Abstract
Molecular modelling suggests that a group of proteins in plants known as the β-hydroxyacid dehydrogenases, or the hydroxyisobutyrate dehydrogenase superfamily, includes enzymes that reduce succinic semialdehyde and glyoxylate to γ-hydroxybutyrate and glycolate respectively. Recent biochemical and expression studies reveal that NADPH-dependent cytosolic (termed GLYR1) and plastidial (termed GLYR2) isoforms of succinic semialdehyde/glyoxylate reductase exist in Arabidopsis. Succinic semialdehyde and glyoxylate are typically generated in leaves via two distinct metabolic pathways, γ-aminobutyrate and glycolate respectively. In the present review, it is proposed that the GLYRs function in the detoxification of both aldehydes during stress and contribute to redox balance. Outstanding questions are highlighted in a scheme for the subcellular organization of the detoxification mechanism in Arabidopsis.
Keywords: aldehyde detoxification, glyoxylate reductase, β-hydroxyacid dehydrogenase, redox balance, succinic semialdehyde reductase
Abbreviations: AKR, aldo-keto reductase; CaM, calmodulin; GABA, γ-aminobutyrate; GABA-T, GABA transaminase; GAD, glutamate decarboxylase; GDC, glycine decarboxylase; GHB, γ-hydroxybutyrate; GLYR, glyoxylate reductase; HIBADH, hydroxyisobutyrate dehydrogenase; ICDH, isocitrate dehydrogenase; NADK, NAD kinase; ROS, reactive oxygen species; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase; SSA, succinic semialdehyde; SSADH, SSA dehydrogenase; TCA, tricarboxylic acid
INTRODUCTION
The occurrence of distinct NADPH-dependent GLYR (glyoxylate reductase) isoforms and activity in the cytosol and chloroplasts of spinach and pea has been documented [1]. Recent research has indicated that Arabidopsis cytosolic GLYR1 and plastidial GLYR2 reduce both glyoxylate and SSA (succinic semialdehyde) to glyoxylate and GHB (γ-hydroxybutyrate) respectively [2–4]. In vitro measurements of recombinant GLYR1 and GLYR2 activities revealed that glyoxylate is preferred over SSA as a substrate, the kinetic mechanism is ordered Bi Bi, involving the complexation of NADPH before glyoxylate or SSA, and NADP is a competitive inhibitor with respect to NADPH. Thus plant GLYRs act as an AKR (aldo-keto reductase), but there is no significant homology with either mammalian and bacterial NADPH-SSA reductase, or other members of the AKR superfamily. There is, however, some sequence identity with known β-hydroxyacid dehydrogenases, such as tartronate semialdehyde reductase and 6-phosphogluconate dehydrogenase, which are members of the HIBADH (hydroxyisobutyrate dehydrogenase) protein family. Furthermore, molecular modelling of GLYR1 via comparison with a well-characterized member of the HIBADH family indicates that a Rossmann fold-like nucleotide-binding site with a three-layer (αβα) sandwich geometry at the N-terminus, orthogonal bundles at the C-terminus, and amino acid residues at the predicted active site are highly conserved (Figure 1). Taken together, these observations suggest that plant GLYR enzymes are members of the HIBADH protein family [2,3], but crystallization of the proteins and active-site mutagenesis are necessary to substantiate this hypothesis.
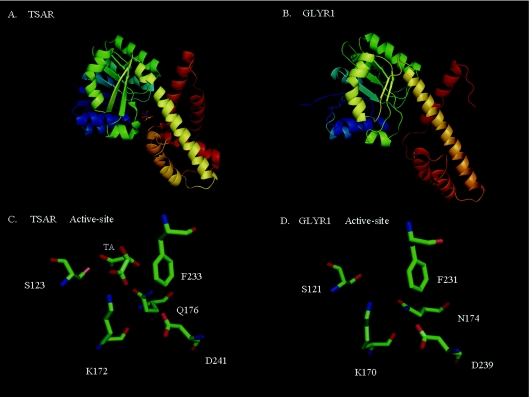
Figure 1. Molecular models of HIBADH family members.
(A) Crystal structure of TSAR (tartronic semialdehyde reductase) and its substrate analogue tartaric acid (TA) (PDB entry: 1VPD). (B) Predicted structure of GLYR1 generated by the fold-recognition server PHYRE (http://www.sbg.bio.ic.ac.uk/phyre/) using TSAR as a template. (C and D) Overlapping positions and identities of the predicted active-site residues for TSAR and GLYR1 respectively. Red, blue and green indicate negative, positive and hydrophobic regions of the residues.
Aldehydes such as glyoxylate and SSA can accumulate in plants under stress, and react with DNA, oxidize membrane lipids, modify proteins or influence the transcription of stress-related genes, thereby causing cellular and developmental problems [5,6]. Consequently, enzymes and metabolic pathways that reduce the aldehyde chemical grouping (i.e. H-C=O) in these compounds to its corresponding alcohol are probably essential for maintaining plant health. Glyoxylate and SSA are best known as intermediates in the metabolism of glycolate and GABA (γ-aminobutyrate) respectively
In the present review, we attempt to place the function of plant GLYRs within a metabolic context. The first three sections discuss the physiological role of the enzymes during stress, with particular emphasis on the turnover of pyridine nucleotides and the metabolism of GABA and glycolate. The final section focuses on a hypothetical scheme for the detoxification of SSA and glyoxylate, and on opportunities for future research.
PYRIDINE NUCLEOTIDES
NAD(P) is ubiquitous in the cell, mediates numerous redox reactions, and influences virtually every metabolic pathway [7–9]. Furthermore, it is involved in the production and consumption of ROS (reactive oxygen species) in plants, and is therefore a key player in signalling events. ROS production is influenced by NADPH/NADP ratios in chloroplasts and mitochondria and NAD(P)H oxidases are key players in ROS generation at the plasma membrane. ROS consumption partly depends on glutathione and ascorbate pools, which are fundamentally maintained by NAD(P)H. NAD(P) plays other defensive and signalling roles, such as production of NO and metabolism of reactive aldehydes.
NAD(P) pools are plastic in plants and undergo significant changes in response to the environment. For example, increasing light causes progressive over-reduction at the reducing side of Photosystem I in the chloroplast, causing NADP levels to decline and thereby favouring the diversion of electrons to oxygen to produce ROS [10]. Yet the chloroplast stromal redox state seems to be constant in spite of irradiance. The cytosolic NAD(P) redox states are independent of light, dark or CO2 availability, yet cytosolic NAD and mitochondrial NAD and NADP are more reduced under photorespiratory conditions [11]. Several other studies indicate that changes in cytosolic and mitochondrial NAD reduction are important in regulating nitrate reduction and nitrogen assimilation [12–14].
Generation of NADPH in the chloroplast depends on the reduction of NADP via ferredoxin NADP reductase in the light, and by the oxidative pentose phosphate pathway dehydrogenases, glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase, in the dark [10,14]. The presence of glucose-6-phosphate and 6-phosphogluconate dehydrogenases in peroxisomes implies that these organelles have the capacity to reduce NADP to NADPH for use in antioxidant protection [15]. In the cytosol and mitochondria, NADP-dependent ICDHs (isocitrate dehydrogenases) play a major role in NADPH pool sizes [11,16]. NADP-ICDH activity in mitochondria is higher in photosynthetic than in heterotrophic tissues, implying an important role during photosynthesis. It has been suggested that mitochondrial NADP-ICDH provides NADPH for redox regulation of the alternative oxidase via thioredoxin [17].
The NADH/NAD ratio in mitochondria is high during photorespiratory conditions due to glycine oxidation via GDC (glycine decarboxylase) activity, which provides more NADH than the TCA cycle (tricarboxylic acid cycle; also known as the citric acid cycle) [11,18]. This higher ratio could stimulate the interconversion of NADH and NADP via transhydrogenase activity [11,19], thereby increasing mitochondrial NADPH levels, inhibiting NAD-ICDH in the TCA cycle, and causing mitochondrial NADP-ICDH to operate in the reverse direction. Consequently, citrate could be exported from the mitochondrion, whereas 2-oxoglutarate could be imported, entering the reverse NADP-ICDH reaction as 2-oxoglutarate dehydrogenase activity is probably low during photorespiration. Thus NADP-ICDH reverse activity might recycle NADPH and 2-oxoglutarate in the mitochondrion during photorespiration. Mitochondrial NADP and NAD pools are more oxidized in the dark [11], and the NADH/NAD ratio is not significantly lower than under non-photorespiratory conditions, which represents oxidation of NADH mainly through the TCA cycle and not through GDC.
The ATP-dependent phosphorylation of NAD(H) is catalysed by NADK (NAD kinase). Genes encoding NADKs have been cloned from human, yeast and bacteria [20–22]. Eukaryotes possess multiple NAD(H)K genes, whereas bacteria apparently have only one. The enzymes are divided into NADKs or NAD(H)Ks depending on their substrate preference for the oxidized and/or reduced forms of NAD [23,24].
Several NADK isoforms are present in plants. In maize, for example, a CaM (calmodulin)-dependent enzyme is found in the cytosol, in the outer membrane of mitochondria and in the chloroplast envelope, whereas a CaM-independent form is located in the chloroplast stroma [25,26]. In Avena sativa L. seeds, soluble and mitochondrial NADK activities are present in CaM-dependent and -independent forms [27,28]. CaM-dependent activity has also been detected in pea seedlings, Solanum lycopersicum L. var. pimpinellifolium (Juslen) Mill and green bean [29,30]. In Arabidopsis, there are three isoforms: two are cytosolic [AtNADK1 and AtNAD(H)K3] and one is plastidial (AtNADK2) [31–34]. Partially purified NADK activity from Arabidopsis displays Ca2+/CaM-dependent activity, whereas recombinant AtNAK1 and AtNADK2 do not, although AtNADK2 has some affinity for CaM [31]. One study has reported that recombinant AtNADK1 can utilize both NAD and NADH in an ATP-dependent manner, although the substrate specificity constant was not determined [24], whereas another reported that AtNADK1 and AtNADK2 utilize NAD, but not NADH [31]. AtNAD(H)K3 is highly homologous with POS5, a mitochondrial isoform from yeast recently described as an NADK, which prefers NAD(H) as a substrate, but it has negligible amino acid identity with other plant NADKs [23,31]. AtNAD(H)K3 can phosphorylate NADH or NAD+ in an ATP-dependent manner, but has a 100-fold greater preference for NADH, and its activity is not stimulated by CaM [32]. AtNADK1 is induced by oxidative stress, such as ionizing radiation and paraquat treatment, and by compatible and incompatible plant–pathogen interactions [24], and NAD(H)K3 is induced by a variety of abiotic stresses and exogenous abscisic acid. NAD(H)K3 apparently plays an important role in cytosolic NADPH levels under abiotic stress [33]. Manipulation of AtNADK2 levels affects chloroplastic NADPH levels, and null mutants are stunted, with a pale yellow colour, and are hypersensitive to abiotic stress [34].
SSA IS AN INTERMEDIATE IN GABA METABOLISM
Glutamate can be metabolized via a short metabolic pathway that bypasses the NAD-dependent 2-oxoglutarate dehydrogenase and the ADP-dependent succinyl-CoA ligase steps of the TCA cycle and allows the glutamate carbon backbone to enter the cycle as succinate [35,36] (Figure 2). This pathway is found in virtually all organisms from bacteria to humans, underscoring its fundamental nature in metabolism. GABA is a four-carbon non-protein amino acid with the amino group on the γ-carbon, rather than the α-carbon. The decarboxylation of glutamate is catalysed via a cytosolic Ca2+/CaM-dependent GAD (glutamate decarboxylase) with the consumption of a proton [35]. The catabolism of GABA apparently involves mitochondrial pyruvate- and glyoxylate-dependent, but not 2-oxoglutarate-dependent, GABA-T (GABA transaminase) activities [37–39], resulting in the production of alanine and glycine respectively. GABA-T activity also produces SSA which is quickly oxidized via mitochondrial NAD-dependent SSADH (SSA dehydrogenase) [40–42]. (GAD, GABA-T and SSADH make up the pathway that is commonly known as the GABA shunt.)
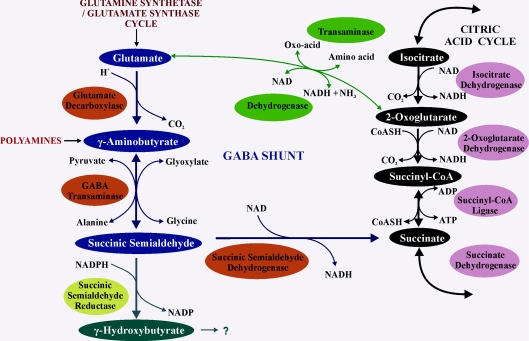
Figure 2. The GABA shunt and its relationship with other metabolic pathways.
The GABA shunt metabolites and enzymes are indicated as blue and orange balloons respectively, and the TCA cycle is shown as black and pink balloons. Glutamate, the precursor to GABA, is supplied predominantly via the glutamine synthetase/glutamate synthase cycle or the TCA cycle. The polyamine putrescine has been proposed as an alternative GABA precursor, although in planta contribution in mature leaves is likely to be minor. There are two branching routes for GABA catabolism via SSA: one to succinate and the TCA cycle and the other to GHB via SSA reductase (a yellow balloon, shown as GLYR in Figure 3), although the products of GHB catabolism in plants are currently unknown.
The GABA shunt provides metabolic flexibility to the cell with the provision of glutamate rather than pyruvate carbon for use in the TCA cycle. The oxidation of glutamate is theoretically less energetically productive than pyruvate, however, because 1 mol each of NADH and ATP are produced at the 2-oxoglutarate dehydrogenase and succinyl-CoA ligase steps respectively, whereas only 1 mol of NADH is produced at the SSADH step in the GABA shunt. Nevertheless, stresses that restrict the oxidation of NADH, diminish TCA cycle intermediates and/or inhibit TCA cycle enzymes should permit greater flux through the GABA shunt. For example, oxygen deficiency inhibits NADH oxidation via the mitochondrial electron transport chain, decreases the glutamate level, and increases the GABA and alanine levels [43–47]. Furthermore, manipulation of TCA cycle enzymes, such as NAD-dependent ICDH or aconitase, alters GABA shunt activity [48,49], and GABA shunt activity can complement reduced expression of succinyl-CoA ligase [49].
GABA accumulation in plants is a general response to various biotic and abiotic stresses and can involve both biochemical and transcriptional processes [35,36,50]. In some cases, the response of GABA is rapid, often within seconds [51,52], suggesting that activation of cytosolic GAD activity by Ca2+/CaM is probably involved. It is known that Ca2+ influx into the cell is increased by touch [53], cold shock [52], heat shock, salinity, drought and osmotic stress [54], and GABA does accumulate. However, at acidic pH GAD is stimulated independently of Ca2+/CaM, and in vivo cytosolic acidification does stimulate GAD activity and GABA production [55,56]. Stresses that disrupt cellular membranes such as wounding, freezing or heat stress, can cause vacuolar contents to be released into the cytosol or inhibit ATPase enzymes which pump protons from the cytosol into the vacuole or apoplast, causing a concomitant decrease in cytosolic pH and stimulation of GAD activity. For example, oxygen deficiency is known to decrease cytosolic pH by 0.3–0.6 pH units and cause GABA to accumulate in soybean, tobacco and Arabidopsis [43,44,47,57]. Transcriptional change in GAD and GABA-T expression can occur in response to low oxygen, water deficit, salinity or Agrobacterium infection [47,58–61].
Evidence is available for the reduction of SSA and accumulation of GHB in Arabidopsis plants subjected to oxygen deficiency, high light, heat, drought and salinity, and for the recovery of GHB levels in tobacco after the removal of oxygen deficiency [42,44,46,47,50,62] (Figure 3). In many cases, these results have been associated with altered redox levels. Furthermore, ssadh-knockout mutants of Arabidopsis exhibit chlorotic lesions and accumulate GHB and H2O2 [62]. GLYR isoforms are also implicated in GHB production because their transcript levels can increase under various stresses, such as cold and heat [46]. GLYR activity is found at all developmental stages and in all tissues, and activity is generally higher in vegetative and reproductive organs than in roots [2]. SSADH-deficient yeast complemented with Arabidopsis GLYR1 grows on 20 mM GABA as the sole nitrogen source and contains elevated levels of GHB [45]. These studies link the GABA shunt to redox homoeostasis in the cell and suggest that SSA-dependent GLYR activity potentially occurs in planta, despite the fact that glyoxylate is the preferred substrate in vitro [2].
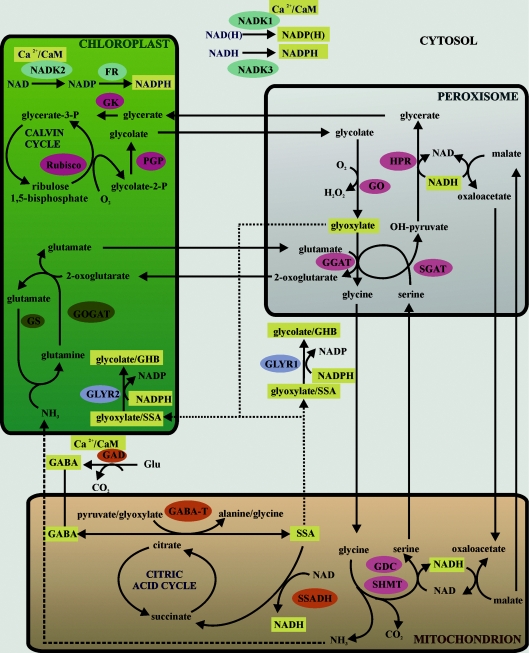
Figure 3. Proposed role for GLYRs during stress.
The subcellular scheme relates recent and hypothetical information about GLYRs and NADKs to pyridine nucleotides and specific intermediates of the GABA and glycolate pathways. There is no consideration of stoichiometry in this scheme. Enzymes of the GABA and glycolate pathways, as well as photorespiratory nitrogen metabolism, are shown as orange, pink and brown balloons respectively. Enzymes associated with aldehyde detoxification and NADPH formation are shown as blue and turquoise balloons respectively. Components whose levels are probably elevated in response to stress are shown as yellow boxes, and potential diffusion pathways are shown as dotted lines. For further discussion and references, please see the text. FR, ferredoxin reductase; GGAT, glutamate:glyoxylate aminotransferase; GK, glycerate kinase; GO, glycolate oxidase; GOGAT, glutamate synthase; GS, glutamine synthetase; HPR, hydroxypyruvate reductase; PGP, phosphoglycolate phosphatase; SGAT, serine:glyoxylate aminotransferase; SHMT, serine hydroxymethyltransferase.
GLYOXYLATE IS AN INTERMEDIATE IN GLYCOLATE METABOLISM
Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase; EC 4.1.1.39) can catalyse the oxygenation of ribulose bisphosphate, resulting in the production of glycolate 2-phosphate, which is metabolized via the glycolate pathway (Figure 3). (This pathway is often referred to collectively as photorespiration.) The rate of oxygenation compared with carboxylation is dependent on the ambient O2 and CO2 concentrations, their Km values and the velocity of the overall reaction [63]. Sharkey et al. [64] estimated that, under ambient conditions, the rate of photorespiratory CO2 release is approx. 25% of the rate of net CO2 assimilation. With rising temperatures, the specificity of Rubisco for CO2 and the relative solubility of O2 to CO2 increase, resulting in enhanced photorespiratory rates. This effect is further augmented by a lowered intercellular CO2 concentration, which occurs upon stomatal closure during drying conditions such as high light and drought [65], thereby imposing strong carbon loss and possibly leading to the depletion of carbohydrates and accelerated senescence. However, the suppression of photorespiration over the long term results in decreased rates of photosynthetic CO2 assimilation, poor plant growth and chloroplast aberrations [66,67].
The photorespiratory pathway is compartmentalized across chloroplasts, peroxisomes and mitochondria (Figure 3). Glycolate 2-phosphate is first hydrolysed in the chloroplast to glycolate, which is oxidized in the peroxisome to glyoxylate. The glyoxylate in turn undergoes transamination to glycine [68,69], which is transported to the mitochondrion for conversion into serine via NAD-dependent GDC and serine hydroxymethyltransferase, with the release of CO2 and NH3. Serine returns to the peroxisome where it is transaminated to hydroxypyruvate, which is reduced to glycerate. Glycerate is then phosphorylated in the chloroplast and the resulting glycerate 3-phosphate enters the Calvin cycle. Because of the transamination of glyoxylate to glycine and the large release of NH3 in the GDC reaction, photorespiratory metabolism is intimately linked to leaf nitrogen metabolism [70].
The energy requirements for fixing CO2 are higher in the presence of photorespiratory metabolism (5.375 mol of ATP and 3.5 mol of NADPH per CO2), than in its absence (3 mol of ATP and 2 mol of NADPH per CO2) [70]. Thus it has been suggested that photorespiration is important for maintaining electron flow to prevent photoinhibition under stress [70]. Photoinhibition occurs when the photochemical processes of photosynthesis do not have enough ADP or NADP to enable continued flow of energy through the electron transport system [71,72]. Electron flow becomes blocked as molecules within the electron transport chain become reduced. Then the electrons of the Photosystem II centre become highly excited as light energy continues to be absorbed; photoinhibition occurs as the excess light energy damages the D1 protein. Photoinhibition is usually temporary because of the turnover rate of the repair mechanisms for the D1 protein. However, if other molecules within the photochemical system are permanently damaged by excess energy, chlorophyll becomes bleached, a process known as photo-oxidation.
The risk of photo-oxidation occurs under conditions such as salt and drought stresses, wherein internal leaf CO2 concentration declines as stomata close to prevent water loss, electron consumption by photosynthesis is reduced, and over-reduction of the electron transport chain occurs [70]. Photorespiration rises in plants under salt stress, as indicated by a higher CO2 compensation point [73], higher light-to-dark ratio of CO2 production [74], higher glycolate oxidase activity, and the formation of specific metabolites such as glycine, serine and glycolate [75]. CO2 assimilation declines more quickly in heterozygous barley mutants with reduced levels of various photorespiratory enzymes than in the wild-type under moderate drought stress, demonstrating increasing control by photorespiratory enzymes on photosynthesis [76]. Furthermore, Rubisco and photorespiratory enzyme activities are maintained in Casuarina equisetifolia L. under drought stress as the internal leaf CO2 concentration decreases, suggesting that photorespiration is likely to increase relative to photosynthesis [77].
Photorespiratory glyoxylate exerts a feedback effect on photosynthesis in heterozygous barley mutated in glutamine synthetase 2, ferrodoxin-glutamate synthase, GDC or serine:glyoxylate aminotransferase [70] (Figure 3). Furthermore, glyoxylate can inhibit the activation of Rubisco in vitro [78], and there is a negative relationship between glyoxylate content in the leaves of heterozygous glutamine synthetase 2 mutants and the activation state of Rubisco [79]. Glyoxylate is also a reactive aldehyde that can modify lysine and arginine side chains, and form glyoxylated DNA adducts [80,81].
As indicated above, in vitro kinetic studies of recombinant proteins revealed that both cytosolic GLYR1 and plastidial GLYR2 catalyse the essentially irreversible, NADPH-based conversion of glyoxylate into glycolate, and can be regulated by the NADPH/NADP ratio [2–4]. Since the Michaelis constants for the two isoforms are apparently in the physiological range (4.5 and 34 μM respectively), it is suggested that the enzymes function in the detoxification of glyoxylate in planta, and, in doing so, provide an alternative sink for excess electrons during the recycling of reducing equivalents. There is not yet any information on expression of the GLYRs under photorespiratory conditions.
HYPOTHETICAL ROLE FOR GLYRs DURING STRESS
Figure 3 summarizes the information discussed above with respect to detoxification of GABA-derived SSA and glycolate-derived glyoxylate in Arabidopsis by mechanisms involving the GLYR isoforms. In the absence of stress, flux through the GABA pathway should be relatively low, whereas photorespiratory flux would be considerable due to the favourable atmospheric CO2/O2 ratio, and glyoxylate produced might contribute to GABA transamination to SSA. In the presence of stress, the cellular Ca2+ or H+ concentration could increase, thereby stimulating GAD activity and resulting in elevated GABA levels, and the oxygenation of ribulose bisphosphate would increase due to stomatal closure, probably resulting in glyoxylate accumulation. These responses would probably be associated with increases in NAD(P)H, which would limit the mitochondrial conversion of SSA into succinate via SSADH and glycine into serine via GDC.
Based on the subcellular localization of the GABA shunt enzymes, it seems likely that GABA produced in the cytosol must be transported into the mitochondrion where it is catabolized to SSA by GABA-T activity. With the limitation of SSADH activity by elevated NADH concentration, SSA probably diffuses to the cytosol and perhaps the chloroplast, where it could be reduced to GHB via GLYR, a process facilitated by elevated NADPH levels. Similarly, some glyoxylate produced in the peroxisome probably diffuses to the cytosol and/or chloroplast to be detoxified by GLYR1 and/or GLYR2 respectively. Thus it is proposed that GLYR activity in plants serves to detoxify aldehydes that are typically generated in two distinct pathways, GABA metabolism and glycolate metabolism, and thereby contributes to redox balance.
The response of the pyridine nucleotides during stress could be attributed in large part to the biochemical control of NAD(H)K. In Arabidopsis, the three isoforms of NADK [cytosolic NADK1 and NAD(H)K3, and chloroplast NADK2] are essential for the phosphorylation of NAD(H) and have been linked to the plant stress response. It is clear that NADK2 utilizes NAD only and that NADK3 prefers NADH, but the preference of NADK1 for NAD and NADH is less certain. Support, although preliminary, exists for activation by Ca2+/CaM of the NADK2 isoform only. Thus further research is required to provide a better picture of the relative importance and the biochemical control of the three NADK isoforms, particularly in view of the Ca2+/CaM activation of GAD, which is located in the cytosol.
To test this scheme, various NAD(H)K expression mutants would be helpful in assessing the impact of altered redox balance on GABA metabolism and GLYR activity. Additional experiments could be conducted with recombinant GLYR1 and GLYR2 to confirm whether the enzymes have the capacity to simultaneously reduce both glyoxylate and SSA, while recycling reducing equivalents. There is also a need to utilize/generate single and double knockouts of GLYR1 and GLYR2 in order to establish their physiological functions during stress. For example, the contribution of GLYR1 and GLYR2 to the balance between energy input through photochemistry and energy utilization through metabolism (i.e. photostasis) [82] could be investigated by determining the levels of GABA/GHB, glyoxylate/glycolate and related amino and organic acids, as well as the redox (including the state of Photosystems) and energy balance and photosynthetic efficiency. If the knockouts are more susceptible to stress than the wild-type, it could be concluded that the GLYR isoforms are involved in photostasis and prevent the accumulation of toxic aldehydes.
AtGLYR2 is localized to the chloroplast; however, its affinity for glyoxylate and SSA is 10-fold lower than that for GLYR1. Typically, the over-reduction of the chloroplast electron transport chain by NADPH accumulation would probably be alleviated by GLYR2, but transgenic plants in which GLYR1 overexpression is targeted to the cytosol or chloroplast could provide additional protection against photoinhibition and other stresses. If so, GLYR1 overexpression could become a strategy for engineering stress resistance in important crop plants.
ACKNOWLEDGEMENTS
We gratefully acknowledge the anonymous reviewers of the manuscript for many useful comments and suggestions.
FUNDING
Work in the authors laboratory in this area is supported by the Natural Sciences and Engineering Research Council of Canada; and the Ontario Ministry of Agriculture Food and Rural Affairs.
References
- 1.Givan C. V., Kleczkowski L. A. The enzymatic reduction of glyoxylate and hydroxypyruvate in leaves of higher plants. Plant Physiol. 1992;100:552–556. doi: 10.1104/pp.100.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoover G. J., Van Cauwenberghe O. R., Breitkreuz K. E., Clark S. M., Merrill A. R., Shelp B. J. Characteristics of an Arabidopsis glyoxylate reductase: general biochemical properties and substrate specificity for the recombinant protein, and developmental expression and implications for glyoxylate and succinic semialdehyde metabolism in planta. Can. J. Bot. 2007;85:883–895. [Google Scholar]
- 3.Simpson J. P., Di Leo R., Dhanoa P. K., Allan W. L., Makhmoudova A., Clark S. M., Hoover G. J., Mullen R. T., Shelp B. J. Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J. Exp. Bot. 2008;59:2454–2554. doi: 10.1093/jxb/ern123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoover G. J., Prentice G. A., Merrill A. R., Shelp B. J. Kinetic mechanism of an Arabidopsis glyoxylate reductase: studies of initial velocity, dead-end inhibition and product inhibition. Can. J. Bot. 2007;95:896–902. [Google Scholar]
- 5.Weber H., Chetelat A., Reymond P., Farmer E. E. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004;37:877–888. doi: 10.1111/j.1365-313x.2003.02013.x. [DOI] [PubMed] [Google Scholar]
- 6.Kotchoni S. O., Kuhns C., Ditzer A., Kirch H. H., Bartels D. Overexpression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006;29:1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x. [DOI] [PubMed] [Google Scholar]
- 7.Moller I. M. Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- 8.Noctor G., Queval G., Gakiere B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 2006;57:1603–1620. doi: 10.1093/jxb/erj202. [DOI] [PubMed] [Google Scholar]
- 9.Noctor G. Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ. 2006;29:409–425. doi: 10.1111/j.1365-3040.2005.01476.x. [DOI] [PubMed] [Google Scholar]
- 10.Foyer C. H., Noctor G. Oxygen processing in photosynthesis: regulation and signaling. New Phytol. 2000;146:359–388. [Google Scholar]
- 11.Igamberdiev A. U., Gardestrom P. Regulation of NAD and NADP-dependent isocitrate dehydrogenases by reduction of levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim. Biophys. Acta. 2003;1606:117–125. doi: 10.1016/s0005-2728(03)00106-3. [DOI] [PubMed] [Google Scholar]
- 12.Rachmilevitch S., Cousins A. B., Bloom A. J. Nitrate assimilation in plant shoots depends on photorespiration. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11506–11510. doi: 10.1073/pnas.0404388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutilleul C., Lelarge C., Prioul J. L., De Paepe R., Foyer C. H., Noctor G. Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol. 2005;139:64–78. doi: 10.1104/pp.105.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollak N., Dolle C., Ziegler M. The power to reduce: pyridine nucleotides: small molecules with a multitude of functions. Biochem. J. 2007;402 doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corpas F. J., Barroso J. B., Sandalio L. M., Distefano S., Palma J. M., Lupianez J. A., Del Rio L. A. A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem. J. 1998;330:777–784. doi: 10.1042/bj3300777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marino D., Gonzalez E. M., Frendo P., Puppo A., Arrese-Igor C. NADPH recycling systems in oxidative stressed pea nodules: a key role for the NADP+-dependent isocitrate dehydrogenase. Planta. 2007;225:413–421. doi: 10.1007/s00425-006-0354-5. [DOI] [PubMed] [Google Scholar]
- 17.Vanlerberghe G. C., Day D. A., Wiskich J. T., Vanlerberghe A. E., McIntosh L. Alternative oxidase activity in tobacco leaf mitochondria-dependence on tricarboxylic-acid cycle-mediated redox regulation and pyruvate activation. Plant Physiol. 1995;109:353–361. doi: 10.1104/pp.109.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atkin O. K., Millar A. H., Gardestrom P., Day D. A. Photosynthesis, carbohydrate metabolism and respiration in leaves of higher plants. In: Leegood R. C., Sharkey T. D., von Caemmerer S., editors. Photosynthesis: Physiology and Mechanisms. Dordrecht: Kluwer; 2000. pp. 153–175. [Google Scholar]
- 19.Bykova N. V., Rasmusson A. G., Igamberdiev A. U., Gardestrom P., Moller I. M. Two separate transhydrogenase activities are present in plant mitochondria. Biochem. Biophys. Res. Commun. 1999;265:106–111. doi: 10.1006/bbrc.1999.1627. [DOI] [PubMed] [Google Scholar]
- 20.Kawai S., Mori S., Mukai T., Hashimoto W., Murata K. Molecular characterization of Escherichia coli NAD kinase. Eur. J. Biochem. 2001;268:4359–4365. doi: 10.1046/j.1432-1327.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 21.Lerner F., Niere M., Ludwig A., Ziegler M. Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun. 2001;288:69–74. doi: 10.1006/bbrc.2001.5735. [DOI] [PubMed] [Google Scholar]
- 22.Outten C. E., Culotta V. C. A novel NAD(H) kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2003;22:2015–2024. doi: 10.1093/emboj/cdg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryotic Cell. 2003;2:809–820. doi: 10.1128/EC.2.4.809-820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berrin J.-G., Pierrugues O., Brutesco C., Alonso B., Montillet J.-L., Roby D., Kazmaier M. Stress induces the expression of AtNADK-1, a gene encoding a NAD(H) kinase in Arabidopsis thaliana. Mol. Genet. Genom. 2005;273:10–19. doi: 10.1007/s00438-005-1113-1. [DOI] [PubMed] [Google Scholar]
- 25.McGuiness E. T., Butler J. R. NAD+ kinase: a review. Int. J. Biochem. 1985;17:1–17. doi: 10.1016/0020-711x(85)90079-5. [DOI] [PubMed] [Google Scholar]
- 26.Sauer A., Robinson D. G. Calmodulin dependent NAD+ kinase is associated with both the outer and inner mitochondrial membranes in maize roots. Planta. 1985;166:227–233. doi: 10.1007/BF00397353. [DOI] [PubMed] [Google Scholar]
- 27.Gallais S., Pou de Crescenzo M. A., Laval-Martin D. L. Characterization of soluble calcium calmodulin-dependent and independent NAD+ kinases from Avena sativa seeds. Aust. J. Plant Physiol. 2001;28:363–371. [Google Scholar]
- 28.Pou de Crescenzo M. A., Gallais S., Leon A., Laval-Martin D. L. Tween-20 activates and solubilizes the mitochondrial membrane-bound, calmodulin dependent NAD+ kinase of Avena sativa L. J. Membr. Biol. 2001;182:135–146. doi: 10.1007/s0023201-0039-8. [DOI] [PubMed] [Google Scholar]
- 29.Delumeau O., Renard M., Montrichard F. Characterization and possible redox regulation of the purified calmodulin-dependent NAD+ kinase from Lycopersicon pimpinellifolium. Plant Cell Environ. 2000;23:1267–1273. [Google Scholar]
- 30.Ruiz J. M., Sanchez E., Garcia P. C., Lopez-Lefebre L. R., Rivero R. M., Romero L. Proline metabolism and NAD kinase activity in greenbean plants subjected to cold-shock. Phytochemistry. 2002;59:473–478. doi: 10.1016/s0031-9422(01)00481-2. [DOI] [PubMed] [Google Scholar]
- 31.Turner W. L., Waller J. C., Vanderbeld B., Snedden W. A. Cloning and characterization of two NAD kinases from Arabidopsis. Identification of a calmodulin binding isoform. Plant Physiol. 2004;135:1243–1255. doi: 10.1104/pp.104.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner W. L., Waller J. C., Snedden W. A. Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress) Biochem. J. 2005;385:217–223. doi: 10.1042/BJ20040292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai M.-F., Wei P.-C., Chen Q.-J., An R., Chen J., Yang S., Wang X.-C. NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J. 2006;47:665–674. doi: 10.1111/j.1365-313X.2006.02816.x. [DOI] [PubMed] [Google Scholar]
- 34.Chai M.-F., Chen Q.-J., An R., Chen Y.-M., Chen J, Wang X.-C. NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol. Biol. 2005;59:553–564. doi: 10.1007/s11103-005-6802-y. [DOI] [PubMed] [Google Scholar]
- 35.Shelp B. J., Bown A. W., McLean M. D. Metabolism and functions of γ-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- 36.Fait A., Fromm H., Dirk W., Galili G., Fernie A. Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci. 2008;13:14–19. doi: 10.1016/j.tplants.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Van Cauwenberghe O. R., Makhmoudova A., McLean M. D., Clark S. M., Shelp B. J. Plant pyruvate-dependent γ-aminobutyrate transaminase: identification of an Arabidopsis cDNA and its expression in Escherichia coli. Can. J. Bot. 2002;80:933–941. [Google Scholar]
- 38.Clark S. M., Di Leo R., Dhanoa P. K., Van Cauwenberghe O. R., Mullen R. T., Shelp B. J. Biochemical characterization, mitochondrial localization, expression and potential functions for an Arabidopsis γ-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J. Exp. Bot. 2009;60:1743–1757. doi: 10.1093/jxb/erp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark S. M., Di Leo R., Van Cauwenberghe O. R., Mullen R. T., Shelp B. J. Subcellular localization and expression of multiple tomato γ-aminobutyrate transaminases that utilize both pyruvate and glyoxylate. J. Exp. Bot. 2009;60:3255–3267. doi: 10.1093/jxb/erp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busch K. B., Fromm H. Plant succinic semialdehyde dehydrogenase. Cloning, purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiol. 1999;121:589–597. doi: 10.1104/pp.121.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Busch K., Piehler J., Fromm H. Plant succinic semialdehyde dehydrogenase: dissection of nucleotide binding by surface plasmon resonance and fluorescence spectroscopy. Biochem. J. 2000;39:10110–10117. doi: 10.1021/bi000589e. [DOI] [PubMed] [Google Scholar]
- 42.Bouché N., Fait A., Bouchez D., Moller S. G., Fromm H. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shelp B. J., Walton C. S., Snedden W. A., Tuin L. G., Oresnik I. J., Layzell D. B. GABA shunt in developing soybean seeds is associated with hypoxia. Physiol. Plant. 1995;94:219–228. [Google Scholar]
- 44.Allan W. L., Peiris C., Bown A. W., Shelp B. J. γ-Hydroxybutyrate accumulates in green tea leaves and soybean sprouts in response to oxygen deficiency. Can. J. Plant Sci. 2003;83:951–953. [Google Scholar]
- 45.Breitkreuz K. E., Allan W. L., Van Cauwenberghe O. R., Jakobs C., Talibi D., André B., Shelp B. J. A novel γ-hydroxybutyrate dehydrogenase: identification and expression of an Arabidopsis cDNA and potential role under oxygen deficiency. J. Biol. Chem. 2003;278:41552–41556. doi: 10.1074/jbc.M305717200. [DOI] [PubMed] [Google Scholar]
- 46.Allan W. L., Simpson J. P., Clark S. M., Shelp B. J. γ-Hydroxybutyrate accumulation in Arabidopsis and tobacco plants is a general response to abiotic stress: putative regulation by redox balance and glyoxylate reductase isoforms. J. Exp. Bot. 2008;59:2555–2564. doi: 10.1093/jxb/ern122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyashita Y., Good A. G. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:92–102. doi: 10.1093/pcp/pcm171. [DOI] [PubMed] [Google Scholar]
- 48.Nunes-Nesi A., Sweetlove L. J., Fernie A. R. Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol. Plant. 2007;129:45–56. [Google Scholar]
- 49.Studart-Guimaraes A., Fait A., Nunes-Nesi A., Carrari F., Usadel B., Fernie A. R. Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated leaves. Plant Physiol. 2007;145:626–639. doi: 10.1104/pp.107.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelp B. J., Allan W. L., Faure D. Role of γ-aminobutyrate and γ-hydroxybutyrate in plant communication. In: Baluska F., editor. Plant–Environment Interactions. Berlin: Springer-Verlag; 2009. pp. 73–84. [Google Scholar]
- 51.Aurisano N., Bertani A., Regianni R. Involvement of calcium and calmodulin in protein and amino acid metabolism in rice roots under anoxia. Plant Cell Physiol. 1995;36:1525–1529. [Google Scholar]
- 52.Cholewa E., Chloewinski A. J., Shelp B. J., Snedden W. A., Bown A. W. Cold-shock-stimulated γ-aminobutyric acid synthesis is mediated by an increase in cytosolic Ca2+ not by an increase in cytosolic H+ Can. J. Bot. 1997;75:375–382. [Google Scholar]
- 53.Knight M. R., Campbell A. K., Smith S. M., Trewavas A. J. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352:524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- 54.Kinnersley A. M., Turano F. J. γ-Aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 2000;19:479–509. [Google Scholar]
- 55.Crawford L. A., Bown A. W., Breitkreuz K. E., Guinel F. C. The synthesis of γ-aminobutyric acid in response to treatments reducing cytosolic pH. Plant Physiol. 1994;104:865–871. doi: 10.1104/pp.104.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caroll A. D., Fox G. G., Laurie S., Phillips R., Ratcliffe R. G., Stewart G. R. Ammonium assimilation and the role of γ-aminobutyric acid in pH homeostasis in carrot cell suspensions. Plant Physiol. 1994;106:513–520. doi: 10.1104/pp.106.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts J. K. M., Hooks M. A., Miaullis A. P., Edwards S., Webster C. Contribution of malate and amino acid metabolism to cytoplasmic pH regulation in hypoxic maize root tips studied using nuclear magnetic resonance spectroscopy. Plant Physiol. 1992;98:480–487. doi: 10.1104/pp.98.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klok E. J., Wilson I. W., Wilson D., Chapman S. C., Ewing R. M., Somerville S. C., Peacock W. J., Dolferus R., Dennis E. S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deeken R., Engelmann J. C., Efetova M., Czirjak T., Müller T., Kaiser W. M., Tietz O., Krischke M., Mueller M. J., Palme K., et al. An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell. 2006;18:3617–3634. doi: 10.1105/tpc.106.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cramer G. R., Ergül A., Grimplet J., Tillett R. L., Tattersall E. A. R., Bohlman M. C., Vincent D., Sonderegger J., Evans J., Osborne C., et al. Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct. Integr. Genomics. 2007;7:111–134. doi: 10.1007/s10142-006-0039-y. [DOI] [PubMed] [Google Scholar]
- 61.Pasentsis K., Falara V., Pateraki I., Gerasopoulos D., Kanellis A. K. Identification and expression profiling of low oxygen regulated genes from Citrus flavedo tissues using RT-PCR differential display. J. Exp. Bot. 2007;58:2203–2216. doi: 10.1093/jxb/erm078. [DOI] [PubMed] [Google Scholar]
- 62.Fait A., Yellin A., Fromm H. GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Lett. 2005;579:415–420. doi: 10.1016/j.febslet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 63.Farquhar G. D., von Caemmerer S., Berry J. A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta. 1980;149:78–90. doi: 10.1007/BF00386231. [DOI] [PubMed] [Google Scholar]
- 64.Sharkey T. D., Kobza J., Seemann J. R., Brown R. H. Reduced cytosolic fructrose-1-6-bisphosphatase activity leads to loss of O2 sensitivity in a Flaveria linearis mutant. Plant Physiol. 1988;86:667–671. doi: 10.1104/pp.86.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noctor G., Veljovic-Jovanovic S., Driscoll S., Novitskaya L., Foyer C. H. Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann. Bot. 2002;89:841–850. doi: 10.1093/aob/mcf096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tolbert N. E., Benker C., Beck E. The oxygen and carbon dioxide compensation points of C3 plants: possible role in regulating atmospheric oxygen. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11230–11233. doi: 10.1073/pnas.92.24.11230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Migge A., Kahmann U., Fock H. P., Becker T. Prolonged exposure of tobacco to a low oxygen atmosphere to suppress photorespiration decreases net photosynthesis and results in changes in plant morphology and chloroplast structure. Photosynthetica. 1999;36:107–116. [Google Scholar]
- 68.Nakamura Y., Tolbert N. E. Serine:glyoxylate, alanine:glyoxylate and glutamate:glyoxylate aminotransferases reactions in peroxisomes from spinach leaves. J. Biol. Chem. 1983;258:7631–7638. [PubMed] [Google Scholar]
- 69.Liepman A. H., Olsen L. J. Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 2001;25:487–498. doi: 10.1046/j.1365-313x.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- 70.Wingler A., Lea P. J., Quick W. P., Leegood R. C. Photorespiration: metabolic pathways and their role in stress protection. Phil. Trans. R. Soc. London Ser. B. 2000;355:1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nilsen E. T., Orcutt D. M. The Physiology of Plants Under Stress, Abiotic Factors. New York: John Wiley & Sons; 1996. Plant carbon balance; pp. 231–277. [Google Scholar]
- 72.Huner N. P. A., Öquist G., Sarhan F. Energy balance and acclimation to light and cold. Trends Plant Sci. 1998;3:224–230. [Google Scholar]
- 73.Fedina I. S., Tsonev T. D., Guleva E. I. The effect of pre-treatment with proline on the responses of Pisum sativuum to salt stress. Photosynthetica. 1993;29:521–527. [Google Scholar]
- 74.Rajmane N. A., Karadge B. A. Photosynthesis and photorespiration in winged bean (Psophocarpus tetragonolobus L.) grown under saline conditions. Photosynthetica. 1986;20:139–145. [Google Scholar]
- 75.Downton W. J. S. Photosynthesis in salt-stressed grapevines. Aust. J. Plant Physiol. 1977;4:183–192. [Google Scholar]
- 76.Wingler A., Quick W. P., Bungard R. A., Bailey K. J., Lea P. J., Leegood R. C. The role of photorespiration during drought stress: an analysis utilizing barley mutants with reduced activities of photorespiratory enzymes. Plant Cell Environ. 1999;22:361–373. [Google Scholar]
- 77.Sanchez-Rodriguez J., Pkrez P., Martinez-Carrasco R. Photosynthesis, carbohydrate levels and chlorophyll fluorescence estimated intercellular CO2 in water-stressed Casuarina equisetijolia Forst and Forst. Plant Cell Environ. 1999;22:867–873. [Google Scholar]
- 78.Campbell W. J., Ogren W. L. Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation in intact, lysed, and reconstituted chloroplasts. Photosynth. Res. 1990;23:257–268. doi: 10.1007/BF00034856. [DOI] [PubMed] [Google Scholar]
- 79.Hausler R. E., Bailey K. J., Lea P. J., Leegood R. C. Control of photosynthesis in barley mutants with reduced activities of glutamine synthetase and glutamate synthase. III. Aspects of glyoxylate metabolism and effects of glyoxylate on the activation state of ribulose-1,5-bisphosphate carboxylase-oxygenase. Planta. 1996;200:388–396. [Google Scholar]
- 80.Schmitt A., Gasic-Milenkovic J., Schmitt J. Characterization of advanced glycation end products: mass changes in correlation to side chain modifications. Anal. Biochem. 2005;346:101–106. doi: 10.1016/j.ab.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 81.Maekawa M., Kawai K., Takahashi Y., Nakamura H., Watanabe T., Sawa R., Hachisuka K., Kasai H. Identification of 4-oxo-2-hexenal and other direct mutagens formed in model lipid peroxidation reactions as dGuo adducts. Chem. Res. Toxicol. 2006;19:130–138. doi: 10.1021/tx050236m. [DOI] [PubMed] [Google Scholar]
- 82.Ensminger I., Busch F., Huner N. P. A. Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiol. Plant. 2006;126:28–44. [Google Scholar]