Abstract
The HSPs (hereditary spastic paraplegias) are genetic conditions in which there is distal degeneration of the longest axons of the corticospinal tract, resulting in spastic paralysis of the legs. The gene encoding spartin is mutated in Troyer syndrome, an HSP in which paralysis is accompanied by additional clinical features. There has been controversy over the subcellular distribution of spartin. We show here that, at steady state, endogenous spartin exists in a cytosolic pool that can be recruited to endosomes and to lipid droplets. Cytosolic endogenous spartin is mono-ubiquitinated and we demonstrate that it interacts via a PPXY motif with the ubiquitin E3 ligases AIP4 [atrophin-interacting protein 4; WWP2 (WW domain-containing E3 ubiquitin protein ligase 2] and AIP5 (WWP1). Surprisingly, the PPXY motif, AIP4 and AIP5 are not required for spartin's ubiquitination, and so we propose that spartin acts as an adaptor for these proteins. Our results suggest that spartin is involved in diverse cellular functions, which may be of relevance to the complex phenotype seen in Troyer syndrome.
Keywords: epidermal growth factor receptor (EGFR), hereditary spastic paraplegia (HSP), microtubule interacting and trafficking domain, mono-ubiquitination, spartin
Abbreviations: AIP, atrophin-interacting protein; DMEM, Dulbecco's modified Eagle's medium; EGF, epidermal growth factor; EGFR, EGF receptor; GFP, green fluorescent protein; HA, haemagglutinin; HECT, homologous with the E6AP C-terminus; GST, glutathione transferase; HSP, hereditary spastic paraplegia; LAMP1, lysosome-associated membrane protein 1; Nedd4.2, neural-precursor-cell-expressed developmentally down-regulated; siRNA, small interfering RNA; α-spartin, polyclonal antibody against spartin; spartinAAAA, mutation of spartin's PPXY motif to AAAA; Vps4, vacuolar protein sorting 4; WWP, WW domain-containing E3 ubiquitin protein ligase
INTRODUCTION
The HSPs (hereditary spastic paraplegias) are genetic conditions in which the main pathology is degeneration of the distal ends of the longest axons of the corticospinal tract, the main pathway that connects the motor cortex of the brain to the spinal cord [1–4]. Study of the HSPs is therefore important to understand the molecular mechanisms involved in axonal maintenance and the dying-back axonopathy seen in these and other more common neurological conditions.
The HSPs are divided into pure forms, in which spastic paraplegia occurs in relative isolation, and complex forms, in which there are other prominent clinical features [1,5]. This clinical diversity is reflected in genetic heterogeneity, and at present more than 30 genetic loci have been mapped, with causative mutations in numerous genes identified [2–4]. One of these is the SPG20 gene, which encodes spartin. Mutation of this gene causes Troyer syndrome, an autosomal recessive complex HSP in which spastic paraplegia is accompanied by severe atrophy of the small hand muscles, dysarthria, mental retardation and mild skeletal abnormalities, including short stature [6]. The condition was identified in the Old Order Amish population, where the causative mutation is a frameshift that is predicted to cause a substitution of 29 amino acids before premature truncation of the protein by 268 residues [6]. This is likely to cause the loss of the protein [7].
To date, studies aimed at determining the subcellular location of spartin have largely relied on over-expression systems. Lu et al. [8] reported that epitope-tagged spartin localized to mitochondria, whereas Bakowska et al. [9] found that epitope-tagged spartin was cytosolic, but was sequestered to endosomes on stimulation with EGF (epidermal growth factor) or on expression of ATPase-defective Vps4 (vacuolar protein sorting 4). ATPase-defective Vps4 has a dominant-negative effect, trapping proteins on endosomes that normally have a transient localization there [10,11]. Bakowska et al. [9] also reported that endogenous spartin is required for efficient endosomal degradation of the EGFR (EGF receptor), supporting a role for the protein at endosomes. In contrast, a very recent report found no recruitment of spastin to dominant-negative Vps4 endosomes. Instead, epitope-tagged, over-expressed spartin was found in cultured cells to be cytosolic, but was recruited to lipid droplets upon induction by oleic acid treatment [12]. Lipid droplets are cytoplasmic structures, bound by a monolayer of polar lipids, which store neutral lipids in their core. They are found in nearly all cell types and play active roles in lipid homoeostasis and a number of other cellular functions [13,14]. In summary, existing localization studies have been of uncertain relevance to endogenous spartin and have given contradictory results.
Spartin is multiply mono-ubiquitinated [9]. Using over-expression systems in a study published during the preparation of this manuscript, Eastman et al. [12] found that spartin interacts with the HECT (homologous with the E6AP C-terminus) family ubiquitin E3 ligase AIP5 [atrophin-interacting protein 5, WWP1 (WW domain-containing E3 ubiquitin protein ligase 1)], although the relevance of this interaction to the endogenous proteins was not demonstrated. HECT ubiquitin ligases contain an N-terminal membrane-binding C2 domain, WW domains that interact with PPXY motifs in binding partners and a C-terminal HECT domain that has ubiquitin binding and ligation properties [15]. The interaction was mediated by binding between a spartin PPXY motif and WW domains in the E3 ligase, and over-expression experiments suggested that AIP5 might be responsible for ubiquitination of spartin [12].
In the present study, we identified the subcellular locations of endogenous spartin, demonstrating that it is present in a cytosolic pool that can be sequestered to endosomes and to lipid droplets. We also identified interactions at the endogenous protein level between spartin and the E3 ubiquitin ligases AIP4 and AIP5. Surprisingly, we show that spartin's PPXY motif, AIP4 and AIP5 are not required for spartin's ubiquitination. Instead, we suggest that spartin has a more complex relationship with these ubiquitin ligases, perhaps functioning as an adaptor for them.
EXPERIMENTAL
Antibodies and markers
Mouse monoclonal anti-α-tubulin (T-9026) and anti-FLAG (A-9594) and rabbit polyclonal anti-actin (A-2066) were from Sigma–Aldrich. Goat polyclonal anti-AIP4 (Sc-11890) and mouse monoclonal anti-LAMP1 (lysosome-associated membrane protein 1; clone H4A3) were from Santa Cruz Biotechnology. Mouse monoclonal anti-AIP5 (H00011059-M01) was from Abnova. Mouse monoclonal anti-EEA1 (early endosome antigen 1; 610457) and anti-GM130 (610823) were from BD Transduction Laboratories. Mouse monoclonal anti-EGFR (Ab-5) and rabbit polyclonal anti-calreticulin (Ab-4) were from Calbiochem. Mouse monoclonal antibodies to polyubiquitinated protein (FK1) and poly- and mono-ubiquitinated proteins (FK2) were from Biomol International. Mouse monoclonal anti-HA (haemagglutinin; code number HA.11) was from Covance. Sheep polyclonal anti-TGN46 (AHP500G) was from AbD Serotec. Mouse monoclonal anti-KDEL (SPA-827) was from Stressgen. Mouse monoclonal anti-Myc (4A6) was from Upstate Biotechnology. Rabbit polyclonal anti-GFP (green fluorescent protein; catalogue number 6556-25) was from AbCam. Mouse monoclonal anti-clathrin (X22) was a gift from Professor Margaret S. Robinson (Department of Clinical Biochemistry, University of Cambridge, Cambridge, U.K.). Rabbit polyclonal anti-spartin was raised against full-length GST (glutathione transferase)–spartin (Harlan SeraLabs). Rabbit polyclonal anti-M6PR was raised as previously described [16]. Peroxidase-conjugated secondary antibodies for Western blotting were obtained from Sigma–Aldrich. MitoTracker and Alexa Fluor®-labelled secondary antibodies for immunofluorescence were from Invitrogen Molecular Probes.
Constructs
The full-length cDNA for spartin was amplified by PCR from the IMAGE clone 4738886. To make GST–spartin, the PCR primers used in the reaction contained restriction enzyme sites compatible with cloning into the pGEX-4T3 expression vector (GE Healthcare), which coded for a 5′ in-frame GST tag. The PCR product was ligated into the expression vector using T4 DNA ligase (New England Biolabs). The sequence of the inserted cDNA and insertion sites was verified by sequencing (Geneservice). For the Myc–spartin and Myc–spartinAAAA (mutation of spartin's PPXY motif to AAAA) constructs, Gateway-compatible primers were used to amplify the spartin cDNA, and the PCR product was then ligated into a pcDNA3-NG-Myc Gateway-compatible vector, using previously described methods [17]. Site-directed mutagenesis of this vector was carried out using the Phusion Site-Directed Mutagenesis kit (Finnzymes). The Myc–Nedd4.2 (neural-precursor-cell-expressed developmentally down-regulated 4.2) constructs were a gift from Dr Adriano Marchese (Department of Pharmacology and Experimental Therapeutics, Loyola University, Chicago, IL, U.S.A.), whereas the AIP4 constructs were a gift from Professor Paul Lehner (Department of Medicine, School of Clinical Medicine, University of Cambridge, Cambridge, U.K.).
Cell culture, transient transfection of DNA and immunofluorescence microscopy
HeLa (human cervical carcinoma), NSC34 (a hybrid murine motor neuron/neuroblastoma cell line) and PC12 (a rat neuroendocrine cell line) cells were maintained in DMEM (Dulbecco's modified Eagle's medium; Sigma–Aldrich) containing 10% FBS (fetal bovine serum; Sigma–Aldrich), 100 units/ml penicillin, 100 μg/ml streptomycin (Sigma–Aldrich) and 2 mM L-glutamine (Sigma–Aldrich). PC12 medium also contained 10% horse serum.
Transient transfections of plasmid DNA constructs were carried out using Effectene® transfection reagent (Qiagen) as previously described [18,19]. Following transfection, coverslips to be processed for immunofluorescence were washed twice in PBS (Sigma–Aldrich). In cases where cytosol extraction was undertaken, cells were washed twice with ice-cold 1× PBS and once with cytosol extraction buffer [25 mM Hepes/KOH, 25 mM KCl, 2.5 mM magnesium acetate, 5 mM EGTA (sodium free) and 150 mM potassium glutamate] before being washed with cytosol extraction buffer supplemented with 0.05% saponin for 30 s.
In most cases, cells were fixed with 3.8% (w/v) formaldehyde in 1× PBS at room temperature (22 °C) for 20 min, and then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) or 0.05% saponin (Sigma; for LAMP1 antibody) at room temperature for 4 min. Cells to be immunostained with anti-EGFR or X-22 were fixed and permeabilized in methanol at −20 °C for 5 min. Further processing was then carried out as previously described in [18]. Coverslips were mounted in anti-Fade Gold medium (Invitrogen Molecular Probes) on a glass slide. Stained samples were viewed under a Zeiss 510 Meta confocal microscope. Images were recorded using LSM Image Analyzer software and data were subsequently processed using Adobe Photoshop and Illustrator programmes.
Protein depletion by siRNA (small interfering RNA) knockdown
Duplex siRNA oligonucleotides targeting human spartin (GGCAAGGAUUGGAAUGUGCAGCUAA and CAAAUACGGAUAUAAUGCAGGAGAA, siRNA1 and siRNA2 respectively, [9]) and AIP4 (GGUGACAAAGAGCCAACAGAGTT) were obtained from Invitrogen. AIP5 On-Target smartpool siRNA, used in a pool containing 4 oligonucleotides (GGUCUGAUACUAGUAAUAA, GAACGCGGCUUUAGGUGGA, GAAAAGCAACGAUAGAUUU and CCAGAUGGAUUGAAGAGUU, catalogue numbers J-004251-07 to -10) and non-targeting control siRNA (#1, catalogue number D-001210-01) were obtained from Dharmacon.
For siRNA transfection, a ‘double hit’ oligonucleotide transfection protocol, using Oligofectamine transfection reagent (Invitrogen), was employed as previously described [19]. Spartin oligonucleotides were used separately at 10 nM concentration, or in combination at a total concentration of 10 nM. The AIP4 oligonucleotide was used at 50 nM and the AIP5 smartpool at a total concentration of 200 nM. Protein depletion was confirmed in each experiment by immunoblotting.
Co-immunoprecipitation
Cells were harvested by scraping them into ice-cold lysis buffer containing 1% Triton X-100, centrifuged at 10000 g for 5 min at 4 °C to remove insoluble material, and then processed as previously described in [18].
Lipid droplet induction and labelling
To induce lipid droplet formation, HeLa cells were cultured in DMEM supplemented with 200 μM oleic acid (Sigma–Aldrich) in 0.3% BSA for 24 h, as previously described in [12]. Lipid droplets were visualized by incubating the cells in PBS with 10 μg/ml BODIPY 493/503 (Invitrogen) for 15 min before fixation.
RESULTS
Spartin is mono-ubiquitinated in human and rat cell lines
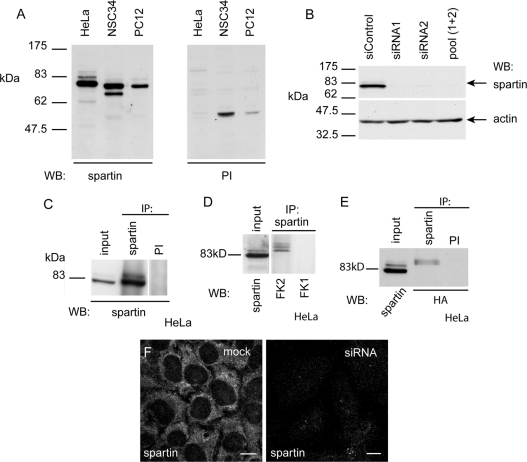
To characterize better endogenous spartin, we raised a polyclonal antibody (termed α-spartin) against a GST-tagged full-length spartin fusion protein. We first verified this antibody for use in Western blotting and immunoprecipitation. In Western blotting with HeLa, NSC34 and PC12 cells, the antibody detected a main band of approx. 80 kDa, consistent with the size predicted for endogenous spartin (Figure 1A). In NSC34 cells, a band of approx. 70 kDa was also seen, which may represent a lower-molecular-mass isoform of the protein. In HeLa cells, the 80 kDa band was greatly diminished after depletion of spartin by siRNA transfection, confirming the specificity of the antibody (Figure 1B). A strong 80 kDa band was also detected on immunoblotting with α-spartin after the antibody was used in immunoprecipitation experiments (Figure 1C), and mass spectroscopic analysis confirmed that spartin protein was present in the immunoprecipitate (results not shown).
Figure 1. A novel anti-spartin antibody confirms that spartin shows multiple mono-ubiquitination.
(A) Western blot (WB) of HeLa, NSC34 and PC12 lysates using α-spartin final bleed (left panel) or pre-immune serum (PI, right panel). (B) HeLa cells transfected with control non-targeting siRNA show a strong ∼80 kDa band on immunoblotting with α-spartin, whereas in cells transfected with each of two siRNA oligonucleotides directed against spartin, or with a pool of the two spartin siRNAs, this band is absent. Actin immunoblotting is shown to verify equal loading. (C) Spartin was immunoprecipitated (IP) from Hela cells by α-spartin, but not by pre-immune serum. (D) Spartin was immunoprecipitated from HeLa cells by α-spartin. Subsequent immunoblotting with FK2 (which recognizes mono- and poly-ubiquitin) detected bands corresponding to the expected size of ubiquitinated forms of spartin, whereas immunoblotting with FK1 (which recognizes only poly-ubiquitin) detected no signal. (E) HeLa cells were transfected with HA-tagged ubiquitin, then immunoprecipitated with α-spartin or with pre-immune serum. Immunoblotting with anti-HA antibody detected bands corresponding to the size of ubiquitinated spartin in the sample immunoprecipitated with α-spartin. (F) Under immunofluorescence confocal microscopy, mock-transfected HeLa cells show a strong cytosolic signal when labelled with α-spartin. However, in cells transfected with an siRNA oligonucleotide directed against spartin, the signal was greatly diminished. Images were obtained using identical confocal microscope settings. Scale bars=10 μm.
It has been reported that spartin is multiply mono-ubiquitinated [9], and in our immunoblotting experiments we saw fainter bands above the main 80 kDa spartin band in HeLa, NSC34 and PC12 cells (Figure 1A). After immunoprecipitation of spartin from HeLa cells, blotting with antibodies recognizing either mono- and poly-ubiquitin (FK2) or polyubiquitin alone (FK1) confirmed that these larger bands represented mono-ubiquitinated forms of spartin (Figure 1D). Similar results were found in NSC34 cells (results not shown). In addition, in HeLa cells transiently transfected with HA–ubiquitin and immunoprecipitated with α-spartin, bands corresponding to the size of ubiquitinated spartin were detected with anti-HA antibody on immunoblotting (Figure 1E). We therefore conclude that spartin is multiply mono-ubiquitinated.
Endogenous spartin can be recruited to endosomes
To resolve the conflicting descriptions of the subcellular location of spartin [8,9,12], we determined the subcellular distribution of endogenous spartin using our α-spartin antibody. In immunofluorescence experiments, endogenous spartin had a widespread diffuse cytoplasmic distribution with typically minimal nuclear staining (Figure 1F). The intensity of the signal was significantly reduced on depletion of spartin by siRNA knockdown, confirming the specificity of the antibody (Figure 1F).
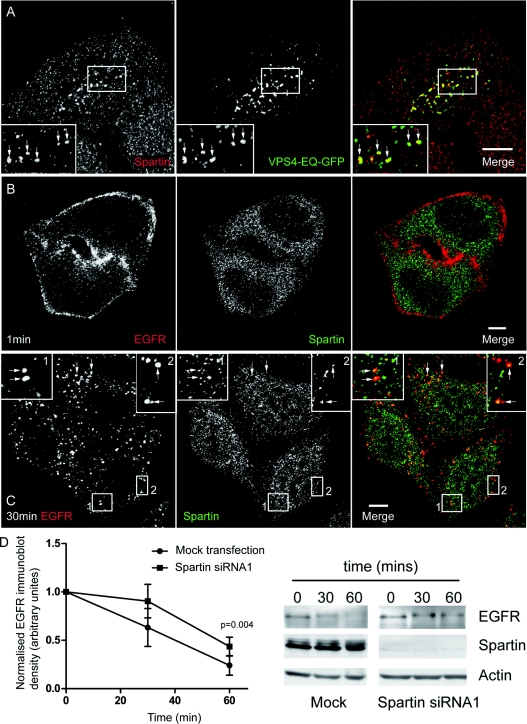
We first co-stained HeLa cells with α-spartin and a variety of organelle markers. There was no, or very limited, co-localization with markers for endosomes, lysosomes, the Golgi apparatus, clathrin-coated vesicles, the endoplasmic reticulum or microtubules (Supplementary Figure S1 at http://www.BiochemJ.org/bj/423/bj4230031add.htm). We next examined whether endogenous spartin could be trapped on Vps4-EQ-positive endosomes, an issue that has been controversial in experiments with epitope-tagged spartin [9,12]. Endogenous spartin was strongly recruited to these (Figure 2A). This suggests that endogenous spartin has a transient association with endosomes, and its removal from them requires Vps4.
Figure 2. Endogenous spartin can be recruited to endosomes.
(A) Endogenous spartin was recruited onto Vps4-EQ-positive puncta in HeLa cells transfected with Vps4-EQ–GFP. Arrows indicate individual puncta that show co-localization. The inset shows a magnified image of the boxed area. (B–C) HeLa cells were incubated with EGF (5 ng/ml in medium) on ice for 1 h, then placed into warm serum-free medium to initiate a synchronized pulse of EGFR endocytosis, and processed for immunofluorescence at the indicated times. At 1 min chase, EGFR was predominantly on the plasma membrane and no co-localization with spartin was present. At 30 min chase, spartin showed co-localization with the endocytosed EGFR. Arrows indicate individual puncta that show co-localization. The insets show magnified images of the boxed areas. In (A–C) and in subsequent micrograph panels, right-hand panels show the merged images; the colour of each marker in the merged image is shown by the colour of its lettering in the non-merged panels. Scale bars=10 μm. (D) Graph showing EGFR immunoblot density in mock-transfected or spartin-depleted cells stimulated with 100 ng/ml of EGF for the times indicated (n=4, one-tailed paired t test). The corresponding immunoblots show a representative experiment.
Upon stimulation of cells with physiological doses of EGF, EGF binds to EGFRs, whereupon the EGF/EGFR complex is internalized via clathrin-mediated endocytosis, then trafficked via early and late endosomes to the lysosomal compartment, where it is degraded [20]. On EGF stimulation, over-expressed epitope-tagged spartin is recruited initially to the plasma membrane and then associates with EGF-positive structures for up to 30 min chase time, after which no co-localization is found with fluorescently labelled EGF [9]. We examined whether endogenous spartin behaved in the same way. We incubated cells that had been serum starved for at least 3 h with EGF (5 ng/ml) on ice for 1 h. We then warmed the cells to 37 °C to trigger a pulse of receptor internalization. As expected, we saw EGFR internalization from the plasma membrane to cytoplasmic vesicular stuctures (Figures 2B and 2C). In contrast with the results obtained previously with epitope-tagged spartin, we found no strong recruitment of endogenous spartin to the plasma membrane at early chase times [0 min (results not shown) and 1 min; Figure 2B], and at these time points there was no obvious co-localization between spartin and EGFR. However, we began to see co-localization between spartin and EGFR at 5 min chase time (results not shown), and this partial co-localization between spartin and EGFR was also present at 30 min (Figure 2C) and 60 min (results not shown).
Spartin has been reported necessary for efficient degradation of EGFR [9]. In HeLa cells stimulated with 100 ng/ml of EGF, we found that depletion of spartin caused a 26% reduction in EGFR degradation after stimulation with EGF for 1 h, compared with mock-transfected cells (Figure 2D; P=0.004; one-tailed paired t test), consistent with the previous report. Together with the immunofluorescence data, this indicates that endogenous cytosolic spartin can be recruited to, and function at, endosomes.
Endogenous spartin is recruited to lipid droplets
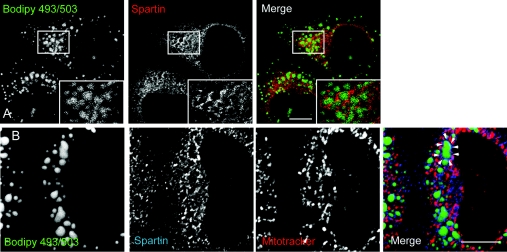
Very recently, epitope-tagged spartin has been reported to be recruited to lipid droplets induced by oleic acid treatment in cultured cells [12]. We examined the relevance of this observation to the endogenous protein. In HeLa cells, we induced lipid droplet formation by adding 200 μM oleic acid to the culture medium for 12–24 h. The spartin signal often surrounded the resulting lipid droplets, and where lipid droplets were clustered, this resulted in a ‘honeycomb’ appearance that was reminiscent of that found with over-expressed spartin (Figures 3A and 3B) [12]. In some cases, spartin decorated the lipid droplet in patches, perhaps suggesting that it is present on specific lipid droplet surface domains (Figures 3A and 3B).
Figure 3. Endogenous spartin can be recruited to lipid droplets.
(A) HeLa cells were treated with oleic acid (200 μM for 24 h) and lipid droplets were labelled by incubation with BODIPY 493/503. Cells were then fixed and labelled for spartin. A spartin signal often surrounded the lipid droplets and, where lipid droplets clustered, a honeycomb appearance was seen (see boxed area; inset shows a magnified image of the boxed area). (B) A cell in which lipid droplet formation was induced in the same way as in (A). The cell was labelled with the mitochondrial marker MitoTracker, as well as with α-spartin and BODIPY 493/503. The spartin signal surrounds many of the lipid droplets; an example is indicated by the arrowheads. The spartin label around the lipid droplets shows minimal co-localization with MitoTracker. Scale bars=10 μm.
We also examined whether there was any co-localization between spartin and mitochondrial markers. To facilitate this examination, we removed cytosolic diffuse spartin staining by pre-fixation treatment with a saponin-based cytosolic extraction buffer. In HeLa cells, removal of cytosolic spartin revealed numerous punctate and tubular structures, and the labelling intensity of these was diminished by spartin siRNA knock-down (Supplementary Figure S2 at http://www.BiochemJ.org/bj/423/bj4230031add.htm). There was co-localization between the mitochondrial marker MitoTracker and the majority of the spartin-positive puncta (Supplementary Figure S2). Although one group has reported localization of epitope-tagged spartin to mitochondria, subsequent studies have not verified this result using a variety of epitope tags [9,12] and in our hands neither Myc- nor GFP-tagged spartin co-localized with mitochondria (results not shown). Lipid droplets have been reported in close association with mitochondria [21], but we found only a minimal overlap between the spartin associated with lipid droplets and mitochondria (Figure 3B).
Spartin interacts with the HECT-family ubiquitin ligases AIP4 and AIP5 via a PPXY motif
Several proteins involved in endocytic processes are ubiquitinated by members of the Hect/Nedd4 family of ubiquitin E3 ligases. A previous proteomics study examining the interaction profile of WW domains suggested an interaction between the WW-domain of the Hect/Nedd4 ubiquitin ligase family member AIP4 and spartin, raising the possibility that AIP4 has ubiquitin ligase activity towards spartin [22]. In addition, a study published during the preparation of the present manuscript used over-expression systems to show that AIP5 also interacts with spartin [12]. We therefore investigated the relevance of these potential interactions to the endogenous protein.
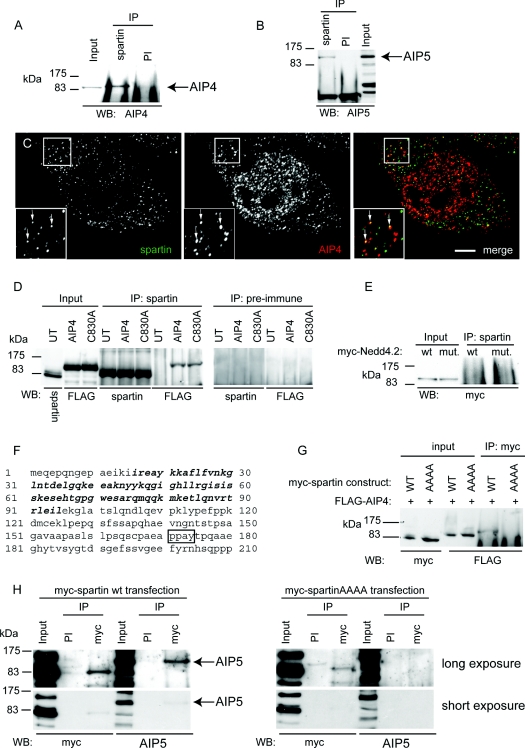
In HeLa cells, endogenous spartin co-immunoprecipitated with endogenous AIP4 and AIP5 (Figures 4A and 4B). Using immunofluorescence microscopy, spartin and AIP4 showed partial co-localization in small puncta (Figure 4C). These experiments strongly support the presence of a physiologically relevant interaction between spartin, AIP4 and AIP5. In the case of AIP4, this interaction did not require AIP4's ubiquitin ligase activity, since both wild-type FLAG-tagged AIP4 and ligase-defective AIP4 co-immunoprecipitated equally well with endogenous spartin (Figure 4D). Within the Hect/Nedd4 family, the interaction of AIP4 and AIP5 with spartin showed some specificity, since we found no interaction between spartin and another member of the Hect/Nedd4 family, Nedd4.2 (Figure 4E).
Figure 4. Spartin interacts with AIP4 and AIP5.
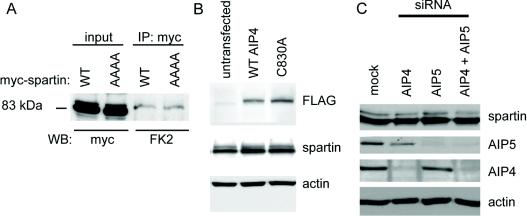
(A) Western blotting (WB) against endogenous AIP4 detects an AIP4-sized band after immunoprecipitation (IP) of HeLa cell lysate with α-spartin, but not with pre-immune (PI) serum. (B) Immunoblotting against endogenous AIP5 detects an AIP5-sized band after immunoprecipitation of HeLa cell lysate with α-spartin, but not with pre-immune serum. (C) Endogenous spartin partially co-localizes with endogenous AIP4, in small puncta. Arrows indicate puncta showing co-localization. The inset shows a magnified image of the boxed area. Scale bar=10 μm. (D) Endogenous spartin was immunoprecipitated from HeLa cells transfected with FLAG-tagged wild-type AIP4 or catalytically inactive AIP4 (C803A). Immunoblotting with anti-FLAG demonstrated that wild-type and catalytically inactive AIP4 were co-immunoprecipitated with spartin. Neither spartin nor the AIP4 proteins were immunoprecipitated by pre-immune serum. UT, untransfected cells. (E) Endogenous spartin was immunoprecipitated from HeLa cells transfected with wild-type (wt) or catalytically inactive (C962S; mut.) Myc-tagged Nedd4.2. Immunoblotting with anti-Myc showed that the Nedd4.2 proteins had not been co-immunoprecipitated with spartin. (F) Sequence of the first 210 amino acids of spartin. The MIT (microtubule-interacting and transport) domain is shown in bold, the PPXY motif is boxed. (G) HeLa cells were transfected with FLAG-AIP4 and either wild-type Myc-tagged spartin (WT) or Myc-tagged spartin in which the PPXY motif had been mutated to AAAA. After immunoprecipitation with anti-Myc, FLAG-AIP4 co-immunoprecipitated with WT Myc–spartin, but not Myc–spartinAAAA. (H) HeLa cells were transfected with wild-type Myc–tagged spartin (left panels) or Myc–spartinAAAA (right panels). After immunoprecipitation with anti-Myc, endogenous AIP5 co-immunoprecipited with wild-type Myc-spartin, but not with Myc–spartinAAAA. An additional short exposure is shown so that immunoblotting results for the input lanes can be properly visualized.
We noted that spartin contains a PPXY motif (residues 171–174; Figure 5F). In many cases, binding of Nedd4 family E3 ligases to target proteins is mediated by interactions between WW domains in the E3 ligase and PPXY motifs in the target protein [15]. On mutation of the PPXY to AAAA, spartinAAAA, the association between epitope-tagged spartin and AIP4 or AIP5 was lost, indicating that spartin's PPXY domain is necessary for these interactions (Figures 4G and 4H).
Figure 5. PPXY, AIP4 and AIP5 are not required for spartin's ubiquitination.
(A) Wild-type (WT) Myc–spartin and Myc–spartinAAAA were immunoprecipitated (IP) from HeLa cells using anti-Myc antibody. Blotting with FK2 (which recognizes poly- and mono-ubiquitin) revealed similar levels of ubiquitination of both forms of spartin. No bands were seen in samples immunoprecipitated with a spurious antibody directed against actin (results not shown). (B) Neither ubiquitination of spartin (as indicated by the density of the higher-molecular-mass ubiquitinated band) nor the concentration of spartin differed between untransfected HeLa cells, HeLa cells transfected with wild-type FLAG-tagged AIP4 (WT AIP4), or with catalytically inactive FLAG–AIP4 (C830A). (C) AIP4 depletion, AIP5 depletion or depletion of both proteins did not affect the concentration of spartin or the strength of the upper ubiquitinated band seen on spartin immunoblotting. In (B) and (C), actin immunoblotting is shown to verify equal loading.
The PPXY motif, AIP4 and AIP5 are not required for ubiquitination of spartin
We next examined whether AIP4 or AIP5 are required for mono-ubiquitination of spartin. We began by investigating whether mutation of spartin's PPXY motif (which is required for binding to AIP4 and AIP5) affected spartin's ubiquitination. We immunoprecipited wild-type Myc–spartin or Myc–spartinAAAA, then immunoblotted with the anti-ubiquitin FK2 antibody. Mutation of the PPXY motif made no difference to ubiquitination of spartin (Figure 5A), suggesting that the interaction of this motif with WW domains of HECT E3 ligases is not required for this.
We determined whether expression of wild-type or ligase-defective AIP4 affected spartin's ubiquitination. After transfection of HeLa cells with FLAG-tagged wild-type or catalytically inactive AIP4, we blotted against spartin and assessed whether the higher-molecular-mass ubiquitinated band was altered. There was no obvious difference in the amount of ubiquitinated spartin, or indeed total spartin, in cells expressing wild-type AIP4, catalytically inactive AIP4 or untransfected cells (Figure 5B). We also examined the effects of AIP4 and/or AIP5 depletion by siRNA knock-down on the higher-molecular-mass ubiquitinated spartin band. We saw no effect on the amount of ubiquitinated spartin, or total spartin, in cells depleted of AIP4, AIP5, or both, compared with control cells, suggesting that neither AIP4 nor AIP5 are required for ubiquitination of spartin (Figure 5C).
DISCUSSION
There has been considerable controversy over the subcellular localization of spartin. Most studies to date have relied on over-expression systems that have given conflicting results. A single study reporting the localization of the endogenous protein used an antibody whose specificity was not verifed by knockdown experiments and gave discrepant results to all of the over-expression studies [23]. To resolve this controversy, we used a novel spartin antibody in immunofluorescence experiments. Importantly, the specificity of this antibody was verified by siRNA knock-down.
Two of the previous localization studies suggested that epitope-tagged spartin has a predominantly diffuse cytosolic localization at steady state [9,12], and our results on the endogenous protein are in agreement with this; much of the spartin immunofluorescence signal could be removed by prefixation treatment with a saponin-based cytosol extraction buffer. However, there was disagreement between the prior studies on whether spartin can be recruited to endosomes. Bakowska et al. [9] reported that epitope-tagged spartin was recruited to dominant-negative Vps4-induced endosomes and could be recruited to endosomes on EGF stimulation. In contrast, Eastman et al. [12] reported no recruitment of epitope-tagged spartin to Vps4 dominant-negative endosomes. We clearly found that endogenous spartin can be recruited to endosomes under some circumstances; spartin was trapped on dominant-negative Vps4-induced endosomes, and EGF stimulation resulted in co-localization between spartin and endosomal structures. In keeping with this observation, we (in the present study) and others have shown that spartin is required for efficient EGFR degradation [9]. Our results from the present study therefore support a functional role for spartin at endosomes. Thus spartin is a member of an ‘endosomal’ group of HSP proteins that includes spastin [19], NIPA1 (Non imprinted in Prader-Willi/Angelman syndrome 1 [24,24a]), maspardin [25] and spastizin [26]. This suggests that endosomal abnormality, perhaps resulting in dysregulation of one or more receptor-mediated signalling pathways important for axonal function, could be an important cause of HSP.
It has been suggested recently that exogenously expressed spartin localizes to lipid droplets and that spartin is involved in lipid droplet turnover [12]. Our results are in agreement with these observations and extend them by showing that endogenous spartin is present on lipid droplets. To date, abnormal lipid droplet biology has not been implicated in the causation of other genetic subtypes of HSP, and further work will be required to assess the importance of this function to the axonal degeneration or other clinical features seen in Troyer syndrome.
Lu et al. [8] reported that N-terminal CFP (cyan fluorescent protein)-tagged spartin co-localized with mitochondrial markers, although we (in the present study) and others [12] have found no localization of exogenous spartin, tagged with either single epitopes or with fluorescent protein domains, to mitochondria. In addition, spartin contains no recognized mitochondrial targeting signal motif. However, after removal of cytosolic spartin, we did find some co-localization of endogenous spartin with mitochondria, and this signal appeared to diminish, but not disappear, on spartin knock-down. Thus additional experiments beyond the scope of the present study will be required to provide a definitive answer as to whether spartin is present in mitochondria.
Spartin is mono-ubiquitinated [9]. Mono-ubiquitination is important at several levels during the endocytosis and degradation of plasma-membrane receptors. Mono-ubiquitination of receptors, e.g. EGFR, can act as a signal to promote their endocytosis and sorting to the late endosomal/lysosomal degradative compartment [20,27]. In addition, much of the cellular machinery involved in this endocytic sorting process is itself mono-ubiquitinated, and some members of this machinery, e.g. eps15, epsin and Hrs, show coupled mono-ubiquitination, a phenomenon in which proteins that contain a ubiquitin-binding domain are themselves mono-ubiquitinated [28,29]. To try to understand better the basis of spartin's mono-ubiquitination, we examined a putative interaction between AIP4 and spartin, suggested by a proteomics study [22]. We found strong evidence of a physiologically relevant interaction between endogenous AIP4 and spartin. We also found an interaction between endogenous spartin and AIP5, an E3 ligase closely related to AIP4. In both cases, this interaction was mediated by a PPXY motif in spartin. These results extend the findings of a paper published during the preparation of the present manuscript, which used over-expression systems to show that spartin interacts with AIP5 via a PPXY–WW domain interaction, without proving the relevance of this interaction to the endogenous situation [12].
Using over-expression systems, Eastman et al. [12] suggested that AIP5 has ubiquitin ligase activity towards spartin. However, our results from the present study indicate that ubiquitination of spartin is unaffected by mutation of the PPXY motif, which has been shown by our experiments and by Eastman et al. [12] to be required for binding to WW-domain containing E3 ligases. In addition, over-expression of wild-type or dominant-negative AIP4, or siRNA-mediated depletion of AIP4, AIP5 or both, all had no effect on spartin's concentration or on its ubiquitination. Our results therefore suggest that neither AIP4, AIP5, nor the capacity to interact with WW-domain containing E3 ligases via its PPXY motif, are necessary for ubiquitination of spartin. It is possible that weak or very transient binding to HECT E3 ligases, below the level detectable by our co-immunoprecipitation experiments, remains after mutation of the classical PPXY, and that, in the absence of AIP4 and AIP5, other HECT family E3 ligases may be sufficient to ubiquitinate spartin. Alternatively, the function of Nedd4-type E3 ligases may be affected by a number of accessory and adaptor proteins that are not necessarily ubiquitinated by the ligase, but which can determine localization of the ligase to a specific subcellular compartment, mediate or stabilize the interaction between the ligase and target proteins, or modify the ligase function of the protein [30]. For example, in the case of AIP4, a number of adaptor proteins have been identified, including Numb and certain Smad proteins, which interact with AIP4 via PPXY–WW domain interactions [30]. We therefore speculate that spartin may be an adaptor protein that regulates the function of AIP4/AIP5 and perhaps other HECT family E3 ligases.
In summary, taken together with the results of other studies, our work from the present study suggests that spartin has a number of roles, including functioning at endosomes and in lipid droplet formation. This complexity of function could be reflected in the complex phenotype seen with Troyer syndrome, with different aspects of the condition being caused by abnormality of spartin's different functions. A full explanation for the disease will require a thorough understanding of spartin's functions at all of these sites and an exploration of the relevance of each function to axonal biology.
Online data
AUTHOR CONTRIBUTION
Thomas Edwards designed the research, performed the research and analysed data. Virginia Clowes performed the research and analysed data. Hilda Tsang and James Connell performed the research. Christopher Sanderson designed the research. J. Paul Luzio designed the research and critically read the manuscript. Evan Reid designed the research, performed the research, analysed data and wrote the manuscript.
ACKNOWLEDGEMENTS
We thank Adriano Marchese for providing Nedd4 constructs and Paul Lehner for AIP4 constructs. We thank Margaret S. Robinson and Matthew Seaman for the gift of antibody reagents. We thank Meredith Ross and Andrew James for helpful discussions. We thank Michael Murphy for helpful discussions and for his critical reading of the manuscript prior to submission.
FUNDING
We thank the Tom Wahlig Stiftung and the UK Familial Spastic Paraplegia Family Group for financial support. E. R. is a Wellcome Trust Senior Research Fellow in Clinical Science [grant number 082381], T. L. E. is supported by a UK Government ORS Award and by the Cambridge Commonwealth Trust, H. T. is supported by a Croucher Foundation Scholarship and V. C. is an MRC Research Training Fellow [grant number G0800435]. T. L. E. and V. C. are Sackler Fellows. This work was also supported by a MRC programme grant [grant number G9310915 (to J. P. L.)] and a Wellcome Trust Strategic Award [grant number 079895 (to Cambridge Institute for Medical Research)].
References
- 1.Harding A. E. Hereditary spastic paraplegias. Semin. Neurol. 1993;13:333–336. doi: 10.1055/s-2008-1041143. [DOI] [PubMed] [Google Scholar]
- 2.Reid E. Science in motion: common molecular pathological themes emerge in the hereditary spastic paraplegias. J. Med. Genet. 2003;40:81–86. doi: 10.1136/jmg.40.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink J. K. Hereditary spastic paraplegia. Curr. Neurol. Neurosci. Rep. 2006;6:65–76. doi: 10.1007/s11910-996-0011-1. [DOI] [PubMed] [Google Scholar]
- 4.Soderblom C., Blackstone C. Traffic accidents: molecular genetic insights into the pathogenesis of the hereditary spastic paraplegias. Pharmacol. Ther. 2006;109:42–56. doi: 10.1016/j.pharmthera.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Harding A. E. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1:1151–1155. doi: 10.1016/s0140-6736(83)92879-9. [DOI] [PubMed] [Google Scholar]
- 6.Patel H., Cross H., Proukakis C., Hershberger R., Bork P., Ciccarelli F. D., Patton M. A., McKusick V. A., Crosby A. H. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat. Genet. 2002;31:347–348. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- 7.Bakowska J. C., Wang H., Xin B., Sumner C. J., Blackstone C. Lack of spartin protein in Troyer syndrome: a loss-of-function disease mechanism? Arch. Neurol. 2008;65:520–524. doi: 10.1001/archneur.65.4.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J., Rashid F., Byrne P. C. The hereditary spastic paraplegia protein spartin localises to mitochondria. J. Neurochem. 2006;98:1908–1919. doi: 10.1111/j.1471-4159.2006.04008.x. [DOI] [PubMed] [Google Scholar]
- 9.Bakowska J. C., Jupille H., Fatheddin P., Puertollano R., Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol. Biol. Cell. 2007;18:1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 11.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 12.Eastman S. W., Yassaee M., Bieniasz P. D. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J. Cell Biol. 2009;184:881–894. doi: 10.1083/jcb.200808041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiele C., Spandl J. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto T., Ohsaki Y., Cheng J., Suzuki M., Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 2008;130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingham R. J., Gish G., Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 16.Reaves B. J., Bright N. A., Mullock B. M., Luzio J. P. The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J. Cell Sci. 1996;109:749–762. doi: 10.1242/jcs.109.4.749. [DOI] [PubMed] [Google Scholar]
- 17.Reid E., Connell J., Edwards T. L., Duley S., Brown S. E., Sanderson C. M. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- 18.Sanderson C. M., Connell J. W., Edwards T. L., Bright N. A., Duley S., Thompson A., Luzio J. P., Reid E. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 2006;15:307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connell J., Lindon C., Luzio J., Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 21.Cohen A. W., Razani B., Schubert W., Williams T. M., Wang X. B., Iyengar P., Brasaemle D. L., Scherer P. E., Lisanti M. P. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 2004;53:1261–1270. doi: 10.2337/diabetes.53.5.1261. [DOI] [PubMed] [Google Scholar]
- 22.Ingham R. J., Colwill K., Howard C., Dettwiler S., Lim C. S. H., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G., et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robay D., Patel H., Simpson M. A., Brown N. A., Crosby A. H. Endogenous spartin, mutated in hereditary spastic paraplegia, has a complex subcellular localization suggesting diverse roles in neurons. Exp. Cell Res. 2006;312:2764–2777. doi: 10.1016/j.yexcr.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Shaw W. R., Tsang H. T., Reid E., O'Kane C. J. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neurosci. 2007;10:177–185. doi: 10.1038/nn1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Tsang H. T., Edwards T. L., Wang X., Connell J. W., Davies R. J., Durrington H. J., O'Kane C. J., Luzio J. P., Reid E. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signaling. Hum. Mol. Genet. 2009 doi: 10.1093/hmg/ddp324. doi:10.1093/hmg/ddp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson M. A., Cross H., Proukakis C., Pryde A., Hershberger R., Chatonnet A., Patton M. A., Crosby A. H. Maspardin is mutated in mast syndrome, a complicated form of hereditary spastic paraplegia associated with dementia. Am. J. Hum. Genet. 2003;73:1147–1156. doi: 10.1086/379522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanein S., Martin E., Boukhris A., Byrne P., Goizet C., Hamri A., Benomar A., Lossos A., Denora P., Fernandez J., et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am. J. Hum. Genet. 2008;82:992–1002. doi: 10.1016/j.ajhg.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welchman R. L., Gordon C., Mayer R. J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell. Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 28.Di Fiore P. P., Polo S., Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell. Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- 29.Hicke L., Schubert H. L., Hill C. P. Ubiquitin-binding domains. Nat. Rev. Mol. Cell. Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 30.Shearwin-Whyatt L., Dalton H. E., Foot N., Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28:617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.