Abstract
Macrophages play an essential role in the resolution of tissue damage through removal of necrotic cells, thus paving the way for tissue regeneration. Macrophages also directly support the formation of new tissue to replace the injury, through their acquisition of an anti-inflammatory, or M2, phenotype, characterized by a gene expression program that includes IL-10, the IL-13 receptor, and arginase 1. We report that deletion of two CREB-binding sites from the Cebpb promoter abrogates Cebpb induction upon macrophage activation. This blocks the downstream induction of M2-specific Msr1, Il10, II13ra, and Arg-1 genes, whereas the inflammatory (M1) genes Il1, Il6, Tnfa, and Il12 are not affected. Mice carrying the mutated Cebpb promoter (βΔCre) remove necrotic tissue from injured muscle, but exhibit severe defects in muscle fiber regeneration. Conditional deletion of the Cebpb gene in muscle cells does not affect regeneration, showing that the C/EBPβ cascade leading to muscle repair is muscle-extrinsic. While βΔCre macrophages efficiently infiltrate injured muscle they fail to upregulate Cebpb, leading to decreased Arg-1 expression. CREB-mediated induction of Cebpb expression is therefore required in infiltrating macrophages for upregulation of M2-specific genes and muscle regeneration, providing a direct genetic link between these two processes.
Keywords: macrophage polarization, muscle regeneration, transcription
The resolution of tissue injury involves a complex interaction between the tissue undergoing repair and the immune system. Immune cells are critical for the removal of necrotic cells and for fending off infectious agents. In addition, they may provide support for stem cells and progenitors as they proliferate and differentiate to repair the inflicted damage. Macrophages may play a key role in this process as the major infiltrating cell population in injured muscle, required for removal of damaged myofibers (1, 2). Macrophages also have an important role in the subsequent regrowth and differentiation of myofibers as depletion of the macrophage population after necrotic cell removal leads to a defect in regeneration (2). This latter function may require the induction in situ of an anti-inflammatory or M2 phenotype. Indeed, the M2 phenotype may be induced in macrophages in vitro through phagocytosis of myofiber debris (2).
M2 macrophage polarization is induced by anti-inflammatory cytokines and growth factors, including IL-4, IL-10, and TGF-β (3). However, there is limited information about the transcriptional control of M2 genes, and a mechanism for their coordinate regulation has yet to be elucidated. Although macrophages lacking the SHIP phosphatase were biased toward an M2 phenotype (4), the downstream transcriptional targets involved are not known. Tumor-associated macrophages with an M2 phenotype activate signaling through IRF3/STAT1 and suppress NF-κB activation (5). Decreased NF-κB signaling is likely to be responsible for the impaired expression of M1 genes, as overexpression of the p50 NF-κB subunit, which lacks a strong transactivation domain, is sufficient to repress M1 gene expression (6). The ability of macrophages to suppress cytotoxic T lymphocyte activity is also impaired by activation of PPARγ (7), indicating that in this context PPARγ activation suppresses M2 polarization. However, the role of PPARγ remains controversial, as PPARγ activation has been observed to promote M2 polarization in adipose tissue (8).
Candidate regulators can be identified by analysis of promoters of M2-specific genes in macrophages, and among these, the Il10 and Arg-1 promoters are regulated by C/EBPβ (9, 10). C/EBPβ is a member of the C/EBP family of basic region-leucine zipper (bZIP) proteins and is known to be important for the antibacterial activity of macrophages (11). However, macrophage expression of genes encoding inflammatory molecules, such as Il1b, Inos, Il6, and Tnfa is also diminished in the absence of C/EBPβ (12), confounding the role of C/EBPβ in specifying the M2 gene program. There is considerable evidence that C/EBPβ is regulated at the proteomic level through competition between interacting transcriptional regulators (see reference 13 for review). In macrophages, C/EBPβ functionally interacts with NF-κB on inflammatory and synergizes with STAT factors on anti-inflammatory promoters, respectively, suggesting that its specificity of action may be mediated by competition between cooperating factors, and in this regard the levels of C/EBPβ could be critical for determining which transcriptional programs are activated. During macrophage activation, Cebpb is transcriptionally induced by the CREB transcriptional activator, another bZIP transcription factor (14), which binds two cAMP response elements (CREs) in the proximal Cebpb promoter (15). However, given the presence of a significant basal level of C/EBPβ protein in resting macrophages, the role of CREB-mediated induction is unclear.
We report here that in activated primary macrophages, CREB-mediated induction of Cebpb expression was dispensable for induction of inflammatory (M1) genes (Il1b, Il6, Il12b, Tnfa), but required for genes characteristic of anti-inflammatory (M2) macrophages (Arg-1, Il10, Il13ra, Msr1). To confirm the role of C/EBPβ in M2 macrophage activation, we generated mice carrying a targeted deletion of two CREB-binding sites in the Cebpb promoter (βΔCre mice), and analyzed their response to skeletal muscle injury. βΔCre mice were defective in resolution of necrotic damage to skeletal muscle. βΔCre macrophages infiltrated injured muscle normally in vivo, but failed to upregulate Cebpb and Arg-1, implicating lack of Cebpb induction and defective M2 polarization in impaired muscle regeneration. These results define a molecular basis for polarized macrophage gene expression, show its significance in tissue repair, and provide a model for C/EBPβ promotion of tissue homeostasis.
Results
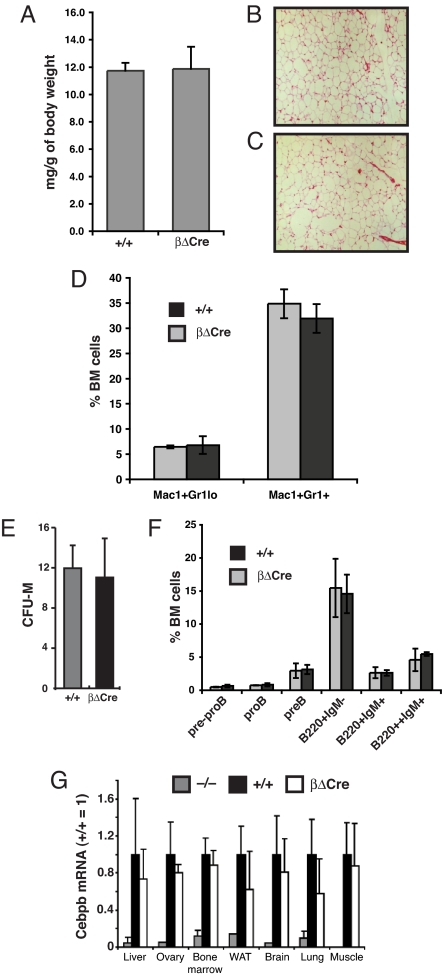
To determine if inhibition of CREB activation prevented Cebpb induction, we established that LPS stimulation caused the recruitment of CREB to the Cebpb promoter in the J774 macrophage cell line (Fig. 1A). In the presence of RO318220, an inhibitor of MSK-1/2, CREB phosphorylation in response to LPS/IFNγ was not induced (Fig. 1B), and Cebpb upregulation was abolished (Fig. 1C), consistent with a requirement for recruitment of activated CREB to the Cebpb promoter to achieve transcriptional induction. We next used homologous recombination in E14.1 ES cells to replace the Cebpb promoter CREs with a sequence composed of five binding sites for the artificial ZFHD transcription factor (16) (Fig. S1). This eliminates binding sites for endogenous transcription factors while maintaining promoter spacing. After germ line transmission, mice were bred to homozygosity for the resulting CebpbΔCRE allele (henceforth βΔCre mice). While Cebpb−/− mice display female infertility (17), βΔCre females were fertile and lactated. Induction of Cebpb expression by CREB promotes adipogenesis in vitro (18); however, both adipose tissue amounts (Fig. 2A) and histological appearance (Fig. 2 B and C) were normal in βΔCre mice. In the hematopoietic system, C/EBPβ is important for B-cell differentiation (19), macrophage function (11), and stress granulopoiesis (20). However, the number and distribution of granulocytic cells (Fig. 2D and Fig. S2), as well as the number of bone marrow (BM) macrophage progenitors (measured as macrophage colony forming cells; Fig. 2E), B cell progenitors, or mature B cells (Fig. 2F and Fig. S3) were not affected by the βΔCre mutation. Examination of Cebpb mRNA levels in adult βΔCre mice showed no significant downregulation compared to wild-type littermate controls in any major C/EBPβ-expressing tissue (Fig. 2G). From this analysis, we conclude that the Cebpb promoter CREs are dispensable for major physiological functions of C/EBPβ during normal development.
Fig. 1.
CREB activation induces Cebpb in macrophages. (A) ChIP of CREB on the Cebpb promoter in J774 macrophages stimulated with LPS. The CRE PCR, specific for a 140-bp DNA fragment that spans the CRE elements on the Cebpb promoter, demonstrates the recruitment of CREB onto the CREs of the C/EBPβ promoter upon LPS treatment. Amplification of a 200-bp fragment in the 3′ UTR was used as a control. (B) IFNγ-primed J774 cells were pretreated with 5 μM Ro 31–8220 or vehicle for 20 min followed by 1 h stimulation with IFNγ/LPS. Cell lysates were processed for immunoblotting with antibody against phospho-CREB (upper panel) followed by stripping and reprobing with antibody against α-tubulin (lower panel). (C) IFNγ primed J774 cells were pretreated with 5 μM Ro 31–8220 or vehicle for 20 min followed by 4 h stimulation with IFNγ/LPS. Relative Cebpb mRNA levels in J774 cells were measured in triplicate by quantitative real-time PCR, normalized to ubiquitin. Data are presented as the mean ± SD (+/+ n = 3; βΔCre n = 3).
Fig. 2.
Role of CREB-C/EBPβ cascade in normal development. (A) Epididymal fat pad weight normalized to total body weight for βΔCre mice and +/+ littermates. (B and C) Eosin hematoxylin staining on epididymal fat pad histological sections from +/+ and βΔCre mice. (D) Frequency of Mac-1+Gr-1lo and Mac-1+Gr-1+ cells from +/+ and βΔCre BM determined by flow cytometry phenotyping. Data are presented as the mean ± SD (+/+ n = 3; βΔCre n = 3). (E) Macrophage colony-forming activity of BM cells were plated in methylcellulose medium containing M-CSF (10 ng/mL). Macrophage colony-forming units were scored after 8 days and are presented as average CFU-M/103 BM cells (+/+ n = 3; βΔCre n = 3). Data are presented as the mean ± SD. (F) Frequency of prepro-B (B220+CD43+AA4.1+CD19−), pro-B (B220+CD43+AA4.1+CD19+), pre-B (B220+CD43−A4.1+CD19+), immature B (B220+IgM−), mature B (B220+IgM+), and recirculating B cells (B220++IgM+) from +/+ and βΔCre BM determined by flow cytometry (+/+ n = 3; βΔCre N = 3). Data are presented as the mean ± SD. (G) Cebpb expression levels in tissues extracted from Cebpb−/−, Cebpb+/+ and βΔCre mice measured by real time PCR (n = 3 for each genotype). Although Cebpb mRNA levels were somewhat lower in tissues derived from βΔCre mice compared to +/+ controls, the differences were not significant. The Cebpb gene is intronless, and Cebpb−/− mice were used to control for influence of genomic DNA contamination on the analysis. WAT, white adipose tissue.
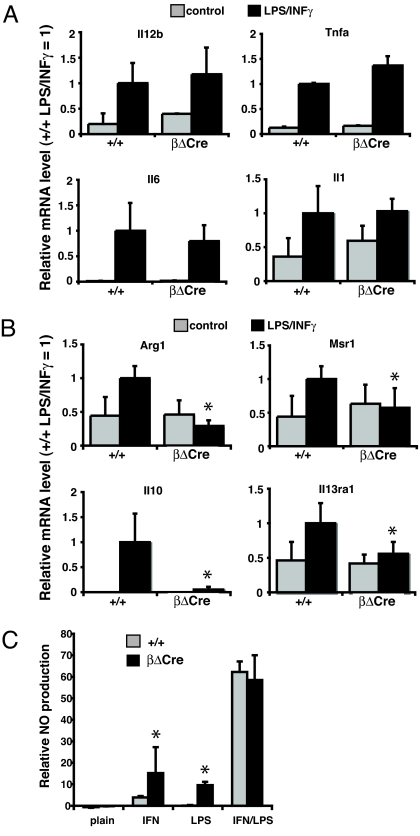
To assess the role of the Cebpb promoter CREs in inflammatory gene expression, primary macrophages were derived from βΔCre and control (+/+) BM in the presence of macrophage-colony stimulating factor (M-CSF). After subsequent activation with LPS/IFNγ, total RNA was isolated, and Cebpb expression analyzed. While significant (3-fold) induction was observed in control macrophage cultures, βΔCre macrophages failed to induce Cebpb expression (Fig. 3A). C/EBPβ induction was also impaired at the protein level in βΔCre macrophages (Fig. 3B), whereas no differences in the levels or kinetics of CREB/ATF1 phosphorylation was observed between +/+ and βΔCre macrophages, indicating that the signal transduction pathways impinging on the Cebpb promoter CREs are functional. Analysis of major proinflammatory C/EBPβ target genes showed that those encoding the central M1-specific cytokines Tnfa, Il1b, Il6, and Il12b were unaffected by the absence of Cebpb upregulation (Fig. 4A). In contrast, analysis of major M2 specific transcripts showed that Il10, Arg-1, Il13ra, and Msr1 were not upregulated in activated βΔCre macrophages (Fig. 4B). Deletion of the Cebpb promoter CREs thus led to a specific and coordinated loss of M2-specific gene expression after LPS/IFNγ activation. Finally, while high level induction of either +/+ and βΔCre macrophages with LPS/IFNγ elicited similar production of nitric oxide (NO), a principal proinflammatory effector molecule, treatment with either agent alone generated higher NO levels in βΔCre macrophage cultures (Fig. 4C), further indicative of their M1 bias.
Fig. 3.
Cebpb promoter CREs are required for induction by LPS/IFNγ. (A) Real-time PCR analysis of Cebpb expression in BM-derived primary macrophages from +/+ and βΔCre mice, treated with IFNγ/LPS as indicated. Data are presented as the mean ± SD (+/+ n = 6; βΔCre n = 6). Significant differences (P < 0.05; Student's t-test) are indicated by asterisk (*). (B) Western blots of C/EBPβ (p33), phospho-CREB (P-CREB), and tubulin (as internal control) from +/+ and βΔCre primary macrophages, either untreated (-) or treated with LPS for the indicated time after pretreatment with IFNγ (+).
Fig. 4.
Defective M2 gene expression in activated βΔCre macrophages. (A) Real-time PCR expression analysis of proinflammatory markers in BM-derived macrophages upon IFNγ/LPS stimulation analyzed as in Fig. 3A (+/+ n = 6; βΔCre n = 6). (B) Real-time PCR expression analysis of anti-inflammatory markers in BM-derived macrophages upon IFNγ/LPS stimulation analyzed as in Fig. 3A (+/+ n = 6; βΔCre n = 6). (C) Relative NO release of thioglycollate-elicited peritoneal macrophages stimulated with IFNγ and/or LPS, as indicated. Data are presented as the mean ± SD (+/+ n = 3; βΔCre n = 3).
To evaluate the pro- and anti-inflammatory C/EBPβ functions in muscle regeneration, a well-established in vivo injury model was used (21). Tibialis anterior (TA) and quadriceps (Q) muscles of one leg were injected with cardiotoxin (CTX), while the contralateral muscles were left uninjured. CTX induces local necrosis (22), followed by an inflammatory response consisting mainly of macrophage invasion (23), and subsequent tissue reconstitution. Control and βΔCre mice developed similar necrotic injuries, as determined by histological analysis at day 2 after CTX injection (Fig. 5 A and B). By day 5, the βΔCre regenerating regions showed increased inflammation, as indicated by the presence of myofibers with eosinophil cytoplasm and accretion of nuclei at the sites of injury (Fig. 5 C and D). At day 10 after CTX injection, where in +/+ mice the injury is largely resolved, macroscopic examination of βΔCre muscle showed a highly fibrotic appearance (Fig. S4). Moreover, at the histological level, multifocal areas of inflammation and severely calcified myofibers were observed among newly formed myofibers in the βΔCre muscles, whereas +/+ muscle was completely regenerated (Fig. 5 E and F). Many of the βΔCre regenerating myofibers appeared smaller (Fig. 5F) than those in control muscles, as confirmed by a decrease in the cross-sectional fiber area (CSA) of βΔCre myofibers (Fig. 5G) and a 20% reduction in the total regenerated area (Fig. 5H) 10 days after CTX injection. Together, these results clearly showed that the regeneration capacity in the βΔCre muscles was decreased and confirmed increased inflammation as an important element of this impairment. To exclude any contribution to the observed phenotype from minor differences in the strain background, these experiments were repeated after backcrossing to C57BL/6 for six generations followed by intercrossing to generate homozygous βΔCre mice; the phenotype remained unaltered (Fig. S5). In addition, no difference in the regenerative timecourse was observed between C57 and 129 mouse strains (Fig. S6).
Fig. 5.
Evaluation of regenerating skeletal muscles in βΔCre mice. (A and B) Trichrome staining of +/+ (A) and βΔCre (B) tibialis anterior muscle shows similar necrosis in both genotypes 2 days after CTX injection. (C and D) Recovery of the injured muscles and regenerating myofibers (containing centralized nuclei) at day 5 postinjury in +/+ (C) and βΔCre (D) muscle. Myofibers with eosinophil cytoplasm are indicated with arrowheads in (D). (E and F) At day 10 postinjury, the existence of numerous small fibers (arrowheads) is evident in the βΔCre injured muscle (F) compared to +/+ muscle (E). Note that, in contrast to control regenerating muscles, βΔCre muscles contained many calcified fibers (arrows). (G) Morphometric analysis of muscle regeneration in +/+ and βΔCre mice. The data show the frequency distribution in the tibialis anterior fiber cross-sectional area (CSA) within the regenerating muscle. (H) Measurement of the total regenerating (marked by centralized nuclei) fiber area in mutant injured muscles. Data are presented as the mean ± SD (+/+ n = 6; βΔCre n = 6). Significant differences (P < 0.05; Student's t-test) are indicated by asterisks (*). (J and K) Regenerated tibialis anterior muscle of +/+ (J) and BMKO (K) mice 10 days after CTX injection. No impairment of regeneration was evident in BMKO muscle compared to +/+ muscle. (L) BM cells from βΔCre mutant or wild-type mice (CD45.1−CD45.2+ allotype) were transferred to lethally irradiated recipient mice (CD45.1+CD45.2+ allotype). Plots show representative FACS analysis of peripheral blood in recipient mice at 4 weeks after transplantation to measure the engraftment of donor (CD45.1−CD45.2+) and recipient (CD45.1+CD45.2+) cells. (M and N) The mice transplanted in (L) were subjected to the CTX injury protocol. Trichrome staining of injured TA muscle sections at day 10 postinjury is shown. Note that the observed defect in muscle regeneration in βΔCre mice was recapitulated in wild-type mice transplanted with βΔCre mutant BM cells (N) while control wild-type mice transplanted with wild-type BM cells showed normal regeneration (M).
Since the mutation in βΔCre mice is ubiquitous, a potential muscle-intrinsic role for C/EBPβ during muscle regeneration could not be excluded. We tested this possibility by conditionally ablating Cebpb gene expression exclusively in skeletal muscles (BMKO mice; generated using a Cre-conditional null allele of Cebpb and the MCK-Cre transgene (24) (Fig. S7). BMKO mice showed normal regeneration after CTX injury (Fig. 5 J and K), consistent with a requirement for C/EBPβ in cells infiltrating the damage, rather than in resident muscle cells. To further determine if BM-derived cells were responsible for the regeneration defect, wild-type mice (CD45.1/2 allotype) were repopulated with +/+ or βΔCre BM (CD45.2 allotype) (Fig. 5L) and subjected to the injury protocol. The presence of βΔCre BM cells was sufficient to impair muscle regeneration (Fig. 5 M and N). Conversely, repopulation of βΔCre mice with +/+ BM was sufficient to rescue the regeneration defect (Fig. S8). Therefore the presence of the βΔCre mutation in the hematopoetic system is both necessary and sufficient for the regeneration defect.
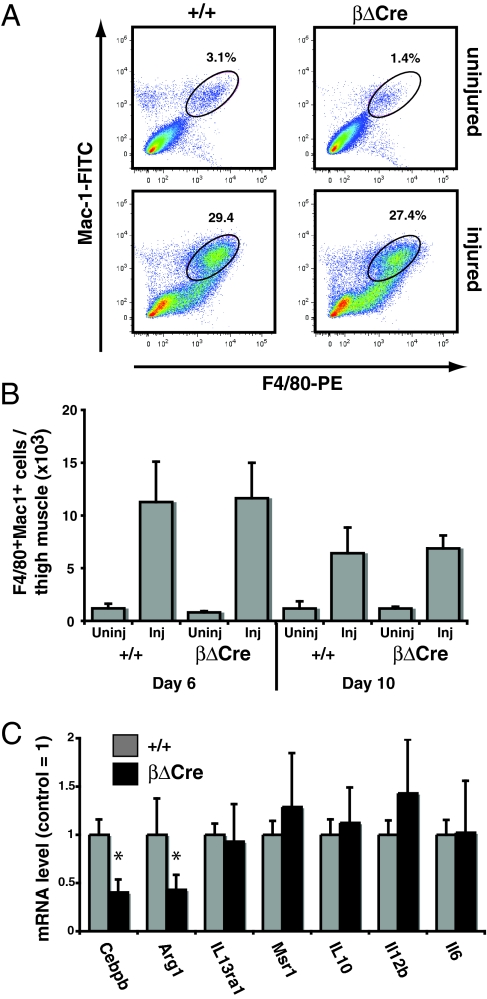
To further investigate the nature of the cells infiltrating damaged muscle, we quantified the amount of macrophages present in injured +/+ and βΔCre muscle by flow cytometry. As expected, injury induced a strong increase in Mac-1+F4/80+ macrophage numbers; however, no difference was observed between βΔCre and +/+ mice (Fig. 6 A and B). We next sorted macrophages from injured muscles 6 days after CTX injection to determine whether Cebpb expression was affected at this stage. Cebpb mRNA was reduced 2.5-fold in βΔCre macrophages compared to controls (Fig. 6C), demonstrating the requirement for CREB-dependent Cebpb upregulation in macrophages during muscle regeneration. Expression of M1-specific genes (Il12b, Il6) was unaltered, whereas expression of the M2-specific Arg-1 was downregulated, consistent with defective M2 polarization of the macrophage population. Deletion of a regulatory element from the Cebpb promoter could potentially affect the expression of neighboring genes. To address this we performed Affymetrix analysis on +/+ and βΔCre macrophages isolated from injured muscle. None of the neighboring genes were deregulated, whereas the expected downregulation of Cebpb was observed (Fig. S9A). Finally, Q-PCR analysis of selected genes with potential relevance for macrophage activation in in vitro-induced +/+ and βΔCre macrophages also did not reveal any significant differences in the expression or regulation (Fig. S9B), consistent with the effect of the βΔCre deletion being solely on the expression of Cebpb.
Fig. 6.
Infiltrating macrophage phenotype in injured βΔCre muscle. (A) Phenotypic analysis from +/+ and βΔCre muscle uninjured and injured 6 days after CTX injection by flow cytometry for the presence of Mac-1+F4/80+ cells. Plots are representative of four independent experiments and numbers represent the frequency of cells in the indicated gates. (B) Total mononuclear cells were isolated from injured and control thigh muscle and analyzed for expression of F4/80 and Mac-1 by FACS. The total number of recovered F4/80+Mac-1+ cells is indicated for each condition (n = 2 for uninjured samples; N> = 5 for injured samples). Error bars indicate standard deviations. (C) F4/80+Mac-1+ cells were sorted from day 6 injured muscle, and gene expression analyzed by real-time PCR (N> = 5/genotype). Data are presented as the mean ± SD normalized to the +/+ value (= 1). Asterisks (*) indicate P < 0.05 (Student's t-test).
Discussion
Macrophage polarization has been proposed to play important roles in tissue repair and cancer. In necrotic muscle injury, infiltrating macrophages are essential for regeneration, as depletion of peripheral monocytes before injury prevents removal of necrotic tissue (2). Depletion of F4/80+ macrophages from regenerating muscle led to incomplete repair and reduced muscle fiber size (2) indicating a direct role for macrophages in promoting fiber formation and growth. Further, muscle fiber regeneration correlated with a transition to an anti-inflammatory macrophage phenotype both in vivo and in vitro (2). Macrophages isolated from primary tumors have also been found to display an anti-inflammatory M2 phenotype and are thought to be recruited and/or M2 polarized by the tumor cells as a means to prevent their destruction by the immune system (25). This may represent subversive use of a mechanism normally activated to resolve tissue injury by shutting down the inflammatory response to damage.
The βΔCre mice described in this report provide a genetic model in which the in vivo role of macrophage polarization may be tested. Our results support a model whereby macrophages recruited to muscle injury are M2 polarized by the regenerating environment to stimulate fiber growth; thus a mutation that specifically impairs M2-specific macrophage gene expression interferes with the later stages of muscle regeneration and fiber replacement, whereas the initial removal of necrotic tissue is maintained. Specifically, the reduction of Arg-1 expression in βΔCre macrophages is likely to result in the re-routing of arginine metabolism away from arginase-mediated polyamine synthesis (which promotes tissue regeneration) toward iNOS-mediated NO production (which promotes degradation of transcripts encoding the myocyte differentiation factor MyoD) (26). An essential CREB-dependent pathway for inducing Arg-1 and polyamine synthesis during axonal regeneration has previously been described (27). Our results are consistent with C/EBPβ acting as an intermediate between CREB and Arg-1 expression in this setting as well, implicating the CREB-C/EBPβ-arginase pathway in multiple cell types to promote tissue repair. Other M2-specific genes (Il13ra1, Msr1, Il10) were not affected in βΔCre macrophages in vivo. This may be because they are not significantly induced even in the wild-type macrophages or more likely because signals not present in our in vitro conditions are operating in injured muscle tissue.
It is notable that macrophage activation does not involve changes in the phosphorylation, DNA binding, or translational control of C/EBPβ, which appears to be in a fully active state in resting macrophages (14). These cells appear poised for rapid deployment of an inflammatory response, which requires only a basal C/EBPβ level before activation. Our findings pinpoint CREB-mediated Cebpb upregulation as a mechanism whereby activated macrophages can coordinate M2-specific gene induction, which may serve to temporally organize pro- and anti-inflammatory responses. Interference in this regulatory mechanism results in the normal induction of M1 proinflammatory genes upon macrophage activation, but impaired upregulation of M2-specific genes. The M2 program thus seems to be specifically sensitive to C/EBPβ levels. Importantly, βΔCre macrophages express lower levels of Cebpb mRNA within the injured muscle, demonstrating that the CREB-C/EBPβ cascade that is normally active during tissue repair has been perturbed.
Although Cebpb expression is generally induced in response to physiological stress, including hypoxia and inflammation in several tissues and cell types, Cebpb−/− mice in which C/EBPβ-dependent gene regulation has been abrogated display a paradoxically improved response to both ischemic stroke (28) and inflammatory steatohepatitis (29), presumably because both the pro- and anti-inflammatory effects of C/EBPβ action have been blocked systemically. More subtle perturbations, such as the one used in the present study, will be necessary to tease out various tissue-specific and temporal roles of C/EBPβ, which are likely to be dependent on relatively small variations in Cebpb expression levels in response to different stimuli. The molecular insights obtained here should allow for therapeutic manipulation of this regulatory pathway to either promote or impair M2 polarization.
Methods
Mouse Strains.
The βΔCre mouse strain was generated by replacement of the Cebpb promoter CREs with ZFHD binding sites using ET recombination. The detailed cloning and ES cell targeting strategy is provided in the SI Methods.
C/EBPβ null mice have been previously described (17) and were obtained from E. Sterneck, National Cancer Institute (NCI), Bethesda, MD. Conditional C/EBPβ knockout mice have been described elsewhere (30). MCK-Cre mice (24) were obtained from R. Kahn, Harvard Medical School, Boston, MA. Mouse strains were maintained on a mixed C57BL/6–129/Ola background, except where specifically stated otherwise. All animal procedures were performed according to Italian national and European Molecular Biology Laboratory (EMBL) institutional guidelines.
Muscle Injury Induction.
For muscle regeneration experiments, animals were anesthetized using 2.5% Avertin. Tibialis anterior (TA) and quadriceps from 2- to 3-month-old control, βΔCre, and BMKO mice were injected with 20 and 40 μL, respectively, of 10 μM CTX (Latoxan). The muscles were collected 2, 5, 6, and 10 days following the injection of CTX. At least three mice per time point were analyzed. Details of histological analysis is found in the SI Methods.
Cell Culture.
To obtain BM-derived macrophages, femurs and tibias were collected from a mouse, crushed in a mortar in presence of 1% FCS/PBS, and filtered. Cells were washed, resuspended, and cultured in differentiation medium consisting of RPMI (Gibco), 20% FCS, 50 μM β-mercaptoethanol, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, and 20 ng/mL macrophage colony-stimulating factor (M-CSF; Sigma). The cells were cultured in differentiation medium for 6 days, after which M-CSF was depleted. For macrophage activation, cells were treated with 100 U/mL IFNγ (PeproTech) in growth medium, with 5% FCS, for 16 h. Next, the cells were stimulated with 100 U/mL IFNγ and 1 μg/mL LPS from Escherichia coli (Sigma) for 4 h, after which RNA was extracted. For the preparation of peritoneal macrophages, mice were killed 3 days after an i.p. injection of 1 mL 3% thioglycollate broth (Sigma). Exudate cells were harvested by washing the peritoneal cavity with 12 mL PBS and subsequently cultured in DMEM, 5% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. For NO production measurements, peritoneal macrophages were plated in triplicate in a 96-well plate at 0.1 million cells/well. The cells were either left untreated or treated with 10 U/mL IFNγ and/or 10 ng/mL LPS for 48 h. NO2- concentration in the medium was measured with the Griess reagents as described (31). NO2- concentrations were normalized to Thiazolyl blue (MTT; Sigma) vital dye staining to correct for variations in cell number and viability. Macrophage colony-forming unit assays were performed as previously described (32). For Western blotting, cells were washed with cold PBS and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 30 mM Na4P205, 1.25 mM NaF, 100 μM Na3VO4), and lysates were processed as described in the SI Methods.
Chromatin Immunoprecipitation (ChIP).
J774 macrophages were grown to subconfluence in 10-cm dishes, and then treated with 1 μg/mL LPS for 15 min. Details of the ChIP protocol are found in the SI Methods.
Cell Preparation.
Injured muscles were placed in warmed DMEM (Gibco), and matrix, fibrotic tissue, and nerves were removed carefully. The muscles were chopped into small pieces, and enzymatic disaggregation was performed, first using freshly prepared 4 mg/mL collagenase (Sigma) for 30 min (37 °C), and then using 1 mg/mL collagenase/dispase (Roche) for 25 min (37 °C). Disaggregation was stopped with 5 mL horse serum (heat-inactivated; Gibco), and after filtration with a 40-μm cell strainer (BD Bioscience), the cells were mixed with cold PBS plus 1% FCS (Gibco). BM cells were obtained as described (32). For staining of cells for flow cytometry see the SI Methods.
Gene Expression Analysis.
RNA isolated from sorted +/+ and βΔCre macrophages 6 days postinjury was subjected to microarray analysis using Affymetrix MOE430.2 arrays. Data were processed as previously described (33). Four biological replicates were performed for each genotype, from which average normalized expression values for Cebpb genomic neighbors were calculated. Real-time PCR conditions are described in Table S1 and in the SI Methods.
Statistical Analysis.
Evalation of statistical significance was done using Student's t-test, except for measurement of total regenerated fiber area, which was done by ANOVA .
Acknowledgments.
We thank Dr. E. Sterneck for providing C/EBPβ genomic clones and Dr. R. Kahn for MCK-Cre mice. This work was supported by the Association for International Cancer Research, the European Commission (EuroCSC STREP), and the European Muscle Development Network (MYORES).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908641106/DCSupplemental.
References
- 1.Lescaudron L, et al. Blood borne macrophages are essential for the triggering of muscle regeneration following muscle transplant. Neuromuscul Disord. 1999;9:72–80. doi: 10.1016/s0960-8966(98)00111-4. [DOI] [PubMed] [Google Scholar]
- 2.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Rauh MJ, et al. SHIP represses the generation of alternatively activated macrophages. Immunity. 2005;23:361–374. doi: 10.1016/j.immuni.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Biswas SK, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 6.Saccani A, et al. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- 7.Van Ginderachter JA, et al. Peroxisome proliferator-activated receptor gamma (PPARgamma) ligands reverse CTL suppression by alternatively activated (M2) macrophages in cancer. Blood. 2006;108:525–535. doi: 10.1182/blood-2005-09-3777. [DOI] [PubMed] [Google Scholar]
- 8.Bouhlel MA, et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6:137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SM., Jr Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Liu YW, Tseng HP, Chen LC, Chen BK, Chang WC. Functional cooperation of simian virus 40 promoter factor 1 and CCAAT/enhancer-binding protein beta and delta in lipopolysaccharide-induced gene activation of IL-10 in mouse macrophages. J Immunol. 2003;171:821–828. doi: 10.4049/jimmunol.171.2.821. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka T, et al. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 12.Gorgoni B, Maritano D, Marthyn P, Righi M, Poli V. C/EBP beta gene inactivation causes both impaired and enhanced gene expression and inverse regulation of IL-12 p40 and p35 mRNAs in macrophages. J Immunol. 2002;168:4055–4062. doi: 10.4049/jimmunol.168.8.4055. [DOI] [PubMed] [Google Scholar]
- 13.Nerlov C. C/EBPs: Recipients of extracellular signals through proteome modulation. Curr Opin Cell Biol. 2008;20:180–185. doi: 10.1016/j.ceb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Bradley MN, Zhou L, Smale ST. C/EBPbeta regulation in lipopolysaccharide-stimulated macrophages. Mol Cell Biol. 2003;23:4841–4858. doi: 10.1128/MCB.23.14.4841-4858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niehof M, Manns MP, Trautwein C. CREB controls LAP/C/EBP beta transcription. Mol Cell Biol. 1997;17:3600–3613. doi: 10.1128/mcb.17.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomerantz JL, Sharp PA, Pabo CO. Structure-based design of transcription factors. Science. 1995;267:93–96. doi: 10.1126/science.7809612. [DOI] [PubMed] [Google Scholar]
- 17.Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang JW, Klemm DJ, Vinson C, Lane MD. Role of CREB in transcriptional regulation of CCAAT/enhancer-binding protein beta gene during adipogenesis. J Biol Chem. 2004;279:4471–4478. doi: 10.1074/jbc.M311327200. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, et al. Impaired generation of bone marrow B lymphocytes in mice deficient in C/EBPbeta. Blood. 1997;90:156–164. [PubMed] [Google Scholar]
- 20.Hirai H, et al. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 21.Mourkioti F, et al. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116:2945–2954. doi: 10.1172/JCI28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sue SC, Chien KY, Huang WN, Abraham JK, Chen KM, Wu WG. Heparin binding stabilizes the membrane-bound form of cobra cardiotoxin. J Biol Chem. 2002;277:2666–2673. doi: 10.1074/jbc.M104887200. [DOI] [PubMed] [Google Scholar]
- 23.McLennan IS. Degenerating and regenerating skeletal muscles contain several subpopulations of macrophages with distinct spatial and temporal distributions. J Anat. 1996;188:17–28. [PMC free article] [PubMed] [Google Scholar]
- 24.Bruning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani A, et al. Infiltration of tumours by macrophages and dendritic cells: Tumour-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Novartis Found Symp. 2004;256:137–145. discussion 146–148, 259–269. [PubMed] [Google Scholar]
- 26.Di Marco S, et al. NF-kappa B-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol Cell Biol. 2005;25:6533–6545. doi: 10.1128/MCB.25.15.6533-6545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, et al. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron. 2004;44:609–621. doi: 10.1016/j.neuron.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 28.Kapadia R, Tureyen K, Bowen KK, Kalluri H, Johnson PF, Vemuganti R. Decreased brain damage and curtailed inflammation in transcription factor CCAAT/enhancer binding protein beta knockout mice following transient focal cerebral ischemia. J Neurochem. 2006;98:1718–1731. doi: 10.1111/j.1471-4159.2006.04056.x. [DOI] [PubMed] [Google Scholar]
- 29.Rahman SM, et al. CCAAT/enhancing binding protein beta deletion in mice attenuates inflammation, endoplasmic reticulum stress, and lipid accumulation in diet-induced nonalcoholic steatohepatitis. Hepatology. 2007;45:1108–1117. doi: 10.1002/hep.21614. [DOI] [PubMed] [Google Scholar]
- 30.Lopez RL, et al. C/EBPα and -β couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat Cell Biol. 2009 doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 31.Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J Immunol. 1988;141:2407–2412. [PubMed] [Google Scholar]
- 32.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 33.Kirstetter P, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]