Abstract
Viruses infecting hyperthermophilic archaea have intriguing morphologies and genomic properties. The vast majority of their genes do not have homologs other than in other hyperthermophilic viruses, and the biology of these viruses is poorly understood. As part of a structural genomics project on the proteins of these viruses, we present here the structure of a 102 amino acid protein from acidianus filamentous virus 1 (AFV1-102). The structure shows that it is made of two identical motifs that have poor sequence similarity. Although no function can be proposed from structural analysis, tight binding of the gateway tag peptide in a groove between the two motifs suggests AFV1-102 is involved in protein protein interactions.
Keywords: archaeal virus, function, structure, fold evolution, gateway tag
Introduction
Double strand DNA viruses from hyperthermophilic Archaea show exceptionally diverse and unusual morphotypes. Moreover, analysis of their genomes revealed that they are unrelated to other known viruses [reviewed in refs. 1–4]. Based on their morphological and genomic properties, they have been classified in seven novel viral families: rod-shaped Rudiviridae,5 filamentous Lipothrixviridae,6–9 spindle-shaped Fuselloviridae,10–12 droplet-shaped Guttaviridae,13 spherical Globuloviridae,14,15 bottle-shaped Ampullaviridae,16 and two-tailed Bicaudaviridae.17 Three more hyperthermophilic archaeal viruses have still not been classified: the icosahedral STIV,18 spindle-shaped STSV119 and PSV.20 The genomes of most isolated archaeal viruses have been sequenced showing that sequence similarities between genes of the different archaeal viral families are generally limited and most predicted genes yield good matches only with those of other members of the same family.4 Despite this wealth of genomic information, the biological knowledge of the biology of these viruses is still very limited and the function of the majority of the proteins encoded by their genomes is unknown. In cases where no functional information can be retrieved from sequence analysis, 3D-structure determination may offer an alternative and provide testable hypothesis on biochemical function. Our decision to embark upon a structural genomics project on proteins from Lipothrixviridae family viruses has two motivations: (1) the discovery of new protein folds (many of the protein sequences are orphans and for the vast majority no reliable structural model can be proposed) and (2) obtaining functional information through 3D-structure determination of the proteins. Here, we present the 3D crystal structure of a small protein (102 amino acids) encoded by acidianus filamentous virus 1 (AFV1-102), which has no sequence homologues and hence no predictable structure and function. We found that AFV1-102 consists of a repeat of a four-stranded β-sheet motif packed against a α helix, together forming a βαβ sandwich. The unexpected presence of the 19 N-terminal residues belonging to the linker between the protein and the 6× histidine purification tag in a groove enclosed by these two motifs suggests that AFV1-102 is involved in protein protein interactions.
Results and Discussion
Overall structure of AFV1-102
AFV1-102 crystallized in space group P6122 and the structure was solved at 1.95 Å resolution. The statistics on data collection and refinement are provided in Table I. Gel filtration chromatography showed that AFV1-102 is a monomer in solution (results not shown). The asymmetric unit contains a single copy of the AFV1-102 protein. AFV1-102 contains two four- stranded β sheets and three α helices forming an overall βαβ sandwich fold [Fig. 1(A,B)]. No structural analogs could be identified using DALI or the SSM server of EBI (http://www.ebi.ac.uk/msd-srv/ssm/). The βαβ sandwich can also be described as an assembly of two motifs with quasi identical structures that are related by pseudo twofold symmetry [Fig. 1(B)]. Each motif is composed of a α helix followed by a four stranded β sheet. One sheet is composed of β1β2β7β8 (motif 1) and the other of β3β4β5β6 (motif 2). AFV1-102 is not a repetition of the two motifs, since the second continuous motif is inserted between the second (β2) and third strand (β7) of motif 1. The main difference between the two motifs consists of an insertion of a third 14 residue helix (α2) between β4 and β5 in motif 2 not present in motif 1 [Fig. 1(C)]. Helix α3 (motif 1) and α1 (motif 2) connect two strands from different β sheets (α1 connects β2 to β3 and α3 connects β6 to β7). Both motifs pack together on their helical sides. Helices α1 and α3 have the same length and pack against the sheets via hydrophobic interactions. Furthermore, both helices are kinked near their C terminus. The residues in the kink of helix α1 form main chain hydrogen bonds with the residues in the connection to β3. In addition, a hydrogen bond between the NH1 of Arg43 located in the linker between β4 and α2, and the main chain carbonyl of Lys25 at C terminus of α1 may further stabilize the kink. The kink in helix α3 is stabilized by very similar interactions: main chain hydrogen bonds between the C terminus of the helix and the α3 β7 connections and a hydrogen bond between the NH1 of the C terminal Arg95 and the carbonyl of a Lys78 at C terminus of α3. Despite very low sequence identity (10%) the two motifs superpose well with an rms deviation of 1.34 Å for 29 aligned Cα positions [Fig. 1(C)]. The identical structures of the two motifs together with the very similar ways their helices are kinked and stabilized suggests that they originate from a common ancestral motif. AFV1-102 probably has evolved by duplication of an ancestral motif and subsequent insertion of motif 2 into motif 1.
Table I.
Data Collection and Refinement Statistics
| SeMet | Native | |
|---|---|---|
| Space group | P6122 | P6122 |
| Unit cell a,b,c (Å) | 62.36, 62.36, 111.27 | 62.89, 62.89, 110.84 |
| Resolution (Å) | 38.75–2.5 (2.64–2.5) | 55.38-1.95 (2.06–1.95) |
| Total number of reflexions | 51,952 (7457) | 347,114 (54,335) |
| Number of unique reflexions | 4860 (666) | 10,006 (1431) |
| Multiplicity | 10.7 (11.2) | 34.7 (38) |
| Rmerge | 0.11 (0.65) | 0.11 (0.59) |
| I/σI | 5.4 (1.1) | 4.4 (0.8) |
| Overall completeness | 100 (100) | 99.5 (100) |
| R/R free | 21.3/27.4 | |
| rmsd bond (Å) | 0.011 | |
| rmsd angle (°) | 1.31 | |
| Ramachandran plot (%) | ||
| Favored | 99.1 | |
| Allowed | 0.9 |
Figure 1.
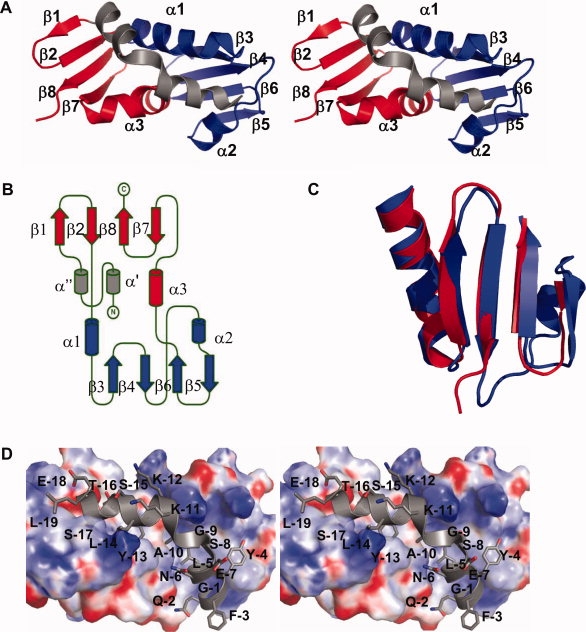
Structure of AFV1-102. (A) Stereo view of the cartoon representation of AFV1-102. The two motifs of the protein are colored in red and blue, the N-terminal tag is colored in gray. The secondary structure elements are indicated. (B) Topology of AFV1-102. Motifs and tag are colored as in A (diagram performed by Topdraw21). (C) Superposition of the two motifs of AFV1-102 in cartoon representation. (D) Focused stereo view on the interaction between the tag (in cartoon and sticks) and AFV1-102 (in electrostatic surface representation with 20% transparency). The residues of the tag are labeled with a one letter code (electrostatic surface calculation performed by APBS program22).
The gateway tag is firmly bound
No structural analogs could be identified for either the intact AFV1-102 protein or the separate motifs using DALI or the SSM server of EBI. Since no homologs are identified for AFV1-102, combining structural and sequence information could not be used to infer biochemical function. However, by serendipity the tag sequence of the genetic expression construct is strongly bound to the core protein in the crystal, suggesting that AFV1-102 is involved in protein protein interactions. The recombinant construction of AFV1-102 that yielded crystals still contains 30 residues at the N terminus encompassing the histidine tag, the attB site from the Gateway recombination system and the Tev protease cleavage site. After refinement of the structure, unexplained strong residual electron density was observed in a canyon lying between the two motifs. We could readily accommodate this residual density by a peptide that corresponds to 19 residues of the tag (sequence LESTSLYKKAGSENLYFQG, numbered from −19 to −1 in the structure). This peptide adopts a kinked helical conformation and packs snugly into the groove [Fig. 1(D)]. Seven residues from the tag (Leu-19, Ser-17, Thr-16, Leu-14, Tyr-13, Tyr-4, Leu-5) make contacts with the protein core, 3 of which are entirely buried (Ser-17, Tyr-13, Leu-5) [Fig. 1(D)]. Leu-19 interacts with Val44 on the edge of the groove; Ser-17 is inserted in a cleft made by Ile23, Leu26, Val28, and Trp49. In addition, the Nɛ1 from Trp49 makes a hydrogen bond with the Oγ of Ser-17. The methyl group of Thr-16 contacts Trp49, Leu52, and Leu72, whereas the Oγ1 and main chain N make hydrogen bonds with Asp48 OD2. Leu-14 mainly contacts Val22, Ile23, Leu26, and the aliphatic chain of Lys25. Tyr-13 is deeply inserted in the hydrophobic core of AFV1-102 in a cleft made by Met19, Val22, Ile23, Trp49, Phe69, and Val73. The OH of Tyr-13 points at the O of Met19 with which it makes a hydrogen bond. Leu-5 contacts Ile8, Ile10, Phe15, Met19, and Leu92. The aromatic cycle of Tyr-4 is not buried but makes contacts with Val3, Ile8, and Phe81 at the edge of the groove. At the C terminus of the tag Gln-2 NE2 makes hydrogen bonds with Ser18 OG and Glu14 O. Tyr-13 occupies a central position in the groove where it is inserted in a very hydrophobic pocket.
The Gateway recombination system is now routinely used for high throughput cloning. This cloning strategy adds a long tag to the target sequence and is usually cleaved after protein purification for structural studies. In our general structural genomics strategy, we decided to keep the tag to avoid the uncertain and time costly protease cleavage steps. Usually protein tags are disordered in the crystal, but this is clearly not the case in the present crystal structure. We could unambiguously determine the conformation of the gateway tag.
To determine the structure of AFV1-102 without the gateway tag, we tried two strategies. We incubated the tagged protein with Tev protease and alternatively we performed the cloning of AFV1-102 with a 6His tag attached at the C terminus. In both case it was not possible to produce soluble protein: the protein precipitated during tag cleavage and the 6His tag version of AFV1-102 was produced as inclusion bodies. This clearly shows that the gateway tag helps to solubilize the protein.
Search for an AFV1-102 partner
The unusual structuring of the tag and the many interactions it engages with the protein, suggests that the groove of AFV1-102 may be used for interaction with a protein partner. A Blast search with the tag-sequence (LESTSLYKKAGSENLYFQG) did not reveal nontrivial hits. However, in one of the proteins encoded by the AFV1 genome (AFV1-94), we identified the presence of a pentapeptide with sequence TSMYK. This peptide is very similar to that part of the gateway peptide (TSLYK) that strongly interacts with the core of AFV1-102. Experiments are underway to test whether AFV1-94 and AFV1-102 interact in solution. Straightforward testing of this interaction was hampered by the insolubility of nontagged AFV1-102.
Methods
Cloning, expression, and purification
The sequence of AFV1-102 was amplified by PCR from cDNA using two primers containing the attB sites of the Gateway recombination system (Invitrogen). The cDNA was cloned into the pDEST17 plasmid using the Gateway technology (Invitrogen).23 A TEV protease cleavage site encoding sequence was inserted between the attB1 and the AFV1-102 gene. Expression was done at 37°C using the E.coli Rosetta (DE3) pLysS strain and the 2xYT medium (BIO 101 Inc.). When the cell culture reached an OD600 nm of 1, overnight induction at 15°C was performed with 0.5 mM IPTG (Sigma). Cells were harvested by centrifugation and resuspended in buffer A (20 mM tris Tris-HCl pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol). Cell lysis was completed by sonication and the lysate was heated for 20 min at 50°C before centrifugation at 20,000 rpm The soluble fraction was loaded on a NiNTA column (Qiagen Inc.) equilibrated with buffer A. The protein was eluted with imidazole and loaded on a Superdex75 column (Amersham Pharmacia Biotech) equilibrated against buffer A with 10 mM β-mercaptoethanol. Selenomethionine substituted protein was produced and purified as the native protein. The homogeneity of the proteins was checked by SDS-PAGE.
Structure resolution
AFV1-102 native and seleno-substituted crystals were grown from a 1:1 μL mixture of protein (14 mg/ml) with 1.5–2M ammonium sulfate, 0.1M HEPES pH 7.5, 2% PEG400, using hanging drop method at 23°C. For cryoprotection, crystals were soaked in a mixture of mother liquor and 30% glycerol. Crystals were then flash frozen at 100 K.
X-ray diffraction data were collected from a SeMet substituted AFV1-102 crystal on beamline BM30A (ESRF) at the Se K-edge. The crystal diffracted to 2.5 Å and belongs to the P6122 space group with two molecules per asymmetric unit, corresponding to 41% solvent content. Native data were collected on the ID29 beamline (ESRF) to 1.95 Å resolution. Crystal belongs to P6122 space group with one molecule per asymmetric unit, corresponding to 41.7% solvent content. Data processing was carried out with the program MOSFLM24 and scaling and merging with SCALA.25
The structure was solved using SAD X ray diffraction data at 2.5 Å resolution. Four Selenium atom sites were found with the program SHELXD26 in the 20–3 Å resolution range. These sites were used for phasing with the program SOLVE.27 After solvent flattening with the program RESOLVE,27 the quality of the electron density map allowed automated construction of 90% of the model. This model was then refined against the 1.95 Å data set with the Arp/Warp28 program that allowed automated building of the missing residues. The model was refined with REFMAC529 and manually corrected using O.30 19 residues of the tag and the 95 first residues are well defined in electron density map and fall within the allowed regions of the Ramachandran plot, as defined by Molprobity.31
Conclusions
The structure of AFV1-102 for which no homologues could be identified reveals a new βαβ sandwich that was created by duplication of two almost identical motifs. No biological function could be deduced from the structure, but very clear binding of the tag peptide into an interdomain groove suggests that AFV1-102 interacts with a protein partner.
Coordinates
Coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 2WB6.
References
- 1.Ortmann AC, Wiedenheft B, Douglas T, Young M. Hot Crenarchaeal viruses reveal deep evolutionary connections. Nat Rev Microbiol. 2006;4:520–528. doi: 10.1038/nrmicro1444. [DOI] [PubMed] [Google Scholar]
- 2.Prangishvili D, Garrett RA. Viruses of hyperthermophilic Crenarchaea. Trends Microbiol. 2005;13:535–542. doi: 10.1016/j.tim.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nat Rev Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 4.Prangishvili D, Garrett RA, Koonin EV. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 2006;117:52–67. doi: 10.1016/j.virusres.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Prangishvili D, Klenk HP, Jakobs G, Schmiechen A, Hanselmann C, Holz I, Zillig W. Biochemical and phylogenetic characterization of the dUTPase from the archaeal virus SIRV. J Biol Chem. 1998;273:6024–6029. doi: 10.1074/jbc.273.11.6024. [DOI] [PubMed] [Google Scholar]
- 6.Janekovic D, Wunderl S, Holz I, Zillig W, Gierl A, Neumann H. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic sulfur reducing archaebacterium Thermoproteus tenax. Mol Gen Genet. 1983;192:39–45. [Google Scholar]
- 7.Bettstetter M, Peng X, Garrett RA, Prangishvili D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology. 2003;315:68–79. doi: 10.1016/s0042-6822(03)00481-1. [DOI] [PubMed] [Google Scholar]
- 8.Vestergaard G, Aramayo R, Basta T, Haring M, Peng X, Brugger K, Chen L, Rachel R, Boisset N, Garrett RA, Prangishvili D. Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J Virol. 2008;82:371–381. doi: 10.1128/JVI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA. 1992;89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stedman KM, She Q, Phan H, Arnold HP, Holz I, Garrett RA, Zillig W. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res Microbiol. 2003;154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 11.Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol. 2004;78:1954–1961. doi: 10.1128/JVI.78.4.1954-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold HP, Ziese U, Zillig W. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology. 2000;272:409–416. doi: 10.1006/viro.2000.0375. [DOI] [PubMed] [Google Scholar]
- 13.Haring M, Peng X, Brugger K, Rachel R, Stetter KO, Garrett RA, Prangishvili D. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology. 2004;323:233–242. doi: 10.1016/j.virol.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Ahn DG, Peng X, Brugger K, Rachel R, Stetter ICC, Garrett RA, Prangishvili D. TTSV1, a new virus-like particle isolated from the hyperthermophilic crenarchaeote Thermoproteus tenax. Virology. 2006;351:280–290. doi: 10.1016/j.virol.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 15.Haring M, Rachel R, Peng X, Garrett RA, Prangishvili D. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol. 2005;79:9904–9911. doi: 10.1128/JVI.79.15.9904-9911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haring M, Vestergaard G, Rachel R, Chen L, Garrett RA, Prangishvili D. Virology: independent virus development outside a host. Nature. 2005;436:1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- 17.Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang X, Chen L, Huang L, Luo Y, She Q, Huang L. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol. 2005;79:8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geslin C, Le Romancer M, Gaillard M, Erauso G, Prieur D. Observation of virus-like particles in high temperature enrichment cultures from deep-sea hydrothermal vents. Res Microbiol. 2003;154:303–307. doi: 10.1016/S0923-2508(03)00075-5. [DOI] [PubMed] [Google Scholar]
- 20.Leslie AGW. Joint CCP4 and EACMB newsletter protein crystallography. Warrington, United Kingdom: Daresbury Laboratory; 1992. p. 26. [Google Scholar]
- 21.Bond CS. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19:311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
- 22.Holst M, Saied F. Numerical solution of the nonlinear Poisson-Boltzmann equation: developing more robust and efficient methods. J Comput Chem. 1995;16:337–364. [Google Scholar]
- 23.Walhout AI, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 24.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Part 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 25.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58(Part 10, Part 2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Part 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56(Part 8):965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Part 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: phi, psi, and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]


