SUMMARY
The Nijmegen breakage syndrome 1 (Nbs1) subunit of the Mre11-Rad50-Nbs1 (MRN) complex protects genome integrity by coordinating double-strand break (DSB) repair and checkpoint signaling through undefined interactions with ATM, MDC1, and Sae2/Ctp1/CtIP. Here, fission yeast and human Nbs1 structures defined by X-ray crystallography and small angle X-ray scattering (SAXS) reveal Nbs1 cardinal features: fused, extended, FHA-BRCT1-BRCT2 domains flexibly linked to C-terminal Mre11- and ATM-binding motifs. Genetic, biochemical, and structural analyses of an Nbs1-Ctp1 complex show Nbs1 recruits phosphorylated Ctp1 to DSBs via binding of the Nbs1 FHA domain to a Ctp1 pThr-Asp motif. Nbs1 structures further identify an extensive FHA-BRCT interface, a divalent MDC1-binding scaffold, an extended conformational switch, and the molecular consequences associated with cancer predisposing Nijmegen breakage syndrome mutations. Tethering Ctp1 to a flexible Nbs1 arm suggests a mechanism for restricting DNA end processing and homologous recombination activities of Sae2/Ctp1/CtIP to the immediate vicinity of DSBs.
INTRODUCTION
The Mre11-Rad50-Nbs1 (MRN) complex coordinates detection, signaling, and repair of cytotoxic DNA double-strand breaks (DSBs). Mutation of MRN subunits in Saccharomyces cerevisiae and Schizosaccharomyces pombe causes genotoxin sensitivity and meiotic defects resulting from defective processing of programmed DSBs (reviewed in Williams et al., 2007). In humans, congenitally acquired hypomorphic NBS1 and MRE11 mutations cause radiosensitivity and chromosome instability syndromes, Nijmegen breakage syndrome (NBS) and ataxia-telangiectasia-like disorder (ATLD), respectively (Carney et al., 1998; Stewart et al., 1999).
MRN possesses critical end-bridging and endonucleolytic activities for homologous recombination (HR) repair of DSBs (Williams et al., 2008). MRN also acts in checkpoint signaling by targeting the ATM/Tel1 kinase to DSBs by binding Nbs1 C-terminus (Falck et al., 2005; You et al., 2005). ATM phosphorylates the histone H2AX C-terminal tail near DSBs to form the γH2AX chromatin modification (Stucki and Jackson, 2006). In mammals, checkpoint adaptor MDC1 binds γH2AX, facilitating the intra-S-phase checkpoint (Goldberg et al., 2003; Lou et al., 2006; Stucki et al., 2005). MDC1 then recruits added MRN-ATM complexes via phosphorylation-dependent interactions with Nbs1 FHA-BRCT1-BRCT2 domains (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008) to amplify or sustain checkpoint signals (Lou et al., 2006). However, no MDC1 homologs are known in lower eukaryotes, so other ancestral Nbs1 FHA-BRCT1-BRCT2 domain functions likely exist.
MRN activities in HR involve the diverged protein known as Ctp1 in S. pombe (Limbo et al., 2007), Sae2 in S. cerevisiae, and CtIP in mammals (Sartori et al., 2007). Sae2 cleaves DNA substrates in vitro (Lengsfeld et al., 2007), consistent with 5′ to 3′ single-strand resection of DSBs in vivo to generate 3′ ssDNA tails for HR (Mimitou and Symington, 2008; Zhu et al., 2008). Ctp1Sae2 is required for removal of proteins linked to the 5′-end of DSBs (Neale et al., 2005), including complexes formed by the anticancer drug etoposide and topoisomerase II (Hartsuiker et al., 2009). Ctp1Sae2 activities appear conserved in mammals, and CtIP is a tumor suppressor in mice (Chen et al., 2005). In human cells, CtIP interacts with MRN (Sartori et al., 2007), and co-precipitates with Nbs1 when co-expressed in insect cells, suggesting CtIP binds Nbs1 (Chen et al., 2008), but the mechanism and significance of this interaction are unknown.
Whereas microscopic and crystallographic analyses of Mre11 and Rad50 were integral in defining MRN DNA bridging and nuclease processing activities (Hopfner et al., 2002; Hopfner et al., 2001; Williams et al., 2008), how Nbs1 physically integrates DNA end sensing and processing with cell cycle checkpoint activation remains a mystery. Here we report combined structural and functional dissection of fission yeast and human Nbs1. X-ray structures and small angle X-ray scattering (SAXS) results show an S. pombe Nbs1 folded core is conserved in human Nbs1, and that disease-associated Nbs1 mutations fall into the folded N-terminal FHA-BRCT1-BRCT2 domains defined by our structures. Our results further uncover the basis for phosphorylation-dependent recruitment of Ctp1 to DSBs by Nbs1. Nbs1 emerges as an extended, flexible binding nexus coordinating interactions between the Mre11-Rad50 core, the checkpoint kinase ATM/Tel1, and the DNA end-processing factor Ctp1.
RESULTS
Mapping Nbs1 Functional Domains
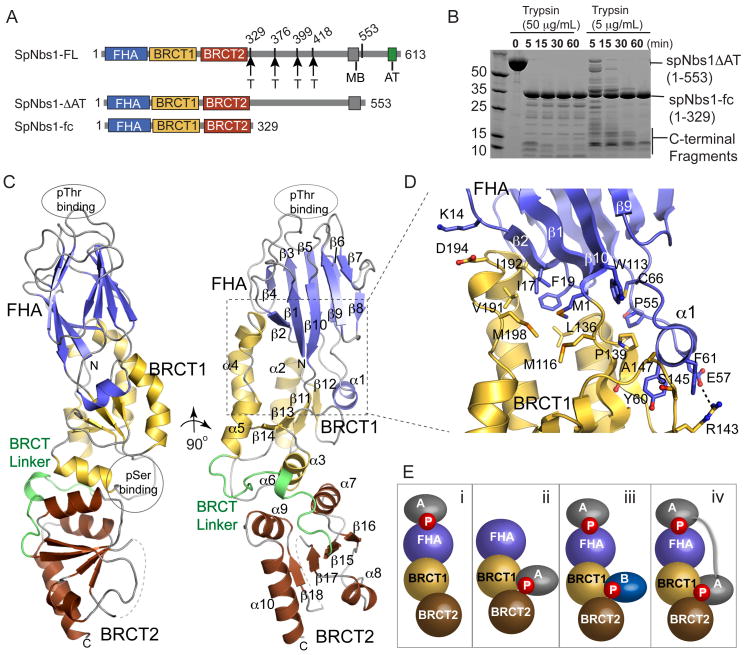
To define functional Nbs1 regions, we expressed a series of truncated S. pombe Nbs1 (spNbs1) proteins and mapped structured domains with proteolysis (Figures 1A and 1B). Full-length spNbs1 expresses at low levels in E. coli, but a 60 residue C-terminal deletion (residues 1-553, Nbs1-ΔAT), removing the ATM/Tel1 binding motif (AT) but retaining an Mre11 binding consensus motif (MB) (Falck et al., 2005; You et al., 2005), facilitates robust recombinant over-expression. Trypsin degradation of Nbs1-ΔAT reveals a stable 37kDa folded core (Nbs1-fc, residues 1-329) encompassing the Nbs1 FHA domain and its two BRCT domains (Figures 1A and 1B). N-terminal sequencing of smaller transiently stable trypsin products maps three additional cleavage sites to a divergent sequence linking BRCT2 and the MB motif. Thus, the N-terminal 55% of spNbs1 comprises a composite FHA-BRCT1-BRCT2 folded Nbs1-fc that is linked to the C-terminal Mre11 and ATM binding regions via a protease-sensitive linker peptide. Like human Nbs1 (Difilippantonio et al., 2005), but unlike S. cerevisiae Xrs2Nbs1 (Shima et al., 2005), the integrity of S. pombe Nbs1-fc is critical for MRN complex DSB repair functions (Figure S1).
Figure 1. Nbs1 N-terminal Domain Structure.
(A) N bs1 domain architecture schematic with flexible trypsin-accessible cleavage sites (“T”), Mre11 binding motif (“MB”) and ATM/Tel1 binding motif (“AT”).
(B) Trypsin degradation (done at 4 °C with indicated trypsin concentrations and 1 mg/mL Nbs1-ΔAT) maps a stable Nbs1 folded core (Nbs1-fc) to residues 1-329. Transiently stable products map added cleavage sites to the Nbs1 central region.
(C) Nbs1-fc adopts an elongated structure with closely interacting FHA (blue), BRCT1 (yellow), and BRCT2 (red) domains.
(D) The Nbs1 FHA-BRCT interface has extensive hydrophobic interactions.
(E) Two phosphoprotein-binding sites in the FHA and at the BRCT1-BRCT2 interface enable four Nbs1 protein-protein binding modes.
Nbs1 Folded Core Architecture and Structure
To examine Nbs1 structure-function, we determined the 2.8 Å resolution X-ray structure of spNbs1-fc with selenomethionine multi-wavelength anomalous dispersion (see Experimental Procedures), (Table S2 and Figure S2). Overall, Nbs1-fc is an elongated, distorted cylinder with dimensions ~35 by ~40 by ~95 Å, composed of joined FHA, BRCT1, and BRCT2 domains (Figure 1C). FHA and BRCT repeat domains mediate phosphorylation-regulated protein interactions, and are ubiquitous amongst DNA repair proteins (Glover et al., 2004; Williams et al., 2004). Like budding yeast Rad53-FHA1 (Durocher et al., 1999) or murine PNK-FHA (Bernstein et al., 2005), the Nbs1 FHA domain folds as a canonical FHA β-sandwich with two opposed 5-stranded anti-parallel β–sheets, and distal variable phosphoprotein-interacting β-strand connector loops (Figure S3).
The tandem Nbs1 BRCT domains adopt BRCT α-β sandwich folds with a central 4-stranded parallel β-sheet surrounded by 3–4 helices, and orient relative to one another with a triple helical hydrophobic inter-BRCT repeat interface. Although similar in structure to hBRCA1 BRCT repeats (Williams et al., 2001), the Nbs1 inter-BRCT repeat angle is offset by ~40 ° relative to BRCA1 tandem BRCTs (Figure S4). Sequence comparisons show that BRCT phosphoprotein interaction motifs (a conserved S/T at the C-terminus of the first β-sheet in BRCT1 (β11-α2 junction in Nbs1), and a TXK motif at the N-terminus of the second helix in the BRCT (α3 in Nbs1) (Clapperton et al., 2004; Williams et al., 2004)) are absent in S. pombe Nbs1, but present in the human, mouse, chicken, and frog Nbs1 homologs (Figure S3).
Despite similarities to known domain structures, key Nbs1 features suggest distinct functions in coordinating DNA repair signaling pathways. Notably, Nbs1 is configured as a divalent binding scaffold to mediate association with phosphorylated binding partners via four possible binding modes (Figure 1E): i) FHA-only binding, ii) BRCT-only binding, iii) simultaneous FHA and BRCT binding for alignment of two distinct binding partners, or iv) recognition of tandem phosphorylation clusters in one target protein via simultaneous FHA/BRCT binding. The phosphoprotein binding sites (spaced ~80 Å apart) could be spanned by an ~22 amino acid spacing (3.8 Å/residue) between phosphorylated residues within a single protein.
A defining Nbs1-fc feature is its extensive FHA-BRCT interface. The FHA N- and C-termini pack at the FHA-BRCT1 interface with N-terminal methionine and the invariant Trp2 buried within the protein core (Figure S5A). Attachment of affinity or epitope tags to the Nbs1 N-terminus (not shown) or a Trp2 to Ala mutation renders spNbs1 non-functional in vivo, suggesting that the FHA-BRCT1 interface is critical for structure and function (Figure S5B). Close FHA and BRCT1 domain juxtaposition results from a short 2-residue linker between the FHA-β10 and BRCT1-β1 strands, and by an Nbs1-specific helical insertion (helix α1) between FHA β-strands β5 and β6 (Figure 1D). At the FHA-BRCT1 boundary, an uninterrupted network of hydrophobic residues from the FHA (Met1, Ile17, Phe19, Cys66, Pro55) and BRCT1 (Met116, Leu136, Pro139, Ala147, Ser145, Arg143), and two flanking salt-bridges (Lys14-Asp194 and Glu57-Arg143) stabilize the FHA-BRCT1 interface. The large buried surface area (~2150 Å2) for the FHA-BRCT1 interface suggests the relative positioning of the FHA-BRCT1-BRCT2 scaffold aligns phosphoprotein interaction sites on opposite faces of Nbs1.
Human Nbs1 Solution Structure
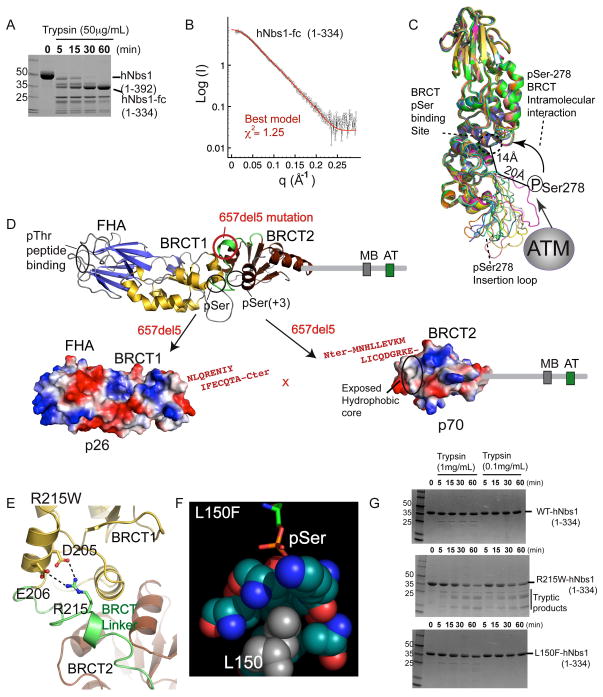
To better understand NBS mutations, we analyzed the human Nbs1-fc structure with SAXS and proteolysis. Human Nbs1 (hNbs1) forms a trypsin-stable composite FHA-BRCT1-BRCT2 core (hNbs1-fc, residues 1-334) similar to spNbs1-fc (Figure 2A). As all parts of a structure contribute to the experimental X-ray scattering, SAXS provides an objective, quantitative measure of structural similarity (Putnam et al., 2007). SAXS data of hNbs1-fc closely matches calculated scattering by the spNbs1-fc X-ray structure (χ2= 2.00), revealing conservation of structure despite low sequence conservation of Nbs1-fc (11% identity, 25% similarity). Yet, structure-based sequence alignments show an insertion missing in the spNbs1-fc model corresponds to an α-helix usually found between the third and fourth BRCT domain β-strands (β17 and β18 of BRCT2), that is linked to an hNbs1 Ser278 phosphorylation site loop (human Nbs1 residues 272–295) (Figure S3). To model these missing regions of hNbs1-fc, we generated 100 hNbs1-fc models based on the spNbs1 structure using MODELLER, and evaluated models by fit to the SAXS data. This approach identified 12 models with superior χ2 fits (χ2<1.3, best model χ2=1.25) that all bear extended accessible pSer278 loop conformations (Figures 2B and 2C). Thus, the overall spNbs1-fc fold is maintained in hNbs1-fc, but is decorated with a solvent-accessible phosphorylated loop. Indeed, based on the BRCT1-BRCT2 interface position and Ser278 accessibility on this loop, we hypothesize phosphorylated Ser278 in hNbs1 is suitably placed to regulate BRCT phosphoprotein interactions in cis through an intermolecular auto-inhibitory mechanism (Figure 2C).
Figure 2. Structural Basis for NBS Mutations.
(A) Trypsin degradation produces a stable hNbs1 folded core (hNbs1-fc, residues 1-334).
(B) Comparison of calculated SAXS curves for hNbs1 models (green and red) with experimental scattering (black) shows hNbs1-fc architecture closely matches the S. pombe structure.
(C) Superposition of 12 homology hNbs1 models with SAXS scattering χ2 fits better than 1.3. ATM Phosphorylation sites in hNbs1 map to surface accessible loops with observed conformational variability between models. Ser278 phosphorylation could mediate auto-regulatory BRCT peptide binding in cis through an interaction with the phosphoprotein-binding cleft.
(D) Nbs1 657del5 frameshift mutation found in 90% of NBS patients generates protein fragments p26 and p70. Sequence in red, resulting from 657del5, shows extra C-terminal residues added to the N-terminal (p26) and C-terminal (p70) fragments.
(E) A homology model for Arg215 in hNbs1 based on the spNbs1-fc structure suggests Arg215 forms a salt bridge from the BRCT linker to a BRCT1 helix. R215W is predicted to disrupt protein folding.
(F) The Nbs1 L150F missense mutation likely disrupts the BRCT pSer binding pocket. Leu150 is found in the hydrophobic core directly contacting pSer ligands, based on the structure of a BRCA1-phosphopeptide complex, RCSB ID 1T2V.
(G) Protease sensitivity of Nbs1 missense mutations. Mutant R215W destabilizes the protein fold whereas L150F shows WT resistance to trypsin digestion.
Nbs1 657del5 Mutation
Nbs1 SAXS and X-ray structures provide molecular insights into the consequences of patient-derived NBS mutations. The 657del5 frameshift NBS predisposition mutation, accounting for ~90% of patient isolated mutations (Digweed and Sperling, 2004), generates a protein truncation plus a cryptic translational start site from which the Nbs1 C-terminus is produced at reduced levels (Maser et al., 2001). This catastrophic molecular disconnect contributes to radio-sensitivity phenotypes, and ultimately to malignancies in NBS patients (Maser et al., 2001). At the protein level, 657del5 causes molecular scission of hNbs1-p95 into two halves: 1) an N-terminal (p26) fragment that fails to interact with other members of the MRN complex and contains the FHA-BRCT1 with an extended 15 amino acid C-terminus, and 2) a C-terminal (p70) fragment with intact Mre11-ATM binding motifs and an extended N-terminal peptide linked to its BRCT2 domain (Figure 2D). Structurally, 657del5 physically uncouples the Nbs1 FHA-and BRCT-mediated phosphoregulatory interactions of Nbs1 from its Mre11 and ATM/Tel1 interaction regions. Besides split FHA and BRCT domains, we hypothesize protein instability and decreased abundance of Nbs1-p70 underlying cancer incidence (Kruger et al., 2007) is caused by solvent exposure of a large hydrophobic patch on BRCT2 that normally forms the BRCT1-BRCT2 interface (Figure 2D).
Nbs1 Missense Mutations
Two missense mutations (R215W and L150F) mapping to Nbs1-fc are linked to NBS (Seemanova et al., 2006) and breast cancer (Heikkinen et al., 2006). The R215W (643C>T) mutation occurs with 657del5 in compound heterozygous individuals with NBS presenting unusually severe neurological abnormities (Seemanova et al., 2006). Arg215 lies in the BRCT1-BRCT2 linker region, and salt-bridges with Asp205 and Glu206 in our hNbs1-fc homology models (Figure 2E). Mutation of hNbs1-fc Arg215 to Trp confers trypsin sensitivity, suggesting R215W decreases Nbs1 protein stability (Figure 2G). Conversely, the L150F breast cancer associated mutation displays no measurable stability defect. However, L150F flanks the Nbs1 BRCT phosphoserine interaction cleft (Figure 2F). By analogy to BRCA1 mutations occluding phosphoprotein-binding pockets, L150F may deform the pSer-binding pocket via a relay of hydrophobic distortion (Williams et al., 2004). Whereas patients with Nbs1 mutations decreasing protein stability and abundance present with classical NBS symptoms, a specific linkage of Nbs1 L150F to breast cancers may reflect critical functional roles for the Nbs1 BRCT interactions in tumor suppression. Thus, Nbs1 disease-causing mutations may either directly impact phosphoprotein-binding as seen for BRCA1 (Williams et al., 2004), or global protein stability as seen for BRCA1 (Williams et al., 2001), XPD (Fan et al., 2008), and superoxide dismutase (DiDonato et al., 2003).
Nbs1 Mutations in the FHA Phosphoprotein-Binding Pocket Impair DSB Repair
hNbs1 binding to hMDC1 depends on both the FHA and BRCT domains (Chapman and Jackson, 2008), probably via bipartite Nbs1 FHA-BRCT interactions (Figure 1E - type IV binding). Consensus phosphoprotein interaction motifs (Williams et al., 2004) are missing in spNbs1 BRCT domains, but present in hNbs1 and higher eukaryotes (Figure S3). As S. pombe also lacks an MDC1 ortholog, the Nbs1 FHA domain may mediate key phosphorylation-dependent interactions with other DSB repair factors through an FHA-only (Figure 1G - class I) interaction.
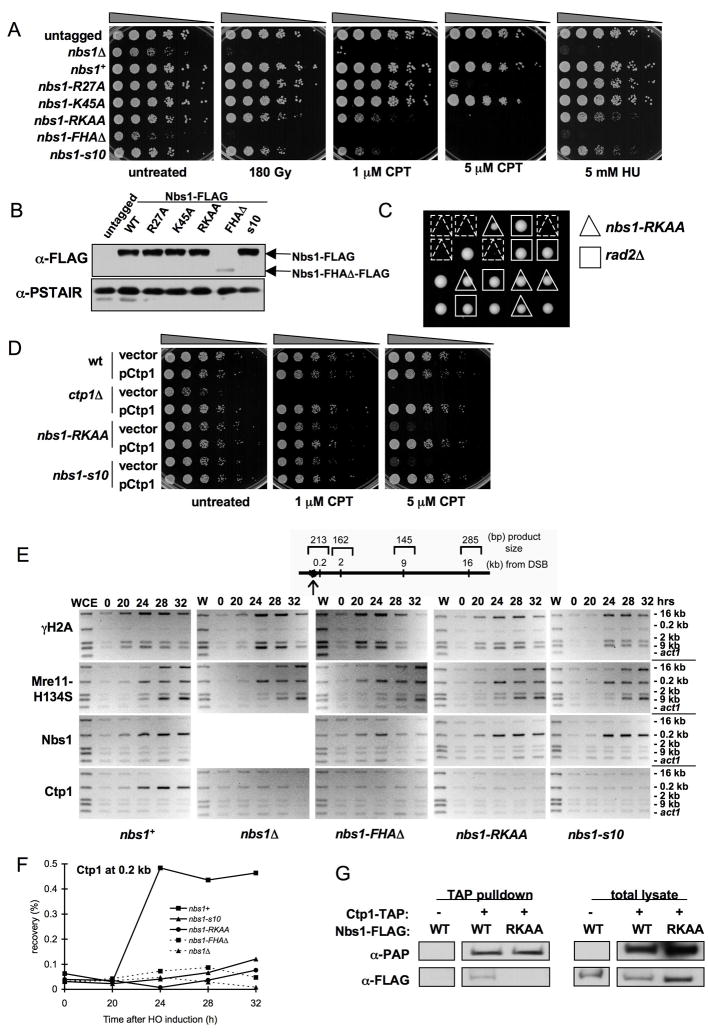
We addressed the functional importance of the FHA phosphoprotein-binding pocket in spNbs1. Arg27 and Lys45 are conserved residues predicted to contact phosphothreonine (pThr) in protein binding targets (Durocher et al., 1999), so we constructed strains replacing one or both residues with alanine. While neither single mutant is sensitive to DNA damage (except at high doses for nbs1-R27A), the double mutant (nbs1-RKAA) is sensitive to ionizing radiation (IR), the topoisomerase I poison camptothecin (CPT), and the DNA replication inhibitor hydroxyrurea (HU) (Figure 3A). These mutations had no effect on Nbs1 abundance (Figure 3B).
Figure 3. An Nbs1 FHA-defective Mutant Causes DNA Damage Sensitivity and Decreases Ctp1 Localization to DSBs.
(A) nbs1-RKAA (R27A/K45A) cells are sensitive to DNA damaging agents.
(B) Immunoblot of Nbs1-FLAG from strains used in (A). Expression of Nbs1-ΔFHA-FLAG is substantially decreased in soluble extracts relative to wild-type (wt). Immunoblot with PSTAIR antisera recognizing Cdc2 is used as a loading control.
(C) Tetrad analysis of a cross between rad2Δ and nbs1-RKAA strains. The rad2Δ/nbs1-RKAA double mutant is synthetic lethal, indicating a key role for the Nbs1 FHA domain in recombination-dependent DNA repair.
(D) Ctp1 is a multicopy suppressor of DNA damage sensitivity observed in the nbs1-RKAA mutant.
(E) ChIP analysis of γH2A, Mre11-H134S, Nbs1, and Ctp1 around a DSB created by HO endonuclease expressed from thiamine-repressible nmt1 promoter. Samples were collected at indicated times after depletion of thiamine from medium. Ctp1 localization at 0.2 kb from the DSB is defective in the nbs1 mutants.
(F) Quantitative real-time PCR was used to measure enrichment of Ctp1 at 0.2 kb from the HO endonuclease-induced DSB.
(G) Ctp1 associates with Nbs1 in an FHA domain-dependent manner. Strains were generated that express TAP-tagged Ctp1 and FLAG-tagged Nbs1 in an mre11-H134S background. IgG Sepharose beads were used to precipitate TAP-tagged Ctp1 and any associated proteins (negative control is a strain lacking Ctp1-TAP).
Interestingly, nbs1-RKAA phenotypes matched those of nbs1-s10 (Figure 3A), a Gly103Asp mutant (Akamatsu et al., 2008) that destabilizes the phosphoprotein-binding loop encompassing Lys45 (Figure S6). Besides causing genotoxin sensitivity, nbs1-s10 has a synthetic lethal interaction with rad2Δ (Akamatsu et al., 2008). Rad2 (FEN-1/RAD27) is a structure-specific endonuclease that processes Okazaki fragments during DNA replication. Unprocessed Okazaki fragments lead to replication fork collapse, causing damage that is repaired by an HR pathway requiring MRN (Akamatsu et al., 2008). Further revealing the importance of the Nbs1 FHA domain in mediating repair of collapsed replication forks, nbs1-RKAA rad2Δ double mutants are inviable (Figure 3C).
Nbs1 FHA Domain Mutations Disrupt Nbs1-Ctp1 Interactions
The nbs1-RKAA and nbs1-s10 phenotypes suggest that Nbs1 FHA domain facilitates a phosphorylation-dependent interaction with a key HR protein. As a high-copy suppressor of nbs1-s10 (Akamatsu et al., 2008), Ctp1 may interact with the Nbs1-FHA, consistent with our finding that ctp1+ overexpression rescues nbs1-RKAA (Figure 3D).
To test if Nbs1 is required for Ctp1 localization to DSBs, we used chromatin immunoprecipitation (ChIP) to measure enrichment of Mre11, Nbs1 and Ctp1 at four sites 0.2–16 kb away from a defined DSB created with HO endonuclease (Limbo et al., 2007). We assayed γH2A as a marker enriched at regions flanking a DSB, and we performed these assays in a mre11-H134S mutant background (Williams et al., 2008), as ablated Mre11 endonuclease activity increases ChIP signals for Mre11, Nbs1 and Ctp1 at DSBs, presumably because defective endonuclease processing of DNA ends increases MRN retention at a DSB. Mre11-H134S, Nbs1, Ctp1 and γH2A were strongly enriched around the DSB in nbs1+ cells (Figure 3E). The γH2A signal was weakest nearest the DSB (0.2 kb), while Ctp1 and Nbs1 signals were strongest at this position. No Ctp1 enrichment at the DSB was detected in nbs1Δ cells (Figure 3E), even though Ctp1 abundance is unaffected by loss of Nbs1 (Akamatsu et al., 2008). Mre11-H134S enrichment was unaffected in nbs1Δ cells (Figure 3E). Truncated Nbs1 in nbs1-FHAΔ cells had a weak or transient interaction with the DSB, but reduced abundance of Nbs1-FHAΔ complicates interpretation of this result. Importantly, Ctp1 enrichment at the DSB was strongly impaired in nbs1-ΔFHA cells. The same defect was seen in nbs1-RKAA and nbs1-s10 mutants, yet the mutant Nbs1 proteins exhibit no defect in associating with the DSB (Figure 3E). Quantitative real-time PCR confirmed diminished recruitment of Ctp1 to the DSB in the nbs1 mutants (Figure 3F).
Although we were previously unable to co-precipitate Ctp1 and Nbs1 (Limbo et al., 2007), their mammalian counterparts associate in vivo (Chen et al., 2008; Sartori et al., 2007). Indeed, by adjusting cell extraction conditions and utilizing an mre11-H134S background, we could detect Nbs1 in a Ctp1 immunoprecipitation when both proteins were expressed at endogenous levels (Figure 3G). Importantly, nbs1-RKAA disrupted this interaction (Figure 3G). Collectively, these results reveal a critical role for the Nbs1 FHA domain in mediating Ctp1 localization to DSBs.
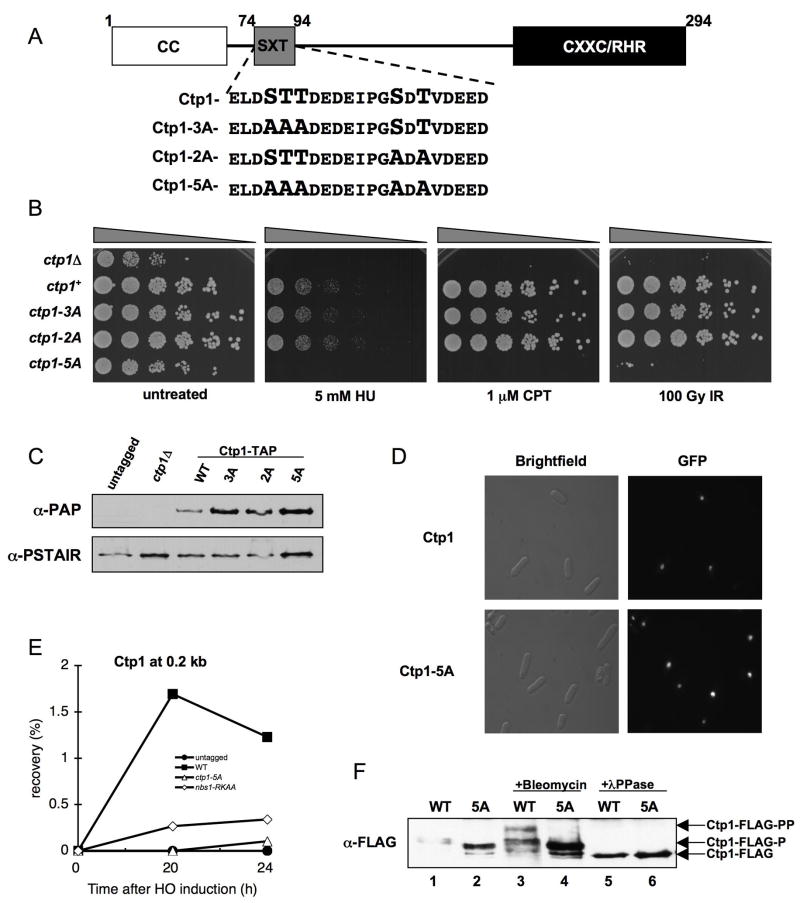
Mutations in Ctp1 SXT Sites Disrupt Interactions with Nbs1
To test if Ctp1 phosphorylation is required for Nbs1 binding, we focused on potential casein kinase 2 (CK2) phosphorylation sites in Ctp1, as CK2 phosphorylates SDT sites in hMDC1 that interact with the Nbs1 FHA in mammalian cells (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) identified two clusters of probable CK2 sites in the Ctp1 N-terminus: the first spanning Ser77, Thr78 and Thr79, and the second containing Ser87 and Thr89 (Figure 4A). Based on their similarity to MDC1 SDT repeats, we designated these clusters as SXT repeats. While mutation of either SXT repeat had no noted effect on DNA damage sensitivity, a mutant removing all Ser/Thr in the repeats (ctp1-5A) was extremely damage sensitive suggesting the SXT repeats are functionally redundant, not cooperative (Figure 4B). The triple threonine mutant (ctp1-T78A T79A T89A) was as DNA damage sensitive as ctp1-5A, while strains with paired Ctp1 Thr-Ala substitutions (ctp1-T78A T89A or ctp1-T79A T89A) are fully functional (Figure S7), so a single Thr within the SXT repeats is sufficient for function. The ctp1-5A mutant grew slowly, as is typical for Ctp1-defective strains (Limbo et al., 2007), but did not reduce Ctp1 abundance or nuclear localization (Figures 4C and 4D), indicating that Ctp1-5A is not grossly destabilized or misfolded. ChIP analysis revealed a striking reduction in Ctp1 enrichment at a DSB in the ctp1-5A mutant, as seen in the nbs1-RKAA background (Figure 4E).
Figure 4. Phosphorylation Motif Required for Ctp1 Activity.
(A) Ctp1 schematic depicting conserved N-terminal coiled coil domain (CC), C-terminal core homology region (CXXC/RHR), and putative phosphorylation motif (SXT). Motif enlargement shows residue substitutions in the ctp1-3A, ctp1-2A, and ctp1-5A strains.
(B) ctp1-5A cells are sensitive to exogenous DNA damaging agents, similar to ctp1Δ cells.
(C) Ctp1-TAP immunoblot from strains used in (B), showing that Ctp1 mutants are expressed at levels at or above the wt amount (PSTAIR immunoblot loading control).
(D) GFP-Ctp1-5A intracellular localization is unaltered relative to GFP-Ctp1-wt.
(E) ChIP analysis of Ctp1 enrichment surrounding an HO endonuclease-induced DSB. Samples were collected at the indicated time after HO-induction. Ctp1 fails to localize to a site 0.2 kb from the DSB in the nbs1-RKAA and ctp1-5A strains.
(F) Basal phosphorylation is altered and damage-induced phosphorylation is abrogated in Ctp1-5A relative to wt. In asynchronous cells, Ctp1 is phosphorylated basally (Ctp1-FLAG-P), shown by increased Ctp1 mobility (Ctp1-FLAG) following λPPase treatment. After exposure to bleomycin, Ctp1 is hyper-phosphorylated, causing a decrease in mobility (Ctp1-FLAG-PP) relative to the basally phosphorylated form.
To test if Ctp1 is phosphorylated on its SXT repeats, we performed SDS-PAGE analyses of FLAG-tagged Ctp1 immunoprecipitated from ctp1+ or ctp1-5A cells. In the absence of DNA damage, Ctp1-5A migrates faster than wild-type Ctp1 (Figure 4F, lanes 1–2), indicating that basal Ctp1 phosphorylation is altered in this mutant. Consistent with this observation, phosphatase treatment causes both wild-type and mutant Ctp1 to migrate faster than either untreated sample, indicating that Ctp1 is basally phosphorylated on SXT repeats and other sites (Figure 4F, lanes 5–6). Exposure to the radiomimetic drug bleomycin induces further Ctp1 phosphorylation that results in a shift to slower mobility, but this shift is ablated in Ctp1-5A (Figure 4F, lane 3–4). This DNA damage-induced hyper-phosphorylation of Ctp1 requires Mre11, Nbs1 and either of the ATM/ATR-like kinases (Tel1/Rad3) in fission yeast (Akamatsu et al., 2008).
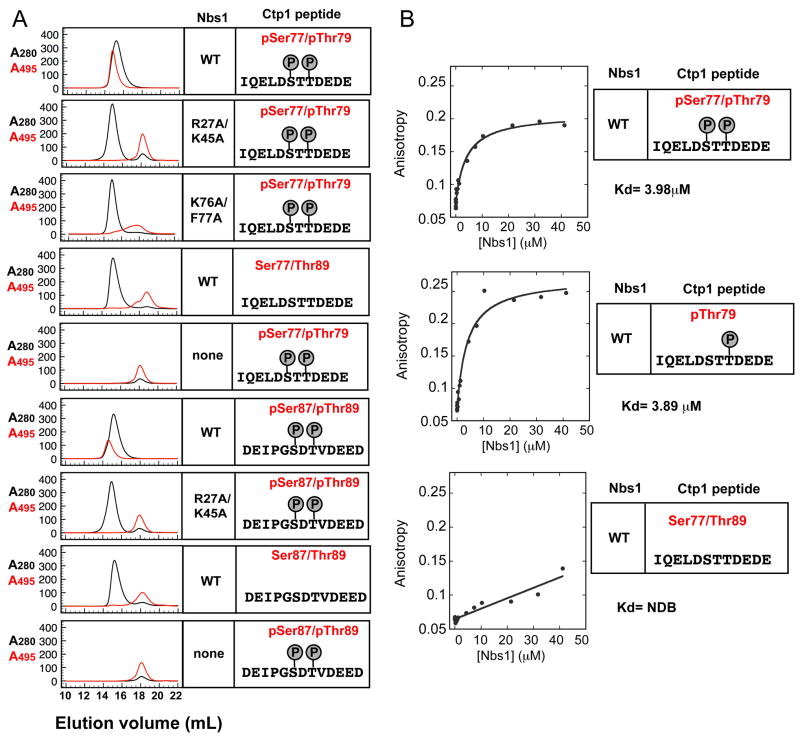
To test if the SXT repeats in Ctp1 mediate direct interactions with the FHA domain of Nbs1, we monitored in vitro association of Nbs1-fc with phosphorylated Ctp1 peptides by size exclusion chromatography (Figure 5). Nbs1-fc directly binds and co-elutes with phosphorylated fluorescein (FITC) conjugated Ctp1 peptides encompassing either of the functionally redundant SXT repeats. These interactions were phosphorylation-dependent and reduced by the nbs1-RKAA mutation.
Figure 5. Nbs1 FHA Domain Binds Phosphorylated Ctp1.
(A) Analytical gel filtration binding analyses of Nbs1-Ctp1 protein-phosphopeptide interactions. Co-elution of FITC-labeled Ctp1 peptides (see sequence insets) absorbing at 495 nm (red trace) with recombinant Nbs1-fc variants (black A280 trace) monitors binding.
(B) Fluorescence polarization binding analyses of Nbs1-Ctp1 peptides.
Nbs1-Ctp1 Complex Structure
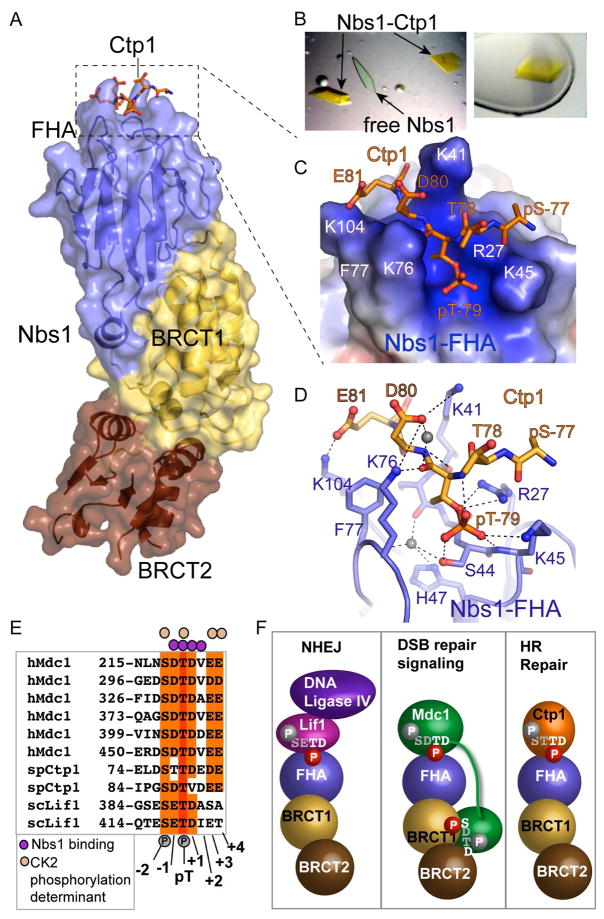
To define Nbs1-Ctp1 interactions, we co-crystallized and determined the 2.15 Å resolution X-ray structure of the first Ctp1 SXT repeat bound to Nbs1-fc (Table S2, Figures 6 and S8). FITC-Ctp1 phosphopeptides enabled us to visually identify yellow Nbs1-fc-Ctp1 complex crystals in a background of peptide-free Nbs1-fc crystals (Figure 6B, Experimental Procedures). Phosphorylated Ctp1 binds in an electropositive groove formed by FHA residues Arg27, Lys45, Phe77, Lys76, Lys104, Ser44, and Lys41 (Figure 6C), and buries 644 Å2 of solvent accessible surface at the Ctp1-Nbs1 interface. Ctp1 Ile72 - pSer77 and residues C-terminal to Glu81 (Asp82 and Glu83) are disordered. In contrast, 2Fo-Fc and Fo-Fc difference maps show electron density for Ctp1 pThr79, positions pThr(−1) (Thr78) through pThr(+2) (Glu81), and pSer77 main-chain, but not the pSer phosphate (Figure S8), underscoring the importance of these residues for Nbs1 binding.
Figure 6. Nbs1-Ctp1 Complex.
(A) Overall structure. Ctp1 binds Nbs1 in a surface groove at the distal N-terminal FHA domain. Bound Ctp1 phosphopeptide (orange) is overlaid upon the Nbs1 surface.
(B) Left: Crystals of the Nbs1-Ctp1 complex (yellow) grow in a background of colorless free Nbs1-fc crystals. Right: Nbs1:Ctp1 co-crystal used for structure determination.
(C) Nbs1 FHA domain forms a positively charged groove (blue electrostatic potential) for binding Ctp1 acidic residues pThr79 and Asp80 (pThr(+1)) and Glu81 (pThr(+2)) in an extended conformation. Ctp1 (orange) is displayed overlaid upon an electrostatic surface representation of the Nbs1 peptide-binding surface.
(D) Nbs1-Ctp1 interaction details.
(E) Sequence alignment of Nbs1 interaction proteins shows key specificity.
(F) Schematic of known Nbs1 protein-phosphoprotein interactions regulating NHEJ, DSB signaling and HR repair. Residues with white and red phosphates are predicted to be key for Nbs1 interactions in the FHA or BRCT-BRCT phosphoprotein binding clefts, whereas grey residues and phosphates in the consensus are predicted to not make direct Nbs1 contacts.
The Nbs1 peptide-binding groove interacts with Ctp1 on three sides (Figures 6C and 6D) and significant rearrangements of the interacting loops occur as they “grasp” Ctp1 (Supplemental Movies S1 and S2). The pThr79 phosphate directly contacts the side chains of Arg27, Ser44, and Lys45. Residues Arg27, Lys45, Ser42, Ile43, Lys76, Ser44, and a tightly bound water molecule mold the pThr interaction site surface (Figure 6D). This pocket appears to specifically bind pThr preferentially over pSer or pTyr: it is too small for pTyr binding and tightly contours around the pThr79 side chain methyl. Redundant contacts by Arg27 and Lys45 with the pThr phosphate explain the mild phenotypes observed for nbs1-R27A and nbs1-K45A single mutants as compared to the nbs1-RKAA double mutant (Figure 3A).
Our structure of the Nbs1-Ctp1 complex, conservation of the Nbs1 FHA phosphopeptide-binding cleft, and homology of the Ctp1 SXT repeats to Nbs1-binding motifs in human MDC1 (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008) and S. cerevisiae Lif1 (Palmbos et al., 2008) together define an Nbs1 FHA-binding consensus sequence motif as a pThr with an Asp at position pThr(+1) (Figures 6D and 6E). Conserved residues Lys76 and Phe77, along with Lys41 and Lys104 recognize Ctp1 Asp80 (pThr (+1)). Conversely, recognition of Ctp1 Glu81 (pThr(+2)) by Nbs1 Lys104 appears specific to the S. pombe Nbs1-Ctp1 interaction. While Phe77 makes van der Waals contacts to the peptide backbone, Lys76 hydrogen bonds to the pThr79 main chain carbonyl and combines with Lys41 in salt-bridges that sandwich the acidic pThr(+1) specificity position (Asp80). Consistent with structural observations, an Nbs1 mutant (Ala76-Ala77) impairs Ctp1 peptide binding, showing the conserved Lys76-Phe77 loop is crucial for the interaction (Figure 5).
Whereas most FHA domains recognize pThr(+3) position, specific interactions with Nbs1-FHA in cells are probably achieved by unique recognition of pThr-Asp motifs. Added specificity for the CtIP-MRN complex interaction in complex eukaryotes may also occur via avidity, as CtIP also binds Mre11 and Rad50 (Sartori et al 2008). Conservation of acidic residues C-terminal to pThr(+2) and phosphorylation at pSer(−2) could reflect kinase recognition-specificity for efficient in vivo phosphorylation, rather than a preferred Nbs1-FHA binding consensus (Figure 6E), as a single or double phosphorylated Ctp1 peptide binds Nbs1-fc with similar affinity (Figure 5B).
Use of a minimalist pThr-Asp Nbs1-FHA interaction consensus binding motif may direct MRN binding to multiple cellular targets. We predict the Xrs2Nbs1-Lif1 interaction facilitating MRN(X) DSB repair by non-homologous end joining in budding yeast (Palmbos et al., 2008) assembles via interactions between the Xrs2Nbs1-FHA domain and Lif1 CK2 phosphoacceptor sites analogous to the Ctp1 SXT repeats (Figures 6E and 6F). By contrast, mammalian Nbs1 binding to clusters of MDC1 SDTD repeats depends on both FHA and BRCT phosphoprotein interaction sites, and phosphorylation of the serine and threonine in the SDTD motif (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008). As BRCT domains typically bind pSer targets with specificity for the pSer(+3) position (Glover et al., 2004; Manke et al., 2003), our data predict that Nbs1-MDC1 complexes assemble through a bipartite interaction between the Nbs1 FHA domain binding the pTD of an MDC1 SDTD motif and the Nbs1 BRCT domains recognizing a neighboring MDC1 pSXXD consensus of a second SDTD repeat (Figures 6E and 6F).
Nbs1 is an Extended Flexible Interaction Hub
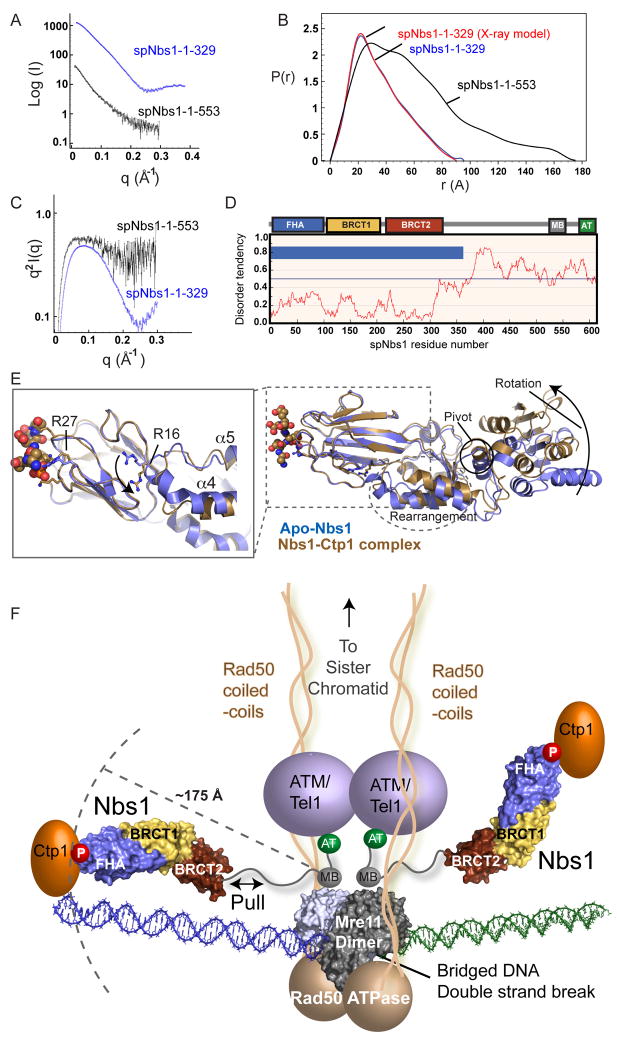
Nbs1 might provide rigid architectures to align its multiple interaction partners. Yet, Nbs1 proteolytic lability suggests its C-terminus is disordered (Figure 1B). To test these possibilities, we examined spNbs1-fc and spNbs1-ΔAT structures in solution with SAXS. While spNbs1-fc SAXS closely matches the X-ray structure, the maximum particle dimension (Dmax) for spNbs1-ΔAT is much larger (~175 Å), indicating a very elongated structure (Figures 7A and 7B). SAXS objectively measures folding: Kratky plots are parabolic for folded structures, asymptotic for unfolded structures, and intermediate for partially folded structures (Putnam et al., 2007). Consistent with an unstructured Nbs1 C-terminus, the spNbs1-ΔAT Kratky plot is intermediate between parabolic and asymptotic curves, while spNbs1-fc has a parabolic plot showing spNbs1-fc is folded consistent with proteolysis and crystal structures (Figure 7C).
Figure 7. Nbs1 is an Extended, Flexible, Effector and Signaling Scaffold.
(A) spNbs1-fc (1–329) (blue) and spNbs1-ΔAT (1–553) (black) proteins were characterized by SAXS.
(B) SAXS electron pair distribution function shows spNbs1ΔAT is elongated in solution.
(C) SAXS Kratky plot indicates spNbs1ΔAT is only partially folded in solution.
(D) S. pombe Nbs1 order-disorder predictions from IUPRED. Predicted ordered (blue bars) and unstructured regions (no blue bars) shows stretches of disorder tendency > 0.5.
(E) Structural overlay of Ctp1-bound (brown) and apo-Nbs1 (blue) crystal forms shows BRCT1-BRCT2 interface rotation and helical rearrangements at the FHA-BRCT1 interface. Left inset: An FHA domain arginine switch (R16) toggles between two conformational states, directly linking the FHA to motions to the BRCT domains.
(F) Model of MRN-Ctp1 complex bound at a bridging DNA DSB. The flexible Nbs1 C-terminus links FHA-bound Ctp1 to an Mre11-Rad50 heterotetrameric core complex bridging a DNA double strand break.
Although its C-terminus may undergo disorder-to-order transitions upon binding Mre11 and ATM, we do not see stable domains (>10 kDa) when Nbs1 is trypsin digested while in complex with Mre11 (Figure S9). Thus, C-terminal Mre11 and ATM interacting regions may be peptide interaction motifs, rather than globular domains. Sequence-based order predictions of Nbs1 homologs (Figures 7D and S10) suggest flexibility is a conserved Nbs1 feature, so inherent pliability of the Nbs1 C-terminus may be needed for function. Furthermore, flexibility extends into Nbs1-fc: the closed, phosphopeptide grasped FHA conformation flips an Arg switch at the FHA-BRCT interface and promotes conformational changes in the BRCT linker region, a ~20° rotation about the BRCT1-BRCT2 interface, and a ~10 Å shift at the BRCT2 C-terminal end in our experimentally determined X-ray structures (Figure 7E).
DISCUSSION
Nbs1 provides the key phosphoprotein sensor and recruitment scaffold for the Mre11-Rad50 complex. Nbs1 forms a folded N-terminal FHA-BRCT1-BRCT2 core, conserved from S. pombe to humans based upon the SAXS results, that uniquely connects an FHA phosphoprotein-binding site with BRCT domains via an extensive interface. The direct fusion of two phosphoprotein-binding domains implies functional crosstalk between binding sites may occur. Interestingly, structural overlays of the apo and peptide-bound crystal structures reveal an Nbs1-fc conformational change that may facilitate FHA-BRCT phosphoprotein binding site communication and pull on the flexible tether to Mre11 (Figure 7E, Supplemental Movies S3 and S4).
Integrating our results with known data supports a testable architectural hypothesis for MRN-Ctp1 (Figure 7F). MRN complex components and Ctp1 co-localize to within ~200 bp (<700 Å), of a defined endonuclease cut DSB in vivo (Figure 3E). Ctp1 is retained at the break site via direct phosphorylation-dependent binding to the N-terminal Nbs1 FHA domain. SAXS and X-ray structures define the reach from the Nbs1 C-terminal Mre11 interaction region to the N-terminal FHA domain as ~175 Å. Prior results also help delineate MRN-Ctp1 complex architecture at a DSB: 1) the Mre11-Rad50 DNA binding head can directly bind and bridge DNA ends (Chen et al., 2001; Williams et al., 2008), 2) the Mre11/Rad50 heterotetrameric core forms a four-lobed DNA binding head composed of dimeric Mre11 and the Rad50 ATPase domains, with Rad50 coiled-coils emanating from the head (Hopfner et al., 2001, Hopfner et al. 2002), and 3) the Mre11-Nbs1 binding interface maps to Nbs1 C-terminal motifs (Falck et al., 2005; You et al., 2005) and an Mre11 dimer surface patch away from the DNA binding cleft (Williams et. al, 2008). Rad50 coiled-coils are positioned to define the reach of the MRN complex to bridge homologous sister chromatids during HR repair (Hopfner et al., 2002; Moreno-Herrero et. al., 2005). Direct synapsis of DNA ends by Mre11 (Williams et al., 2008), and the tethering length of the Nbs1 connection between Ctp1 and Mre11 defined here support a mechanism for localizing both the MRN complex and Nbs1-linked Ctp1 to regions directly flanking a DSB (Figure 7F). Consistent with this composite structural model, archaeal Mre11/Rad50 core complex (Hopkins and Paull, 2008) or eukaryotic MRN/Ctp1Sae2 5′-3′ resection activity is limited to regions proximal to a DSB (Mimitou and Symington, 2008; Zhu et al., 2008).
Flexible Nbs1 molecular associations may accommodate large-scale MRN complex conformational changes, proposed to regulate ATM activation and dimer-to-monomer transitions (Lee and Paull, 2005). Like the flexible linkage of PCNA to ligase for DNA replication (Pascal et al., 2006), the malleable nature of Nbs1 and the Rad50 coils (Moreno-Herrero et al., 2005), provides a molecular basis for concerted MRN DNA break sensing and repair activities while preserving the integrity of a structurally dynamic chromatin template. With its apex FHA binding motif and conformational switch encompassing the FHA-BRCT interface, the mobile Nbs1 hub appears suitable to promote integration of repair sensing and effector activities of MRN by interface exchange and handoff interactions with multiple partners, as proposed for PCNA (Chapados et al., 2004) and BRCA2-Rad51 partners (Shin et al., 2003).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant S. pombe Nbs1 variants were expressed from pET29b as C-terminal 6-His tagged proteins in E. coli Rosetta2 (DE3) cells (Novagen) grown at 16 °C in Terrific Broth. After Ni-NTA affinity chromatography, proteins were purified by Superdex 75 (GE Healthcare) (spNbs1-fc, residues 1-329) or Superdex 200 (Nbs1-ΔAT, residues 1-553) gel filtration, and anion exchange chromatography. G103D, R27A/K45A and K76A/F77A mutations were made with Quickchange (Stratagene).
Nbs1-fc Crystallization
spNbs1-fc crystals were grown at 8 °C by hanging drop vapor diffusion. Nbs1-fc crystals were grown by mixing 1 μL protein solution (10 mg/mL spNbs1-fc in 500 mM NaCl, 20 mM Tris-HCl pH 7.5, 0.1% β–mercaptoethanol) and 1uL of crystallization solution A (2 M NaCl, 100 mM sodium acetate pH 5.5). Plate morphology was improved with crush dilution seeding into drops with 1 μL crystallization solution B (1.2–1.4 M potassium formate, 100 mM sodium acetate pH 4.6, 5% w/v polyethylene glycol 8000) and 1 μL protein solution. For data collection, crystals were transferred to cryo-protectant (crystallization solution B plus 26% ethylene glycol) and flash cooled in liquid nitrogen.
Nbs1-Ctp1 Complex Crystallization
Peptides were synthesized at the Scripps Research Institute core facility. Colorless peptide-free and yellow peptide-bound (FITC-conjugated) crystals grew in the same drop. 10 mg/mL spNbs1-fc in 40 mM NaCl, 20 mM Tris 7.5, 0.1% β-mercaptoethanol was mixed with FITC-Ctp1-1 (FITC-KKIQELD(pS)T(pT)DEDEI-nh2) at a 4:1 Ctp1-1/spNbs1-fc ratio. Crystals were grown at 8 °C by hanging drop vapor diffusion by mixing 1 μL of Nbs1-Ctp1 complex with 1 μL crystallization solution C (18% Polyethylene glycol 3350, 100 mM MES pH 6.1–6.5, 260 mM potassium thiocyanate). Crystals were washed in cryoprotectant (crystallization solution C with 26% glycerol) and yellow peptide-containing crystals were flash cooled in liquid nitrogen for data collection.
X-ray Diffraction data collection phasing and processing
Data processing, phasing, model building and refinement are detailed in Supplemental data. Final refined models of apo Nbs1 at 2.8 Å resolution (R= 23.6, Rfree=28.5) and Nbs1/Ctp1 complex at 2.2 Å resolution (R=21.6, Rfree=26.4) exhibit excellent geometry (Table S2).
Accession Numbers
Coordinates and structure factors for Nbs1-fc (3HUE) and Nbs1-fc/Ctp1 (3HUF) complex are in the Protein Data Bank.
Peptide Binding Studies
FITC-conjugated peptide binding to Nbs1-fc variants was monitored by co-elution in gel filtration or fluorescence polarization for sample concentrations detailed in the Supplemental Material.
Small Angle X-ray Scattering (SAXS) Data Collection and Processing
SAXS data were collected at SIBYLS beamline 12.3.1 at the Advanced Light Source, Lawrence Berkeley National Laboratory. SAXS scattering data was collected for 2.5, 5.0, and 10.0 mg/mL samples for Nbs1-fc, 2 mg/mL samples of spNbs1-ΔAT, and 10 mg/mL hNbs1-fc. SAXS analysis was done as described in the Supplemental Data.
Strain construction
Strains used are listed in Table S1.
Antibodies and Western Blotting
α-PAP, α-FLAG, and α-PSTAIR antibodies (Sigma) were used for Western blotting of whole-cell extracts or immunoprecipitates as described (Akamatsu et al., 2008; Limbo et al., 2007).
Cell Extract Preparation
Exponentially growing cells were exposed to bleomycin as described (Akamatsu et al, 2008). Whole-cell extracts were prepared by disrupting in lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 2.5 mM EDTA, 0.002% NP-40, 50 mM NaF, protease inhibitor tablet (Roche, complete Mini)) with a bead beater.
HO Endonuclease ChIP Assay and Real-time PCR
Chromatin immunoprecipitation assays were done as described (Limbo et al., 2007) with strains detailed in the Supplemental Material.
Nbs1-Ctp1 Co-Immunoprecipitation
Soluble whole-cell extracts were made from exponentially growing cells by disrupting in low-salt lysis buffer B1: (50 mM Tris pH 8.0, 50 mM NaCl, 2.5 mM EDTA, 0.002% NP-40, 50 mM NaF, protease inhibitor tablet (Roche, complete Mini)) with a bead beater. Ctp1-TAP and associated proteins were precipitated from extracts with a 50% suspension of IgG Sepharose (GE Healthcare), washed 3 times with B1 buffer, resolved on 8% SDS-PAGE, and examined by Western blotting.
Fluorescence Microscopy
Ctp1 cDNA was cloned as N-terminal GFP fusion proteins under control of the thiamine-repressible nmt1 promoter (pREP41). Cells were then grown in the absence of thiamine for 20 hours to induce GFP-Ctp1 expression. Images were collected as described (Limbo et al., 2007).
Ctp1 Phosphorylation Assay
Ctp1-FLAG immunoprecipitation from whole-cell extracts and subsequent λPPase treatment of precipitates were done as described (Akamatsu et al., 2008).
Supplementary Material
Acknowledgments
Fellowship support was provided to R.S.W. (Canadian Institutes of Health Research, the Alberta Heritage Foundation for Medical Research, and the Skaggs Institute for Chemical Biology), G.E.D. (National Institutes of Health NRSA), and Y.Y. (The Uehara Memorial Foundation). Work on Mre11 complex and Ctp1 in the authors’ laboratories is supported by National Cancer Institute grants CA117638, CA92584 and CA77325. SIBYLS beamline (BL12.3.1) efforts at the Advanced Light Source of Lawrence Berkeley National Laboratory are supported by U.S. Department of Energy program IDAT (for Integrating crystallography and X-ray scattering) under Contract Number DE-AC02-05CH11231. We thank SIBYLS staff G. Hura and M. Hammel for expert SAXS data collection assistance, B. Chapados, C. Chahwan for discussions, and E. Getzoff and G. Williams for comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008 doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- DiDonato M, Craig L, Huff ME, Thayer MM, Cardoso RM, Kassmann CJ, Lo TP, Bruns CK, Powers ET, Kelly JW, et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J Mol Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, Yang YP, Petukhova GV, Eckhaus M, Feigenbaum L, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:1207–1217. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen K, Rapakko K, Karppinen SM, Erkko H, Knuutila S, Lundan T, Mannermaa A, Borresen-Dale AL, Borg A, Barkardottir RB, et al. RAD50 and NBS1 are breast cancer susceptibility genes associated with genomic instability. Carcinogenesis. 2006;27:1593–1599. doi: 10.1093/carcin/bgi360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT. The P. furiosus Mre11/Rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. Embo J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger L, Demuth I, Neitzel H, Varon R, Sperling K, Chrzanowska KH, Seemanova E, Digweed M. Cancer incidence in Nijmegen breakage syndrome is modulated by the amount of a variant NBS protein. Carcinogenesis. 2007;28:107–111. doi: 10.1093/carcin/bgl126. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 complex to a double-strand break requires Interactions With Yku80 and the Xrs2 FHA domain. Genetics. 2008;180:1809–1819. doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemanova E, Sperling K, Neitzel H, Varon R, Hadac J, Butova O, Schrock E, Seeman P, Digweed M. Nijmegen breakage syndrome (NBS) with neurological abnormalities and without chromosomal instability. J Med Genet. 2006;43:218–224. doi: 10.1136/jmg.2005.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Suzuki M, Shinohara M. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics. 2005;170:71–85. doi: 10.1534/genetics.104.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. Embo J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.