Abstract
DsrA is an 87-nucleotide regulatory RNA of Escherichia coli that acts in trans by RNA–RNA interactions with two different mRNAs, hns and rpoS. DsrA has opposite effects on these transcriptional regulators. H-NS levels decrease, whereas RpoS (σs) levels increase. Here we show that DsrA enhances hns mRNA turnover yet stabilizes rpoS mRNA, either directly or via effects on translation. Computational and RNA footprinting approaches led to a refined structure for DsrA, and a model in which DsrA interacts with the hns mRNA start and stop codon regions to form a coaxial stack. Analogous bipartite interactions exist in eukaryotes, albeit with different regulatory consequences. In contrast, DsrA base pairs in discrete fashion with the rpoS RNA translational operator. Thus, different structural configurations for DsrA lead to opposite regulatory consequences for target RNAs.
Keywords: natural antisense, RNA turnover, RNA–RNA interactions, structural dynamics, translation
RNA plays a variety of regulatory roles in the cell, in addition to its central function in translation processes. These activities are collectively termed riboregulation (1). RNA–RNA interactions provide a basis for sequence-specific RNA regulation of other RNAs in both prokaryotes and eukaryotes (2, 3). DsrA, an 87-nt untranslated RNA in Escherichia coli, is one such regulatory RNA. DsrA contains regions of sequence complementarity to at least five different genes, hns, argR, ilvIH, rpoS, and rbsD (4), and it has been demonstrated to regulate two of these genes, hns and rpoS, by RNA–RNA interactions (4, 5).
H-NS is an abundant nucleoid-structuring protein with global transcriptional repressor functions (6, 7). DsrA antagonizes H-NS function by decreasing the levels of H-NS protein in the cell (4). In contrast, the translation of RpoS, the stationary phase and stress-response sigma factor (8), is enhanced by DsrA, especially at low temperatures (9). Thus, DsrA has opposite effects on these two targets, both mediated by RNA–RNA interactions, with global regulatory consequences for the transcriptional state of the cell. Whereas the mechanism of DsrA action at hns is not known, DsrA binds the translational operator of rpoS (4, 5) to open a stable stem-loop of rpoS RNA (10), enabling access to the Shine–Dalgarno sequence and thus enhancing translation.
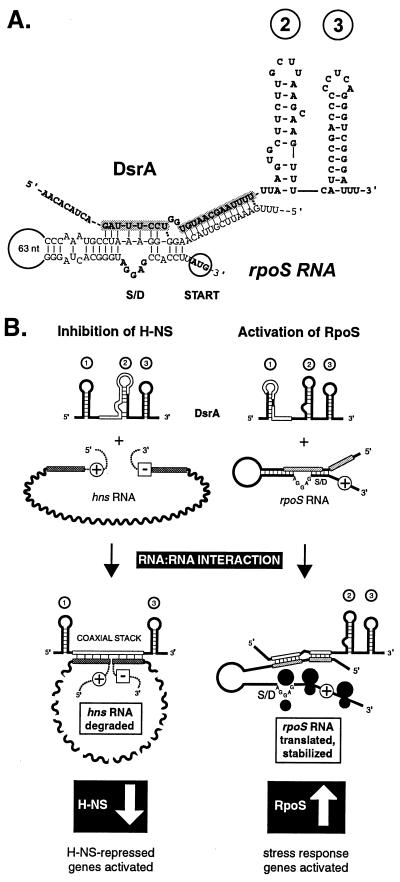
Structure predictions based on thermodynamic criteria suggest that DsrA consists of three stem-loops, the third of which is the transcription terminator of DsrA (Fig. 1A; ref. 11). The hns complementary region, in the center of the molecule, resides within the predicted second stem-loop, whereas the rpoS complementary region occupies the predicted first stem-loop and the base of the second stem (4, 5). However, no additional structural data are currently available for DsrA. Mapping of the different regions of complementarity on the structure of DsrA is important for understanding how such a small RNA molecule interacts with multiple targets, and how such interactions might be regulated.
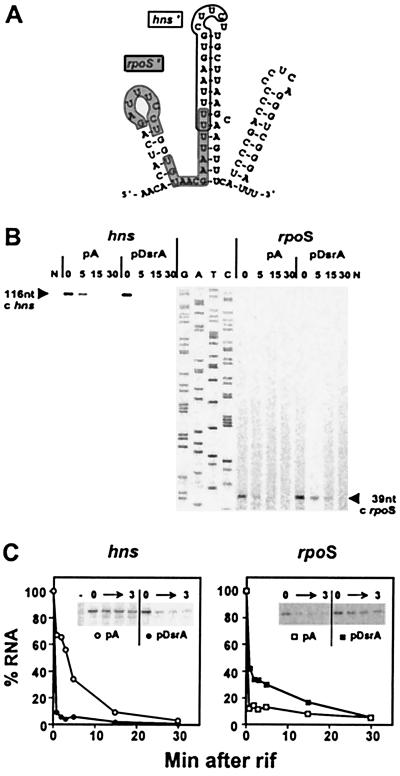
Figure 1.
Primer extension analysis to monitor RNA stability. (A) Computer-predicted structure of DsrA RNA (11). The rpoS-complementary region (rpoS′) is shown on a gray background; the hns-complementary region (hns′) is outlined (4). (B) Primer extension analysis of hns and rpoS mRNAs. RNA from cells containing a plasmid (pDsrA) was compared with a vector control (pA). Numbers indicate the time (min) after rifamycin addition (0 min). N, no-RNA control. A DNA sequencing ladder (G, A, T, C) was used as a mobility marker. (C) RNA stability plots. cDNA levels from primer extension analyses were quantified by PhosphorImager and plotted relative to the zero time point for each experiment. Each data point represents the average of two experiments. The decrease in hns mRNA half-life in the presence of excess DsrA is >8-fold. The increase in rpoS mRNA stability in the presence of excess DsrA is ≈3-fold. Gels inset into the graphs are primer extension analyses of 0–3 min time points.
Here we show that the basis of DsrA regulation at hns involves enhanced turnover of hns mRNA, perhaps by blocking H-NS translation, whereas the translational stimulation of RpoS involves stabilization of rpoS mRNA. The secondary structure of DsrA, determined in the absence and presence of hns RNA in vitro, suggests a distinct DsrA–rpoS interaction involving stem-loop 1, whereas a two-part DsrA–hns interaction involves stem-loop 2. The DsrA–hns RNA interactions are proposed to circularize hns mRNA and define a structural basis of DsrA activity at hns. Interestingly, similar structures can be drawn for DsrA:argR and DsrA:ilvIH RNA–RNA interactions. Taken together, these data show DsrA to be a dynamic RNA molecule that changes its structure and interactions to play different regulatory roles.
Materials and Methods
Strains, Plasmids, and Culture Conditions.
E. coli M182 (4, 12) served as the reference strain. Plasmids pA (pACYC184) and pDsrA are described elsewhere (9, 13). The plasmid pT7hns was provided by Qingyun Liu (Harvard Medical School, Boston, MA). All cells were grown in TBYE medium (14) at 30°C, with antibiotics (200 μg/ml ampicillin or 25 μg/ml chloramphenicol) as appropriate.
Computational Analyses.
The sequences of hns, argR, and ilvIH were compared with DsrA sequences by using the programs gap and findpatterns in the Seqlab Suite of sequence analysis programs (Genetics Computer Group, Madison, WI). Sequences of DsrA from Salmonella typhimurium (GenBank accession no. AF090431) and Klebsiella pneumoniae (11) were aligned manually on the E. coli DsrA model secondary structure described here.
RNA Stability Primer Extension Assay.
Cells were grown to an OD650 of 0.2–0.3 and rifamycin was added to 300 μg/ml. Aliquots were taken immediately before and at time intervals after rifamycin treatment. RNA was extracted and primer extension analysis performed as described (14) by using 5′-end-labeled gene-specific primers W166 (hns; ref. 12) and W1246 (rpoS; 5′-GTTGTTCGGCAACATTTTGTTAGC-3′). Primer labeling and DNA sequencing molecular size standards are as described (4). A stable transcript (ompA) was used as an internal control by primer extension with oligonucleotide W912 (5′-GCGCCTCGTTATCATCC-3′; data not shown).
DsrA Substrate Construction.
A “double ribozyme” plasmid (15) was constructed to generate a DsrA RNA substrate in vitro with precisely defined 5′ and 3′ ends. The sequence “GGAA” was added to the 5′ end of DsrA to facilitate labeling. DsrA was amplified by PCR with two oligonucleotides, one containing the sequence of a hammerhead ribozyme (W1187, 5′-CCAAGGGCCCGGGAGATGATGTCCCTGATGAGTCCGTGAGGACGAAACGGTACCCGGTACCGTCGGAACACATCAGATTTCC-3′), and the other containing the VSs1 substrate portion of a trans-acting VS ribozyme variant (W1186, 5′-CGCGGATCCTCGGGGCGACGACGCCCTTAAAATCCCGACCCTGAGG-3′). The PCR product was cloned by using the vector pBSII SK(−) (Stratagene) by digestion with ApaI and BamHI. Candidate clones of the double ribozyme plasmid pBS-DsrA-DRz were screened by restriction analysis and verified by DNA sequencing.
DsrA and hns RNA Preparation In Vitro.
Plasmid templates were linearized by digestion with restriction enzymes as follows: pBS-DsrA-DRz with BamHI, pAvaRzA-3 (VS-AvaRz, ref. 17) with SspI, and pT7hns with BamHI and BglII. Linearized templates were gel purified. The hns RNA was transcribed separately, whereas DsrA-DRz and the VS-AvaRz were cotranscribed (15), by using T7 RNA polymerase (BRL) according to the manufacturer's instructions. DNA templates were removed by DNase I (BRL) digestion at 37°C for 10 min and RNA was extracted and ethanol precipitated. The double-ribozyme RNA was incubated in VS reaction buffer (50 mM Tris·HCl, pH 8.0/2 mM spermidine/25 mM KCl/20 mM Mg Cl2; ref. 17) at 42°C for 1 h to promote ribozyme reactions. RNAs were desalted over Sephadex G-50 equilibrated in diethyl pyrocarbonate-treated water (DEPC-water) (18). The hns RNA was concentrated by centrifugation under vacuum, resuspended in 40 μl of DEPC-water, and quantified by spectrophotometry at 260 nm. The integrity of RNAs was monitored by fractionation on denaturing polyacrylamide gels, and visualized by staining with ethidium bromide after soaking the gel in 1× TBE running buffer (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.3) to remove urea. The hns RNA was used without further purification.
The 3′ cyclic phosphate of DsrA produced during the second ribozyme reaction (15) was removed by treatment with T4 kinase in T4 kinase buffer without ATP (ref. 19; R. A. Collins, personal communication), before denaturing polyacrylamide gel purification. RNA was eluted from excised bands into DEPC-water by heating at 60°C for 1 h (R. A. Collins, personal communication). After centrifugation, the supernatant was desalted, aliquoted, and concentrated as above.
Labeling of Purified DsrA.
DsrA (5 pmol) was 5′ end-labeled with [γ-32P]ATP (NEN) and T4 kinase at 55°C after heating (70°C for 1 min) and snap-cooling on ice. An equivalent amount of DsrA was 3′ end-labeled with [α-32P]5′,3′ bis-phosphate (pCp; NEN) and T4 RNA ligase as described (20) except that the ATP concentration was 0.1 μM (16, 21). Labeled RNA was extracted, and unincorporated nucleotides were removed by desalting. Radioactive incorporation was measured by Cerenkov scintillation counting. RNA was aliquoted, concentrated as above, stored dry, and rehydrated before use.
Nuclease Cleavage Assays.
Labeled DsrA (2.5 × 104 cpm) was digested under denaturing conditions as described (22) except that RNase U2 was used in the buffer described by the manufacturer (Amersham Pharmacia). For denaturing conditions, 0.5 units of RNase T1, 2.5 units of RNase U2, or 0.5 ng of RNase A were used. For nondenaturing conditions, RNA was resuspended in the appropriate buffer, heated (70°C for 1 min), and snap-cooled on ice. Where appropriate, 85 ng/μl hns RNA was added before renaturation. Reactions were incubated for 10 min at 16°C and stopped by the addition of one volume of 95% formamide/dye mix (18) premixed with 1/10 volume of 100 mM aurin tricarboxylic acid (Sigma) in 50 mM Tris⋅HCl pH 8.0. For nondenaturing conditions, RNase T1 (0.05 units) was used in T1 sequencing buffer without urea (22), RNase V1 (0.024 units) was used in the buffer recommended by the manufacturer (Amersham Pharmacia), and S1 nuclease (0.115 units) was used in the buffer supplied by the manufacturer (BRL).
Results
DsrA Affects the Turnover of Its Target mRNAs.
Previously, DsrA was shown to decrease the levels of H-NS protein, but not steady-state hns mRNA levels (4). Because H-NS represses its own transcription (6), a decrease in H-NS protein should lead to increased hns mRNA levels. How, then, does DsrA block synthesis of H-NS? Curiously, DsrA decreases the steady-state mRNA levels of a Δhns null mutant, in which autoregulation is abolished (4). Therefore, we hypothesized that DsrA exerts an effect on hns RNA turnover. To test the effects of DsrA on RNA turnover, the initiation of RNA synthesis was inhibited with rifamycin and RNA levels were assayed by primer extension analysis. When overproduced from a plasmid (pDsrA), DsrA substantially decreased hns mRNA stability relative to vector (pA) controls (Fig. 1B Left). Quantitation of mRNA levels taken from 1 to 30 min after addition of rifamycin shows that hns mRNA levels dropped immediately after rifamycin addition, with a sharp decrease in mRNA half-life, from ≈4 min to <0.5 min (Fig. 1C Left), a greater than 8-fold difference.
DsrA acts at rpoS by enhancing RpoS translation (9) via RNA–RNA interactions that antagonize the rpoS translational operator (4, 5, 10). Here we show that DsrA overproduction also increases rpoS mRNA stability (Fig. 1B Right). The levels of rpoS mRNA were stabilized relative to the control. Although the relative stabilization was difficult to determine because of the short half-life of rpoS mRNA, we estimate an approximately 3-fold increase in rpoS stability (Fig. 1C Right), in sharp contrast to the effect of DsrA on hns mRNA (Fig. 1C Left).
Computational Analysis Supports a Putative Second DsrA–hns Interaction.
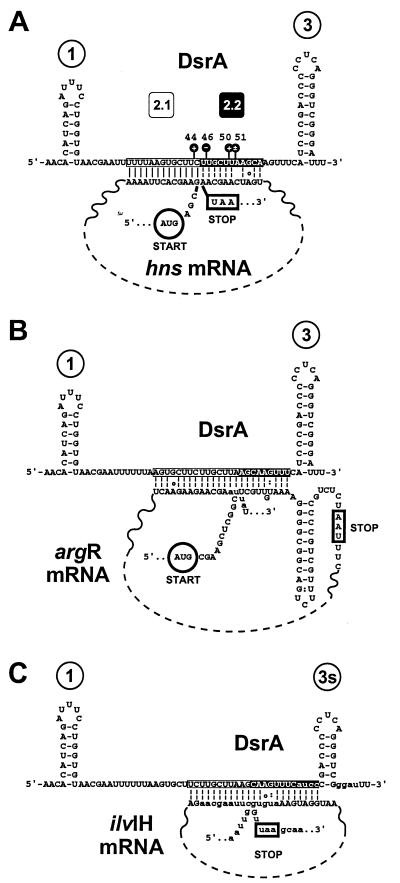
The DsrA-complementary region within hns described (Figs. 1A and 2A, boxed sequence) was initially found by computer alignment and verified by compensatory mutagenesis (4). A computational reexamination of the hns coding sequence revealed a second potential interaction of hns with DsrA (Fig. 2A, white letters on a black background). Interestingly, whereas the previously described complementary sequence is near the start codon for hns translation, the second region is near the hns stop codon (Fig. 2A). Base pairing between DsrA and the two regions of hns would form a contiguous coaxial stack, looping out the middle part of hns mRNA (Fig. 2).
Figure 2.
DsrA–RNA interactions in the middle stem-loop. (A) Computer sequence alignment of the hns coding sequence depicted as a coaxial stack with regions of DsrA. The two regions of DsrA complementary to hns are depicted as boxed (region 2.1) and as white letters on a black background (region 2.2). Nuclease susceptibility (black lollipops) and numbers over stem-loops are as in Fig. 3. (B and C) Other RNAs modeled as coaxial stacks. The argR (B) and ilvIH (C) RNAs of E. coli with DsrA complementarity. Because of overlap in the DsrA complementary sequences, there is potential for different pairings within coaxial stacks (lowercase letters). Vertical lines represent base pairs verified by compensatory mutagenesis (4); dashed vertical lines represent base pairs predicted by informatic or biochemical analyses; a colon (:) represents a G:U base pair; ○ represents a G:A base pair. Start codons are circled; stop codons are boxed.
The putative interaction between DsrA and the hns stop codon region encouraged us to reexamine other genes by computer sequence alignment with DsrA. As with hns, DsrA has the potential to pair with both argR and ilvIH mRNAs near the start codon (ref. 4; Fig. 2 A–C, boxed sequences). Strikingly, computational analysis revealed that in analogy to hns, DsrA also contains extended sequences with potential to base pair with both the argR and ilvI stop codon regions (compare Fig. 2 A–C, white letters on a black background). Similarly to hns mRNA, coaxial stacks can be modeled on the two-part DsrA:argR and DsrA:ilvIH mRNA sequence complementarities (Fig. 2 B and C), lending a common theme to all three potential RNA–RNA interactions involving the DsrA middle stem-loop. In all three cases, the stop codons are near the putative DsrA interactions. Although the effects of DsrA on argR and ilvlH are not known, the theme of base pairing interactions with the start and stop codon regions by the central region of DsrA could represent a general mode by which DsrA regulates its target genes.
Structure of DsrA Inferred from Nuclease Footprinting.
DsrA interacts with the mRNAs of both hns and rpoS by RNA–RNA interactions that involve distinct regions of DsrA (4, 5). To determine the structural basis of these phenomena, the conformation of DsrA in solution was deduced by RNase footprinting (23) with single-strand-specific endonucleases, RNase T1 and S1 nuclease, and the double-strand-specific RNase V1. For this purpose, we produced a DsrA substrate with precisely defined 5′ and 3′ ends via a double ribozyme construct with DsrA in the middle (15). Cleavage by nucleases can induce conformational rearrangements that lead to secondary cleavages, complicating RNA structural analysis (23). Accordingly, it was necessary to compare nuclease footprinting of both 5′- and 3′-labeled substrates to obtain a consensus structure (Fig. 3 A and B).
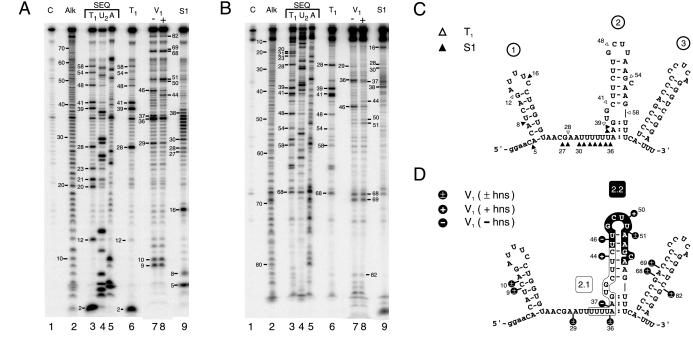
Figure 3.
Nuclease footprinting of DsrA. DsrA substrate RNA produced and end-trimmed in vitro was labeled at the 5′ or 3′ end, then treated with ribonucleases. Digestion products are numbered by phosphodiester bond position from the 5′ end. (A) Footprinting of 5′-labeled DsrA. (B) Footprinting of 3′-labeled DsrA. Lane numbers are given below, with reactions specified above. Lane C, untreated control RNA; Alk, alkaline hydrolysis ladder; lanes 3–5, sequencing reactions performed under denaturing conditions; and lanes 6–9, specific nuclease footprinting reactions performed under nondenaturing conditions. Lanes 3 and 6, RNase T1; lane 4, RNase U2; and lane 5, RNase A; lanes 7 and 8, double-strand-specific RNase V1 reactions performed in the absence or presence, respectively, of unlabeled hns RNA; and lane 9, S1 nuclease. (C and D) Predicted secondary structure of DsrA based on single-strand- and double-strand-specific nuclease susceptibility. (C) Single-strand-specific nuclease susceptibility. ▵, RNase T1; and ▴, S1 nuclease. (D) Double-strand-specific nuclease susceptibility. The hns-complementary region characterized (4) is shown boxed (region 2.1) whereas the newly proposed hns-complementary sequence (region 2.2) is shown in white letters on a black background. Cleavage sites are indicated by black lollipops. +, Cleavage only in the presence of hns RNA; −, cleavage only in the absence of hns RNA; ±, cleavage occurs in the presence or absence of hns RNA; and :, partial double-strand character.
RNase T1 cleaves after unpaired guanosine residues, whereas residues within helices are unaffected (Fig. 3 A and B, compare lanes 3 and 6; Fig. 3C). For example, nucleotide positions 39 and 41 are especially sensitive to RNase T1 when DsrA is in either denatured or native form, suggesting that this region is single stranded under native conditions. In contrast, positions 20, 21, and 23 are cleaved only under denaturing conditions, and must therefore be double stranded in solution. The structural information derived from these analyses is summarized in Fig. 3C (▵).
RNase V1 cleaves double-stranded RNA of 4–6 bp, although not all such sequences are cleaved (23, 24). Digestion with RNase V1 confirms the existence of several regions of double-stranded structure within DsrA, notably stem-loop 1 (Fig. 3A lane 7, nucleotides 9–10; Fig. 3D) and stem-loop 3 (Fig. 3 A, B, and D, nt 68, 69, and 82; additional data not shown), confirming the computed thermodynamic predictions. Comparison of T1 and V1 nuclease cleavage patterns suggests the structure for stem-loop 2 depicted (Fig. 3C, positions 39 and 41, and Fig. 3D, positions 37 and 46), with an unstructured stretch between stem-loops 1 and 2, except at position 29 (compare Fig. 3 C and D).
We further characterized DsrA secondary structure by treatment with S1 nuclease, which cleaves single-stranded RNA without sequence preference (23). The S1 nuclease data confirmed the loop region for stem 1, because cleavage at position 16 (Fig. 3 A and B, lanes 9) is consistent with RNase T1 and V1 analyses and thermodynamic predictions (Fig. 3 C and D). DsrA was also cleaved by S1 nuclease in the predicted single-strand region between stems 1 and 2 (Fig. 3 A and B, lanes 9, nucleotide positions 27–38), with the exception of position 29. This corresponds precisely with V1 nuclease cleavage at position 29, indicating double strandedness, although the pairing partner at position 29 is not known.
There is an apparent contradiction between the S1 nuclease and RNase V1 cleavage data for positions 36 and 37 (compare Fig. 3 C and D). Positions 37–38 are therefore likely to be at least transiently double stranded (Fig. 3 C and D, colons between residues). It is well known that S1 nuclease is extremely active and can cleave regions of double-stranded nucleic acid that transiently unpair or “breathe” (18, 25).
A 3′-labeled DsrA substrate behaves similarly to the 5′-labeled substrate, verifying this analysis (Fig. 3B). In summary, the consensus secondary structure map (Fig. 3 C and D) confirms previously predicted stem-loops 1 and 3 (11), whereas suggesting a different structure for stem-loop 2. Furthermore, the data suggest a dynamic or partial double-strand character at the base of stem-loop 2.
Change in DsrA Structure Induced by hns RNA in Vitro.
As mentioned, DsrA forms a RNA–RNA interaction with hns mRNA, decreasing the levels of H-NS protein (4), evidently by enhancing RNA turnover (Fig. 1). To examine the structure of this DsrA–hns RNA complex, excess unlabeled hns RNA was mixed with end-labeled DsrA and the complex was treated with RNase V1 to look for variations in double-stranded RNA regions. Whereas one main region of double-stranded RNA exists in the central part of DsrA in the absence of hns RNA (Fig. 3A, lane 7, nt position 46), two enhanced nuclease-susceptible regions of double-stranded RNA form on addition of hns RNA (Fig. 3 A and B, compare lanes 7 and 8, nt positions 44 and 50–51). An equivalent result was seen by using 3′-labeled substrate (Fig. 3B, compare lanes 7 and 8, nt positions 44 and 50–51; additional data not shown). The changes in DsrA associated with the hns RNA interaction are summarized (Fig. 2A and 3D, black lollipops). As expected, the DsrA–hns RNA interaction at position 44 of DsrA is within the previously described complementary region (Fig. 3D, region 2.1, boxed sequence; compare Fig. 1A; ref. 4). Interestingly, the hns-induced double-stranded complex at positions 50–51 lies outside of this region, suggesting a second DsrA–hns RNA interaction that includes the loop region of stem-loop 2 (Fig. 3D, nt positions 50–51), in agreement with computer predictions (Fig. 2A, and Fig. 3D, region 2.2).
Phylogenetic Analysis.
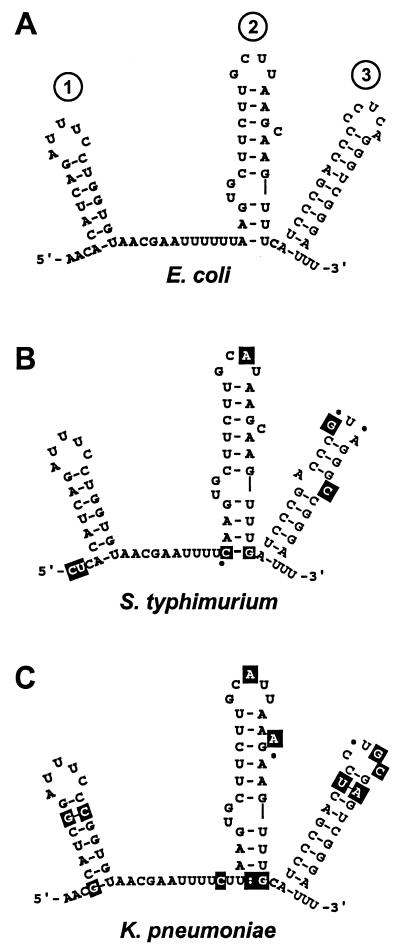
DsrA sequences are available from three species, E. coli, S. typhimurium, and K. pneumoniae. These sequences and their predicted structures (Fig. 4) are consistent with our structural analyses. Notably, sequence variations at the bottom of stem-loop 2 that extend the stem by one base pair are suggested in both the S. typhimurium and K. pneumoniae sequences (Fig. 4 B and C, white letters on a black background). Additional changes in both S. typhimurium and K. pneumoniae sequences relative to the inferred E. coli structure mostly occur in regions predicted to be single stranded, or when in regions predicted to be double stranded, are compensated by second-site mutations that would restore base pairing (Fig. 4 B and C, white letters on black background). This is the case for 23 base changes of S. typhimurium and K. pneumoniae DsrA relative to E. coli DsrA. The only exception to these structural constraints on sequence differences from E. coli DsrA is the single-nucleotide substitution that creates a bulge in the third stem-loop of S. typhimurium DsrA (Fig. 4B).
Figure 4.
Phylogenetic structure analysis. Structure models of DsrA are given for E. coli (A), S. typhimurium (B), and K. pneumoniae (C). Base changes relative to the E. coli sequence are shown as white letters in a black box, and dots indicate deletions.
DsrA Interactions with rpoS RNA.
The interaction of E. coli DsrA with rpoS mRNA is also consistent with the new DsrA structure model derived from RNA footprinting analysis (Fig. 3 C and D, and Fig. 5A). Here, in contrast both to the previous DsrA structure model (Fig. 1A) and to the DsrA–hns mRNA interaction (Fig. 2A), stem-loop 2 is predicted to remain intact, whereas stem-loop 1 would melt out and, together with the single-strand stretch between stem-loops 1 and 2, contact rpoS mRNA (Fig. 3 C and D, and Fig. 5). Thus, formation of RNA–RNA interactions with DsrA targets would involve distinct annealing processes and different DsrA conformations.
Figure 5.
Models of regulation. (A) DsrA shown with the rpoS translational operator. The structure of the rpoS translational operator (10) is shown with the new DsrA structure model emphasizing the integrity of stem-loop 2. The DsrA–rpoS RNA interaction also is described elsewhere (4, 5). (B) DsrA forms a complex with hns mRNA (Left) and with rpoS mRNA (Right). The hns- and rpoS-complementary regions in DsrA are outlined in white. The start codons of hns and rpoS (plus sign in circle) and the stop codon of hns (minus sign in box) are shown. The DsrA–hns RNA interaction is shown below as a coaxial stack, with the second DsrA stem-loop melted and the first and third stem-loops intact. In contrast, the DsrA complex with rpoS mRNA has the first DsrA stem-loop melted out and the second and third stem-loops intact. Stem-loops are numbered in circles.
Discussion
DsrA RNA acts as an orchestrator of cellular gene expression by simultaneously modulating the activities of at least two global transcriptional regulators, H-NS and RpoS (Fig. 5B), with potential for direct interactions with the RNAs of at least three more genes, argR, ilvIH, and rbsD (4). A revised secondary structure model for DsrA, based on compelling RNA footprinting and phylogenetic data (Figs. 3 and 4), accounts for its differential regulation of hns and rpoS. Remarkably, DsrA is a modulator of both RNA stability (Fig. 1) and translation (4, 9). DsrA decreases H-NS protein levels, whereas it increases RpoS levels, with a corresponding decrease in hns mRNA stability in vivo (>8-fold) and an increase in stability of rpoS mRNA, approximately 3-fold (Fig. 1). As discussed below, RNA stability may reflect the translational status of the RNA.
DsrA Structural Dynamics.
DsrA performs disparate functions by forming alternative structures in complex with target mRNAs (Fig. 2, 3, and 5). Whereas footprinting confirmed stem-loops 1 and 3 of the structure originally proposed for DsrA (11), the analysis predicts a different structure for stem 2 (Fig. 3). Additionally, a coaxial stack proposed between hns RNA and DsrA is postulated as a basis for DsrA regulation of the hns message (Fig. 2). However, whereas compensatory mutagenesis confirmed the 2.1 base-pairing region (4), similar experiments have not proven the region 2.2 pairing (R.L., D. Smith, and M.B., unpublished results). Whereas this RNA–RNA interaction is clearly seen in vitro, the conditions are not yet understood for its formation in vivo.
Interestingly, the DsrA–hns RNA interaction at the hns stop codon (Fig. 2A) is paralleled by potential RNA–RNA interactions at the argR and ilvI stop codons, which would represent second interactions for DsrA on these mRNAs (Fig. 2 B and C). All three of these proposed RNA–RNA interactions at stop codons contain a nonWatson–Crick G:A base pair (Fig. 2, open circles), which has been seen in other RNA–RNA interactions (26), particularly in coaxial stacks with single mismatches (27). The formation of a coaxial stack is expected to stabilize both parts of these RNA–RNA interactions (Fig. 2).
A coaxial contiguous helix between hns and DsrA would loop out the central portion of hns mRNA (Fig. 2A). In this model, the first and third stem-loops of DsrA remain intact whereas the central stem-loop melts during formation of the DsrA–mRNA interaction. The effect of this RNA–RNA interaction at hns would be to enhance the turnover of hns mRNA (Fig. 1 B and C), perhaps by creating ribonuclease-sensitive sites in hns mRNA, or by preventing translation, which would otherwise protect the mRNA from degradation (Fig. 5B Left). In this regard, however, we find that RNase III is dispensible for DsrA activity at hns (R.L. and M.B., unpublished results). Cleavage at the 5′ or 3′ end could facilitate degradation (28), whereas cleavage at the 5′ end could abolish translation by eliminating initiation. Such a degradation model would prevent translation and thereby offset increases in hns RNA levels caused by reduced H-NS protein levels, because of H-NS autoregulation (4). The proximity of both hns and argR start and stop codons also suggest inhibition of translation. Conversely, in eukaryotic systems, interactions between the 5′ and 3′ ends of mRNAs are proposed to enhance translation, although the requirement for circularization in vivo has not been fully elucidated (29).
In contrast to the interaction with hns, it is the first stem-loop of DsrA that is complementary to rpoS, and which forms the RNA–RNA interaction with rpoS mRNA (Fig. 5A, and Fig. 5B Right; refs. 4, 5). Elucidation of the structure of DsrA (Fig. 3) suggests that stem-loop 2 can remain intact while base paired to rpoS mRNA (Fig. 5 A and B). Although it is not yet known whether DsrA stabilizes rpoS directly or indirectly, we favor the hypothesis that enhanced translation reduces mRNA turnover (30).
What Governs the Dynamic Interaction of DsrA with Its Targets?
Loops and single-strand stretches of RNA mediate initiation of RNA–RNA interactions (31). Sequences within DsrA complementary to both rpoS and hns reside in regions of DsrA that appear either in loops or single-stranded regions (Figs. 3 C and D, 4, and 5A), suggesting that “kissing complexes” might form between DsrA and target mRNAs, leading to the RNA–RNA interaction. The separation of two disparate functions of DsrA (Fig. 1) into distinct structural elements (Figs. 3–5) forms a basis both for strand–strand interactions and potential regulation of these interactions (Fig. 5B).
RNA-binding proteins are also likely to be involved in the regulatory activity of DsrA. Many small RNAs require a protein component for activity, and function as ribonucleoprotein particles, as in the case of tmRNA (32), RNase P, and eukaryotic small nuclear ribonucleoproteins (33). The RNA chaperone Hfq (HF-I) has been shown to be required for opening of the S. typhimurium rpoS translational operator (10), and enhances DsrA action at rpoS RNA (D. Sledjeski, personal communication). Hfq involvement has also been proposed for the activity of OxyS RNA at rpoS (34). Whereas some hfq strains accumulate DsrA (D. Sledjeski, personal communication), in our strains hfq-null mutations markedly decrease levels of DsrA RNA, complicating analysis of the role of this protein with DsrA in vivo (R.L. and M.B., unpublished results).
What advantage is there in having a RNA, not a protein, as a regulator? Because of the specificity of base pairing interactions, DsrA can target multiple independent sites with economy, because the small RNA requires little energy to produce and is extremely stable, with a half-life of > 30 min (R.L. and M.B., unpublished results). Because of its small size and stability, DsrA could rapidly accumulate in response to different environmental signals, to enhance or inhibit turnover and/or translation of existing specific RNAs, and persist to maintain this altered state.
The plethora of roles played by RNA, widely distributed and acting in diverse ways, speaks not only to the flexibility of RNA as a biopolymer but also to an evolutionary history rooted in a RNA or RNA–protein world (33). Riboregulation is a form of regulation that, at minimum, requires only RNA–RNA interactions. RNA governing turnover and/or translation of RNA by changing both its conformation and that of its targets would be a form of regulation that we could expect to see in such a RNA or ribonucleoprotein world.
Acknowledgments
We thank D. Smith and S. Lawrence for expert technical help, A. Ferré-D'Amaré and R. Collins for providing plasmids and ribozyme suggestions, O. Uhlenbeck for suggestions on 3′ labeling, B. Cousineau and D. Sledjeski for helpful discussions, B. Cousineau, S. Gottesman, S. Hanes, P. Masters, and S. Woodson for suggestions on the manuscript, the Molecular Genetics Core facility at Wadsworth Center for oligonucleotide synthesis and DNA sequencing, M. Carl for preparation of the manuscript, and J. Dansereau for help with figures. This work was supported by National Institutes of Health Grants GM39422 and GM44844 to M.B.
Footnotes
See Commentary on page 9824.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170281497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170281497
References
- 1.Eddy S. Curr Opin Genet Dev. 1999;9:695–699. doi: 10.1016/s0959-437x(99)00022-2. [DOI] [PubMed] [Google Scholar]
- 2.Moss E G, Lee R C, Ambros V. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 3.Wassarman K M, Zhang A, Storz G. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 4.Lease R A, Cusick M E, Belfort M. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majdalani M, Cunning C, Sledjeski D, Elliot T, Gottesman S. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams R M, Rimsky S. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 7.Atlung T, Ingmer H. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 8.Hengge-Aronis R. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 9.Sledjeski D D, Gupta A, Gottesman S. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 10.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sledjeski D, Gottesman S. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 13.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belfort M, Ehrenman K, Chandry P S. Methods Enzymol. 1990;181:521–539. doi: 10.1016/0076-6879(90)81149-o. [DOI] [PubMed] [Google Scholar]
- 15.Ferré-D'Amaré A R, Doudna J. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCoy M I, Gumport R I. Biochemistry. 1980;19:635–642. doi: 10.1021/bi00545a005. [DOI] [PubMed] [Google Scholar]
- 17.Guo H C T, Collins R A. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Weber T R. Doctoral thesis. Univ. of Illinois at Urbana-Champaign; 1985. [Google Scholar]
- 20.England Y E, Uhlenbeck O C. Nature (London) 1978;275:560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- 21.Hinton D M, Baez J A, Gumport R I. Biochemistry. 1978;17:5091–5097. doi: 10.1021/bi00617a004. [DOI] [PubMed] [Google Scholar]
- 22.Kuchino Y, Nishimura S. Methods Enzymol. 1989;180:154–163. doi: 10.1016/0076-6879(89)80099-0. [DOI] [PubMed] [Google Scholar]
- 23.Moine H, Ehresmann B, Ehresmann C, Romby P. In: RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 77–115. [Google Scholar]
- 24.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiegand R C, Godson G N, Radding C M. J Biol Chem. 1975;250:8848–8855. [PubMed] [Google Scholar]
- 26.Nagaswamy U, Voss N, Zhang Z, Fox G E. Nucleic Acids Res. 2000;28:375–376. doi: 10.1093/nar/28.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkhard M E, Turner D H, Tinoco I., Jr . In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 233–264. [Google Scholar]
- 28.Arraiano C M, Cruz A A, Kushner S R. J Mol Biol. 1997;268:261–272. doi: 10.1006/jmbi.1997.0962. [DOI] [PubMed] [Google Scholar]
- 29.Wells S E, Hillner P E, Vale R D, Sachs A B. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 30.Kushner S R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 849–860. [Google Scholar]
- 31.Zeiler B N, Simons R W. In: RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 437–464. [Google Scholar]
- 32.Karzai A W, Susskind M M, Sauer R T. EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gesteland R F, Cech T R, Atkins J F. The RNA World. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 1–709. [Google Scholar]
- 34.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]