgccgggagctttgtgagaaggccattgaagtggggagagaaaaccg-3′, (4) 5′-ttttcttctttgaagtaggagttgccaattcgagcatatgcgtDggcaatcaKtcgatagtcctctcggtttt ctctccccacttc-3′, (5) 5′-tcctacttcaaagaagaaaagtacaaggatgccatccatttctataacaagtctctggcagagcaccgaacc -3′, and (6) 5′-cccttgctccttcaggattttctccgcctgttggcacttttttagcacctttggggttcgg tgctctgccagag-3′. At each position of randomization, an equimolar mixture of specific bases was added, where B denotes G,C,T; D denotes A,G,T; and K denotes G,T. Three sites were randomized [Fig. 2(E)]: “A” which corresponds to position 39 in TPR2A (263 in HOP), “B” which corresponds to position 74 in TPR2A (298 in HOP), and “C” which corresponds to position 77 in TPR2A (301 in HOP). Oligonucleotides 1 and 2, 3 and 4, and 5 and 6 were joined by Klenow extensions, and this was followed by a series of two PCR amplifications. The first PCR amplification fused oligonucleotides 3–6 using the primers: 5′-cagcggtatactttgaaaag-3′ and 5′-taataaaactttcattgctccttcaggattttc-3′. A final PCR amplification joined the two remaining fragments using the primers 5′-taataaggatccaagcaggcactgaaag-3′ and 5′-taataaaactttcattgctccttcaggattttc-3′. The library of inserts was then double digested with Bam HI and Hind III and ligated into a modified pProEx-HTA vector (GibcoBRL, Gaithersburg, MD) to create genes with an N-terminal His6-tag followed by a TEV cleavage site.23 This vector has a shifted Bam HI restriction site to eliminate a 5 residue N-terminal extension that would result from cloning into the unmodified vector. The final constructs were sequenced to verify their identity.
To assemble the GST-peptide fusion library, the C-terminal 24 residues of Hsp90 were cloned into a modified pGEX4T3 vector (GE Healthcare). The oligonucleotides: 5′-taataaggatccccgaattccgaaaacctgtattttcagggctccagtgctgctgtaactgaagaaatgccacccc-3′ and 5′-attattgcggccgcttaMaHtacttcttccatgcgtgatgtgtcgtcatctccttcaaggggtggcatttcttcag were synthesized, where M denotes an equimolar mixture of A and C, and H denotes A,C, and T. The oligonucleotides were assembled by Klenow extension and ligated at the Bam HI and Not I restriction sites to create genes with N-terminal GST followed by thrombin and TEV cleavage sites, and the peptide with a free C-terminus. The final constructs were sequenced to verify their identity.
The gene encoding T-Mod(MMY)-mEGFP24 was created using standard molecular biology protocols and confirmed by sequencing.
Protein expression and purification
The plasmids were transformed into E. coli BL21(DE3). Overnight cultures were diluted 1:100 in 1 L of Luria broth, followed by growth at 37°C to an OD600 of 0.4–0.6. Expression was induced with 0.6 mM IPTG, and the cells were grown for an additional 5 h at 30°C or 15 h at 18°C. The cells were harvested and the pellets were frozen at −20°C until purification.
To purify the TPR proteins, the pellets were thawed by resuspending in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol) supplemented with one tablet of complete EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). The suspension was sonicated, and the lysate cleared by centrifugation for 1 h at 35,000g. The proteins were then purified using Ni-NTA resin (Qiagen) according to the manufacturer's protocols.
To purify the GST fusion proteins, the pellets were thawed by resuspending in Dulbecco's Phosphate Buffered Saline (GibcoBRL) supplemented with 5 mM β-mercaptoethanol and EDTA-free protease inhibitor cocktail. The suspension was sonicated and lysed as described earlier. The cleared lysate was purified using glutathione agarose resin (Pierce, Rockland, IL) according to the manufacturer's protocols and eluted with 50 mM Tris pH 8.0, 50 mM glutathione.
For both the TPR and GST-peptide fusion purifications, the fractions containing protein were pooled, concentrated, and then dialyzed into 50 mM Tris pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol. All protein concentrations were determined by measuring UV absorption at 280 nm, using extinction coefficients calculated from amino acid composition.25 All experiments were carried out using His-tagged TPR fusions with the exception of the crystallography experiments, for which the His tag was cleaved.
Peptide synthesis
Fmoc-protected amino acids, preloaded Fmoc-Phe-Wang resin, and all related reagents were purchased from EMD Biosciences unless noted otherwise. All coupling steps were carried out for 30 min, which according to the Kaiser test gave complete coupling. Each deprotection step was performed twice for 5 min and was verified by the Kaiser test.
The MEEVF peptide was synthesized manually on a 250 μmole scale using standard Fmoc chemistry. Briefly, preloaded Fmoc-Phe-Wang resin was used so that the final peptide would have a free C-terminus, and at each coupling step four molar equivalents of Fmoc protected amino acids were added with four molar equivalents of PyBOP and six molar equivalents of DIPEA. Following synthesis, the N-terminus was acetylated with 2 mL 1:1 acetic anhydride:triethylamine in 10 mL 1:1 dichloromethane:dimethylformamide for 1 h. The peptide was cleaved from the resin upon treatment with 95% TFA, 2.5% water, and 2.5% EDT. The final, deprotected peptide was precipitated upon addition of diethyl ether, dried, and then resuspended in 4M guanidine hydrochloride supplemented with a minimal amount of acetonitrile. The peptide was purified by reverse phase HPLC on a Vydac C18 column using an acetonitrile gradient. Peaks corresponding to pure peptides were lyophilized and their identity confirmed by MALDI-MS.
Circular dichroism measurements
Circular dichroism spectra were acquired using 6 μM protein samples in PBS using an AVIV Model 215 CD spectrophotometer (AVIV Instruments, Lakewood, NJ).
Far-UV CD (190–260 nm) spectra were recorded at 25°C to confirm proper folding of the TPRs. Thermal denaturation curves were recorded by monitoring ellipticity at 222 nm while heating from 4°C to 98°C in 1°C increments with an equilibration time of 1 min at each temperature. Melting temperatures (Tm) were estimated as the temperature at which 50% of the protein was unfolded.
Surface plasmon resonance assays
SPR binding assays were performed on a Biacore 3000 Instrument (Biacore, Uppsala, Sweden) using a Ni-NTA chip. All measurements were performed with HBS-EB (10 mM HEPES pH 7.4, 150 mM NaCl, 50 μM EDTA, 0.005% (v/v) Surfactant P20) as running buffer. Each cycle was composed of two segments, immobilization and analysis. For the immobilization segment, all steps were performed over only flow channel 2 at a flow rate of 10 μL/min. 10 μL of 500 μM NiCl2 in HBS-EB was injected, followed by 30 μL of 50 nM His-TPRs to give an immobilization level of 1500–2000 response units. For the analysis segment, all measurements were performed at 40 μL/min over flow channels 1 and 2 to allow for background subtraction. Here, following immobilization, 2 injections of 40 and 120 μL HBS-EB were injected followed by dissociation periods of 40 and 200 s, respectively, to ensure a stable baseline. This was followed by a 130 μL injection of GST-peptide fusion in KINJECT mode. After a 200 s dissociation period, regeneration was achieved by a 120 μL injection of 0.35M EDTA, pH 8.0. The needle and internal fluidics cartridge were washed before the following cycle began. For each GST-peptide fusion, a series of increasing concentrations was run using the identical dilution of TPR.
For all measurements, the response in channel 1 was subtracted to correct for nonspecific binding to the chip surface. To calculate the equilibrium dissociation constants (Kd), the average response values at equilibrium (Req) were plotted versus concentration. The curves were fit to a one-site binding model using SigmaPlot (Systat Software, Point Richmond, CA) using the equation:
where Req is the average equilibrium response, Rmax is the equilibrium response at saturation, Kd is the dissociation constant, and [P] is protein concentration.
Isothermal titration calorimtetry
Isothermal titration calorimetry (ITC) experiments were performed on an ITC200 calorimeter, and all data analysis was conducted using Origin 7 software with the ITC200 add-on package (Microcal, Northampton, MA). Proteins were purified as described above, concentrated, and dialyzed exhaustively into 50 mM Tris pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol. The protein concentrations were quantified by absorption at 280 nm in triplicate, and peptide concentration was estimated by phenylalanine absorption and confirmed by amino acid analysis (Keck Facility, Yale University). For protein–protein titrations, the cell was loaded with the GST-peptide fusions, typically at 50 μM, and the syringe was loaded with 0.8–1.1 mM TPR. For the peptide titration, 700 μM peptide was titrated into 40 μM TPR. All experiments were performed with a 0.5 μL initial injection, which was discarded in data analysis, followed by twenty 2 μL injections with 250 s spacing. The syringe was stirred at 1000 rpm with thermostating at 25°C. The binding isotherms were fit to a one-site binding model to obtain the thermodynamic parameters.
Crystallization of T-Mod (VMY), data collection, solution, and refinement
T-Mod(VMY) was expressed and purified as described earlier and the hexahistidine tag was cleaved with AcTEV protease (Invitrogen). The sample was then passed back over Ni-NTA resin to remove the His tag and TEV protease. The protein was concentrated and mixed in a 1:1.3 molar ratio with MEEVF peptide to give a final T-Mod(VMY) concentration of 22 mg/mL. The crystallization conditions for the complex were obtained by screening around reported conditions for the cognate protein–peptide pairs3 using the hanging drop vapor diffusion method. Clusters of thin stacked plates appeared in 5 days in reservoir solution of 100 mM Tris pH 8.5, 30% PEG MME 2000, 5–10 mM NiCl2, and 10% xylitol. The crystals were frozen directly under cryostream. Diffraction data was obtained using a Rigaku MicroMax-007 rotating anode. Reflections with spacing between 50 Å and 2.1 Å were recorded on a Mar345 Image plate detector. The data was integrated, scaled, and merged in HKL2000 (Table I).26
Table I.
Data Collection and Refinement Statistics
| Data Collection Statistics | |
| Space group | P21 |
| Unit cell | a = 37.6 Å, b = 66.6 Å, c = 48.6 Å, β = 107.21° |
| Wavelength (Å) | 1.54178 |
| Resolution (Å) | 50–2.2 (2.8–2.2) |
| Signal/noise (1/σ) | 25.8 (4.9) |
| Completeness (%) | 98.6 (87.4) |
| Rsyma (%) | 5.3 (18.7) |
| Redundancy | 5.7 |
| Unique reflections | 11,529 |
| Refinement statistics | |
| Number of molecules per asymmetric unit | 2 |
| Number of atoms | |
| Total | 2332 |
| Protein | 2195 |
| Peptide | 48 |
| Waters | 85 |
| Ni | 4 |
| RMSD bond lengths | |
| Bond lengths (Å) | 0.0009 |
| Bond angles (°) | 1.2 |
| Average B value | 14.8 |
| RWorking (%) | 18.4 |
| RFreeb (%) | 24.4 |
| Ramachandran plot (2 complexes per asymmetric unit) | |
| Residues in the most favored regions | 255/260 |
| Residues in the additional allowed regions | 5/260 |
| Residues in the generously allowed regions | 0 |
| Residues in the disallowed regions | 0 |
The values in parenthesis are for the highest resolution bins.
Rsym = 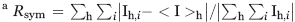 , where <I>h is the average intensity of symmetry-related reflections.
, where <I>h is the average intensity of symmetry-related reflections.
Rfree was calculated the same as Rwork but on 5% of randomly selected data not used in refinement.
The structure was solved by molecular replacement using Phaser.27 The TPR2A-MEEVD peptide complex (PDB accession code 1ELR)3 was used as a search model with the peptide removed. Rigid body refinement was carried out in REFMAC5.27 The peptide was modeled into strong and continuous Fo − Fc density calculated from the Phaser solution. After the peptide was built, a simulated annealing composite omit map was calculated using CNS28 to verify the conformation of the peptide. This map together with a solvent flattened, NCS averaged, prime and switch map calculated from RESOLVE29 was used to adjust the model. Rounds of restrained refinement in REFMAC5 and model adjustment in COOT30 were then performed. NCS restraints were released and a TLS model31 was used in the late stages of refinement. Water molecules were added using ARP/wARP27 and validated using Fourier difference maps. Final R factors for the model are Rworking = 18.4% and Rfree = 24.4%.
Western blots using T-Mods as the “primary antibody”
For Western blotting, to allow for precise control of the amount of the different GST-peptide fusions in each lane, BL21(DE3) extracts were supplemented with purified protein. To prepare the extracts, cell pellets from 10 mL cultures were resuspended in 1-mL B-Per (Pierce) and lysed by shaking with occasional vortexing for 10 min. The lysates were cleared by centrifugation and the soluble supernatant was reserved. In parallel, soluble lysate and water were supplemented with purified GST fusion protein at the appropriate concentrations to load 1.75 μg of fusion protein per well in 20 μL to minimize variation in loading the wells. The samples were separated on 18% SDS-polyacrylamide gels. One gel was stained with Coomassie Brilliant Blue to visualize total protein, and the others were transferred onto a PVDF membrane (Millipore). The membranes were blocked in 5% nonfat milk in TBS-T(20 mM Tris pH 8.0, 150 mM NaCl, 0.1% Tween-20) overnight at 4°C with shaking. The blots were washed in TBS-T and incubated with purified 30 μM His-TPRs in TBS (20 mM Tris pH 8.0, 150 mM NaCl) for 4 h at room temperature with shaking. The blots were probed at room temperature for 1.5 h with a 1:200 dilution of anti-His antibody (His probe H-3, Santa Cruz Biotechnology) in TBS-T supplemented with 0.5% nonfat milk, followed by a 1 h incubation with a 1:2000 dilution of alkaline-phosphatase conjugated goat anti-mouse antibody (sc-2058, Santa Cruz Biotechnology) in TBS-T supplemented with 0.5% nonfat milk. The blots were then developed with NBT/BCIP substrate (GibcoBRL) according to the manufacturer's suggested protocols.
Western blots using T-Mods for one-step detection
For one-step detection of the protein of interest, identical protocols were used for the transfer and blocking steps. Membranes were then incubated in 30 μM TPR-GFP fusion in TBS for 1.5 h, washed three times in TBS, and visualized using a UV transilluminator at 365 nm.
Affinity purification of MEEVF tagged proteins using T-Mods
Overnight cultures of BL21(DE3) expressing the His-TPRs and GST fusions were prepared as discussed earlier. Cultures were aliquoted into 150-mL fractions, the cells were harvested by centrifugation, and the pellets stored at −20°C until further analysis. The cells were resuspended in 4-mL lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol) supplemented with protease inhibitors and lysed by sonication. The lysates were cleared by centrifugation and filtering through a 0.4 micron filter. The His-TPR was bound to 800 μL equilibrated Ni-NTA resin and washed thoroughly. The GST-fusion extract was then applied to the column and passed over the resin four times. The resin was then washed with 15 mL wash buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol). For the TPR2A-Hsp90 purification, elution was accomplished with five 1.5 mL applications of high salt buffer (50 mM Tris pH 8.0, 1M NaCl, 5 mM β-mercaptoethanol). For the T-Mod(MMY)-MEEVF purification, the resin was washed with 15 mL wash buffer followed by five 1.5 mL applications of high salt buffer. Elution was accomplished with five 1.5 mL applications of wash buffer supplemented with 100 μg/mL purified MEEVF peptide. Fractions were analyzed by SDS-PAGE and the gels were stained with Coomassie Brilliant Blue.
Coordinates
Coordinates have been deposited with the Protein Data Bank and assigned the PDB accession code of 3FWV.
Acknowledgments
We thank Aitziber Lopez Cortajarena, Robielyn Ilagan, Lenka Kundrat, and Fang Yi for valuable discussions and comments on the manuscript. We thank Tijana Grove for advice in peptide synthesis and Ann Valentine for helpful discussions regarding the ITC experiments.
References
- 1.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 2.Binz HK, Pluckthun A. Engineered proteins as specific binding reagents. Curr Opin Biotechnol. 2005;16:459–469. doi: 10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 4.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 5.Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 6.Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. Embo J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortajarena AL, Regan L. Ligand binding by TPR domains. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortajarena AL, Kajander T, Pan W, Cocco MJ, Regan L. Protein design to understand peptide ligand recognition by tetratricopeptide repeat proteins. Protein Eng Des Sel. 2004;17:399–409. doi: 10.1093/protein/gzh047. [DOI] [PubMed] [Google Scholar]
- 9.Cortajarena AL, Yi F, Regan L. Designed TPR modules as novel anticancer agents. ACS Chem Biol. 2008;3:161–166. doi: 10.1021/cb700260z. [DOI] [PubMed] [Google Scholar]
- 10.Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 11.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 12.Damodaran S, Song KB. The role of solvent polarity in the free energy of transfer of amino acid side chains from water to organic solvents. J Biol Chem. 1986;261:7220–7222. [PubMed] [Google Scholar]
- 13.Eisenberg D, McLachlan AD. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 14.Kellis JT, Jr, Nyberg K, Fersht AR. Energetics of complementary side-chain packing in a protein hydrophobic core. Biochemistry. 1989;28:4914–4922. doi: 10.1021/bi00437a058. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 16.Chan HS, Dill KA. Solvation: how to obtain microscopic energies from partitioning and solvation experiments. Annu Rev Biophys Biomol Struct. 1997;26:425–459. doi: 10.1146/annurev.biophys.26.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J Biol Chem. 2002;277:19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 18.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319–333. [Google Scholar]
- 19.Aoki T, Takahashi Y, Koch KS, Leffert HL, Watabe H. Construction of a fusion protein between protein A and green fluorescent protein and its application to western blotting. FEBS Lett. 1996;384:193–197. doi: 10.1016/0014-5793(96)00289-x. [DOI] [PubMed] [Google Scholar]
- 20.Aoki T, Miyashita M, Fujino H, Watabe H. A flexible single-step detection of blotted antigen using a fusion protein between protein A and green fluorescent protein. Biosci Biotechnol Biochem. 2000;64:1547–1551. doi: 10.1271/bbb.64.1547. [DOI] [PubMed] [Google Scholar]
- 21.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Zhu W, Kodadek T. Selection and application of peptide-binding peptides. Nat Biotechnol. 2000;18:71–74. doi: 10.1038/71951. [DOI] [PubMed] [Google Scholar]
- 23.Kajander T, Cortajarena AL, Regan L. Consensus design as a tool for engineering repeat proteins. Methods Mol Biol. 2006;340:151–170. doi: 10.1385/1-59745-116-9:151. [DOI] [PubMed] [Google Scholar]
- 24.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 25.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]


