Abstract
Ribosome-inactivating proteins (RIPs) catalyze the hydrolytic depurination of one or more adenosine residues from eukaryotic ribosomes. Depurination of the ribosomal sarcin-ricin tetraloop (GAGA) causes inhibition of protein synthesis and cellular death. We characterized the catalytic properties of saporin-L1 from Saponaria officinalis (soapwort) leaves and demonstrate robust activity against defined nucleic acid substrates and mammalian ribosomes. Transition state analogue mimics of small oligonucleotide substrates of saporin-L1 are powerful, slow-onset inhibitors when adenosine is replaced with the transition state mimic 9-deazaadenine-9-methylene-N-hydroxypyrrolidine (DADMeA). Linear, cyclic and stem-loop oligonucleotide inhibitors containing DADMeA and based on the GAGA sarcin-ricin tetraloop gave slow-onset tight-binding inhibition constants (Ki*) of 2.3 to 8.7 nM at physiological conditions and bind up to 40,000-fold tighter than RNA substrates. Saporin-L1 inhibition of rabbit reticulocyte translation was protected by these inhibitors. Transition state analogues of saporin-L1 have potential in cancer therapy that employs saporin-L1 linked immunotoxins.
Ribosome inactivating proteins (RIPs) are N-glycohydrolases that catalyze the depurination of adenosine A4234 from the highly conserved sarcin-ricin loop of the 28S eukaryotic ribosomal subunit RNA (1). Depurination inhibits the binding of elongation factor 2 to the ribosome, halts protein synthesis, and causes cell death (2). RIPs with broad polynucleotide:adenosine glycosidase activity can target other ribosomal sites and non-ribosomal substrates including DNA, RNA and poly(A) (3, 4). Saporin-L1 toxin, a RIP from the leaves of the Saponaria officinalis plant can release adenine from poly(A), herring sperm DNA, tRNA, E. coli rRNA, and globin mRNA at physiologic pH (5, 6). Fifteen saporin isoforms have been characterized from Saponaria officinalis including 9 seed, 3 leaf, and 3 root RIPs. These isoforms differ in ribosome translation inhibition activities and nascent cellular toxicity (7). Analysis of 50 type I and II RIPs revealed only saporin-L1 with the ability to release adenine from RNA of MS2, TMV, and AMCV viruses at physiologic pH, a catalytic activity unique in the RIP family of enzymes (3).
Our goal is to develop transition state analogue inhibitors of ribosome inactivating proteins. These may find use in rescue therapy, by preventing the vascular leak syndrome sequelae common in clinical trials of RIPs linked to antibodies directed against cancer epitopes. Such inhibitors could provide extracellar protection against circulating toxins. We model these inhibitors on the transition state of ricin A-chain, the only RIP where transition state information is available. Kinetic isotope effect studies established that ricin A-chain hydrolysis of 10-mer RNA and DNA stem-loop substrates occurs through leaving group activation and forms ribooxacarbenium ion transition states (8 - 10). Small RNA stem-loops are substrates for RIPs and stem-loop substrate mimics of the sarcin-ricin loop serve as inhibitor scaffolds. Substrate stem-loops contain a GAGA tetraloop for RIP recognition and alternating C-G base pairs for stem stability and loop folding (Figure 1). Transition state analogues for ricin A-chain featured protonated 1-azasugars to mimic the oxacarbenium ion intermediate and leaving groups with an elevated pKa at the depurination site (8). Ricin A-chain shows robust catalytic activity on stem-loop RNA substrates at pH 4 but is inactive with these substrates at neutral pH. Transition state analogues of ricin A-chain are nanomolar inhibitors at pH 4 but did not protect ribosomes from ricin A-chain action at neutral pH. Therefore, we searched for another RIP with robust catalytic activity at physiological pH values and with the ability to be inhibited by transition state analogues, a prelude to use as an anticancer agent when linked to an appropriate recognition motif (11). Powerful inhibitors of the toxin can then provide an extracellular, circulating rescue agent to prevent the post-therapy vascular leak syndrome commonly associated with RIP immunochemotherapy (12).
Figure 1.
Substrate and inhibitor constructs for saporin-L1 assays and inhibition. Top: Transition state mimic DADMeA (9-DA), a 9-deazaadenine N-hydroxypyrrolidine sugar. A-10 RNA stem-loop substrate contains the GAGA tetraloop motif and alternating C-G base pairs for stem structure and loop folding. A-10 (9-DA) 2′-OMe contains DADMeA at the target RIP depurination site of the GAGA tetraloop and contains 2′-OMe modified bases (excluding DADMeA). Middle: Three 14-mer constructs containing DADMeA including A-14 (9-DA) 2′-OMe, RNA, or DNA scaffolds. Cyclic oxime G(9-DA)GA 2′-OMe or DNA constructs are tetramers with 5′- to 3′-oligonucleotide ends closed by a synthetic linker (16). Bottom: Linear inhibitor. Tetramer G(9-DA)GA 2′-OMe, trimer G(9-DA)Gs3 2′-OMe, dimer s3(9-DA)Gs3 2′-OMe and monomer s3(9-DA)s3 where s3 denotes a propyl phosphate.
Saporin-L1 has not been kinetically characterized with small substrates or inhibitors. Here we characterize its kinetic properties on small stem-loop substrates, on mammalian ribosomes, and report novel transition state analogue inhibitors, all at physiological pH values. Kinetic analysis takes advantage of a sensitive and continuous assay for adenine linked to luciferase-based light production (13). Saporin-L1 catalyzes the depurination of adenines from A-10 (an RNA stem-loop 5′-CGCGAGAGCG-3′ mimic of the sarcin-ricin loop), linear and covalently closed circular constructs related to A-10 and from mammalian 80s ribosomes, all at physiologic pH (Figure 1).
The TS mimic 9-deazaadenine-9-methylene-N-hydroxypyrrolidine (DADMeA) replacement for adenosine in the RIP recognition GAGA tetraloop motif inhibits saporin-L1 catalysis (Figure 1). We synthesized inhibitors against saporin-L1 including monomer to 14-mer oligonucleotides employing the DADMeA transition state mimic (Figure 1). Stem-loop inhibitor constructs, cyclic tetramer 5′- to 3′- covalently closed circular GAGA tetraloops, and monomeric inhibitors were also applied to saporin-L1. A minimal saporin-L1 inhibitor scaffold (Figure 1) with 2′-OMe containing compounds designed to give enhanced nuclease stability provided inhibition of saporin-L1 to low nanomolar Ki values and also protected ribosomes from saporin-L1 action in rabbit reticulocyte lysates. (Figure 1).
Experimental Procedures
Materials
Oligonucleotide A-10 was purchased from Dharmacon (Lafayette,CO). 5′-DMT protected 9-DA azasugar was synthesized and purified as previously described.[8] DNA/RNA synthesis reagents were purchased from Glen Research (Sterling, VA) and ChemGene Co. (Ashland, MA). HPLC purifications were preformed on a Waters 626 pump with a 996 photodiode array detector with Millennium software. Firefly luciferase ATP assay kit (ATPlite) was purchased from Perkin Elmer (Waltham, Massachusetts). Phosphatase inhibitors (PhosSTOP) were purchased from Roche Applied Science (Indianapolis, IN). RNase inhibitor (SuperRNasin) was purchased from Ambion (Austin, TX). Ricin A-chain and Saporin-S6 was purchased from Aldrich Chemical Corp. (Ashland, MA). For translation assays, Flexi® Rabbit Reticulocyte Lysate System, luciferase assay system, and rabbit reticulocyte lysate (untreated) was purchased from Promega (Madison, WI). Buffers and enzyme preparations were checked for RNase activity using RNaseAlert from Ambion (Austin, TX). DEPC treated water (0.1% DEPC stirred for 20 min followed by 30 min autoclave treatment) was used in all enzymatic reactions and buffers. All other reagents used were purchased in the highest purity available from Fisher Scientific (Pittsburgh, PA) or Aldrich Chemical Corp. (Ashland, MA). Concentrations of adenine and oligonucleotides were measured using a NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA). Inhibitor concentrations were determined spectrophotometrically including the published millimolar extinction coefficient of 8.5 at 275 nm at pH 7 for 9-deazaadenosine (14). Enzyme concentrations of saporin-L1 and saporin-S6 were determined with the BCA protein assay kit from Pierce (Rockford, IL). Luminescence measurements were accomplished on a GloMax 96-well luminometer from Promega (Madison, WI).
Saporin-L1 Isolation
Saporin-L1 was isolated from the leaves of Saponaria officinalis (common soapwort) as described previously with with modifications described below (7). Freshly harvested leaves (10 g) were frozen and ground with a pestle under liquid nitrogen. The powder was suspended in 80 mL extraction buffer (10 mM Na2HPO4 pH 5.5 (titrated with citric acid), 175 mM NaCl, 2.5 mM MgCl2, 1 mM CaCl2, one tablet of complet protease inhibitor (Roche), 1% (w/v) poly(vinylpolypyrrolidone), 1% of cellulase, 0.5% hemicellulase, and 150 units pectinase). The mixture was stirred at room temperature for 3 hours and then acidified to pH 4.0 with acetic acid. Triton X-100 was added to 0.5% (v/v) and the mixture was stirred for an additional 1 hour. The digested and lysed leaf mixture was then filtered through cheese cloth and centrifuged at 25,000 g for 30 minutes. The supernatant was loaded on SP-sepharose FF resin (Amersham) pre-equilibrated in 10 mM sodium phosphate pH 4.5. The column was extensively washed with 10 mM sodium phosphate pH 7.4 and the crude saporin-L1 containing fraction was eluted with the same buffer containing 1 M NaCl. The elute was dialyzed against 10 mM sodium phosphate pH 7.4 titrated to pH 4.5 with acetic acid and loaded onto carboxymethyl-FF (three 1 mL columns, Amersham) pre-equilibrated in buffer [10 mM sodium phosphate pH 7.4]. The columns were washed extensively with buffer to achieve pH equilibration. Saporin-L1 was eluted with a 50 min linear gradient of 0-300 mM NaCl in 10 mM sodium phosphate pH 7.4 at 1 ml/min and was identified as an ∼30 kD band by SDS-PAGE. Saporin-L1 fractions were combined, titrated to pH 4.5 with acetic acid, and loaded onto heparin HP (three 1 mL columns, Amersham) pre-equilibrated in buffer. Saporin-L1 was eluted with a 50 minute linear gradient of 0-800 mM NaCl in 10 mM sodium phosphate pH 7.4 (1ml/min). Saporin-L1 was eluted as the last major peak in the chromatogram and was identified by SDS-PAGE. The >80% pure Saporin-L1 was concentrated by spin Amicon concentrator and purified to >95 % homogeneity with a BioSep-SEC-S 2000 column (Phenomenex) equilibrated in 20 mM sodium phosphate pH 7.4 and eluted at 1 mL/min (Supporting information). The gel filtration purification step was needed to remove trace DNAase and RNAase activities from Saporin-L1. Saporin-L1 was concentrated to ∼1 mg/mL and stored at 4 °C. The yield was ∼ 0.5 mg saporin-L1 from 10 g of leaf material. Saporin-L1 isolation from Saponaria officinalis seeds is described in Supporting information. Commercial saporin-S6 was purified with the heparin chromatography step as described above.
Saporin-L1 N-terminal Sequencing and Mass Analysis
Purified saporin-L1 from the leaves of Saponaria officinalis was verified by N-terminal sequencing at the Rockefeller University Proteomics Resource Center (New York, NY). The N-terminal sequence was VIIYELNLQG which matched previous reports for the Saporin-L1 isoform (7). The mass of saporin-L1 was measured on a MALDI-TOF mass spectrometer in the linear positive ion mode with external calibration. Protein samples (∼20 μM) were desalted with a ZipTip (Waters) as described by the manufacturer and eluted onto a 100 well gold plate with 1 μL of matrix solution (20 mg/mL sinapic acid in 70 % acetonitrile/H20 with 0.1 % TFA). The mass of saporin-L1 (28,749 Da) isolated from the leaves of Saponaria officinalis was comparable to previous reported masses for saporin-L1 leaf and vacuolar isoforms (28,740 to 28,765 Da) (Supporting information) (15).
Synthesis of Oligonucleotides Inhibitors
Cyclic oligonucleotides and 1-aza sugar phosphoramidites were synthesized and purified as reported previously (16). Stem-loop oligonucleotides were synthesized on a 1 μmol scale with DMT-on mode using an Expedite 8909 DNA/RNA synthesizer following standard synthesis protocols for β-cyanoethyl phosphoramidite chemistry with acetyl protected cytosine phosphoramidite and 5-benzylthio-1H-tetrazole as activator. Cleavage from solid support and base deprotection of a 1 μmol synthesis was accomplished in 1.5 mL AMA reagent (1:1 concentrated NH4OH to 40% aqueous methylamine) for 45 minutes at 37 °C. The reaction mixture was centrifuged, the supernatant collected, and the resin was washed twice with 3:1:1 ethanol:acetonitrile:water. Combined supernatant and washes were evaporated to dryness under vacuum. 2′-O-TBDMS deprotection of the A-14 (DADMeA) RNA (1 μmole) was accomplished using 250 μL of anhydrous TEA HF/NMP solution (1.5 mL of N-methylpyrrolidinone, 750 μL of TEA, and 1.0 mL of TEA-3HF) heated to 65 °C for 2 h (17). This reaction mixture was diluted with 2 mL of 0.5 M NH4OAc and evaporated to dryness under vacuum.
HPLC purification of the 5′-trityl stem-loop oligonucleotides was accomplished to >95% purity on a Waters Delta-Pak (7.9 mm × 300 mm) semipreparative C18 reversed phase column at 3.5 mL/min in 20 mM NH4OAc/5% CH3CN with a linear 0-40% gradient of CH3CN in 25 min. Trityl protected oligonucleotides were the major peak and eluted at ∼25 min. The major late eluting fraction was evaporated to dryness under vacuum. The pellet was dissolved in 1 mL of 80% acetic acid in water, incubated at 30 °C for 1 hour, and the solution was evaporated to dryness under vacuum. HPLC purification of the final oligonucleotide was accomplished to >95% purity on a Waters Delta-Pak (7.9 mm × 300 mm) semipreparative C18 reversed phase column at 3.5 mL/min in 50 mM triethyl ammonium acetate pH 7.0 with a linear 0-80% gradient of 50% aqueous methanol in 40 min. The final product was evaporated to dryness in a speed vac concentrator and resuspended in sterile RNAase free water.
Linear inhibitors were synthesized in DMT-off mode on an Expedite 8909 synthesizer in otherwise identical conditions to stem-loop oligonucleotides. After deprotection in AMA, the oligonucleotides were purified by HPLC to >95 % purity as described for stem-loops in 50 mM triethyl ammonium acetate pH 7.0 with a linear 0-50% gradient of 50% aqueous methanol in 40 to 60 min.
Stem-loop, cyclic, and linear oligonucleotide structures were confirmed using a MALDI-TOF mass spectrometer as described previously (16). Observed and calculated masses for the final compounds are in Supporting information. Prior to use in inhibition assays, stem-loop oligonucleotides were heated to 95 °C for 1 minute and cooled on ice.
Saporin-L1 Kinetic Assay
Saporin-L1 kinetics on substrate A-10 RNA (5′-CGCGAGAGCG-3′) were determined using a continuous coupled assay for quantifying free adenine by linking it to the production of light from luciferase (13). In brief, an adenine detection buffer was prepared in bulk (50 mL of charcoal-filtered solution containing 100 mM tris-acetate pH 7.7, 2 mM phosphoenolpyruvic acid, 2 mM sodium pyrophosphate, 2 mM 5-phospho-D-ribosyl-1-pyrophosphate (PRPP), 15 mM NH4SO4, 15 mM (NH4)2MoO4, and phosphatase inhibitors in RNAase free water) and stored at -80 °C in 1 mL aliquots. Prior to use in adenine assays, coupling enzymes were prepared by adding 10 mM MgSO4, 8 units of APRTase, 8 units of phosphoenolpyruvate dikinase, 200 μL D-luciferin/luciferase (ATPLite) reagent and 1 μL of SuperRNasin (Ambion) per 1 mL of coupling enzymes buffer. One unit of enzyme activity was defined as the amount that forms one μmole of product per min at 20 °C.
Varying concentrations of A-10 RNA (5′-CGCGAGAGCG-3′) were prepared in 1:1 diluted coupling enzymes in a 96-well luminometer plate and reactions were initiated with 300 pM saporin-L1 (50 μL total reaction). Luminescence was measured with a luminometer in kinetic acquisition mode for several minutes. Adenine standards were prepared in identical assay conditions. The initial rates of adenine formation were calculated by converting luminescent rate (lumens/second) to enzymatic rate (pmol adenine/min/pmol enzyme) calibrated from the adenine standard curve. Kinetic parameters kcat and Km were calculated by fitting initial rates to the Michaelis-Menten equation and inhibitor dissociation constants were obtained by fitting the data to the equation for competitive inhibition with the assumption that substrate and inhibitor act as competitive inhibitors.
Rabbit ribosomes (80S) were purified from rabbit reticulocyte lysate by sucrose cushioin centrifugation (13). Saporin-L1 (300 pM) was analyzed for kinetic parameters with ribosomes as substrate as described for A-10 RNA substrate. Ribosome concentration was determined by depurinating (to completion) two stock concentrations with 500 nM RTA and comparing the final luminescence to the adenine standard curve fit. RTA releases 1 mol adenine from 1 mol ribosome and thus provides a method of quantitation.
Saporin-L1 Inhibition Assays
Saporin-L1 inhibition constants for stem-loop, circular, and linear oligonucleotides were determined in a competition assay using RNA A-10 substrate with quantitative analysis of adenine release as described for kinetic assays. Varying concentrations of inhibitor were pre-incubated with 300 pM saporin-L1 for 10 min in 1× continuous assay buffer at 20 °C. Reactions were initiated by the addition of A-10 (∼80 μM) and light generation (RLU) was measured in a luminometer over several minutes to obtain the initial rates (lumens/sec). The maximum rate of catalysis (kcat) was calculated from the Michaelis equation as described in kinetic assays. In cases where slow-onset inhibition was observed, Ki* was used to define the inhibition. Pre-incubation of inhibitor with saporin-L1, followed by initiation of the reaction with substrate provided a direct measure of Ki*. Values for the inhibition constant (Ki*) were calculated by fitting post slow-onset rates to the equation for competitive inhibition, ν = kcat[S]/[[S] + Km(1 + I/Ki*)], where ν is the initial reaction rate, [S] is the substrate concentration, Km is the Michaelis constant for A-10, and kcat is the initial rate at A-10 saturation. For tight inhibition, when the concentration of inhibitor was ≤5 times the enzyme concentration, a correction was made for free inhibitor concentration. The free inhibitor concentration was determined by the relationship I = It - (1 - νi/νo)Et, where It is total inhibitor concentration, νi and νo are the inhibited and uninhibited steady-state rates, respectively, and Et is the total enzyme concentration.
Protein translation assays
Saporin-L1 inhibition of protein translation was determined using a reticulocyte lysate translation system to express luciferase from mRNA as described by the manufacturer. For IC50 determination, 30 μL translation reactions in triplicate with varying concentrations of saporin-L1 were incubated at 37 °C at 1.5 hr. A 10 μL aliquot was sampled and luminescence was measured with a luciferase detection kit (Promega) according to the manufacturer's protocol in a 96-well plate format on a luminometer. Percent translation relative to control was plotted versus the log of saporin-L1 concentration and fit to a dose-response curve for the calculation of IC50.
For EC50 determination, triplicate reactions of 2.1 nM saporin-L1 with increasing inhibitor concentrations were pre-incubated at room temperature for 10 min in 5 μL of buffer (20 mM tris-acetate pH 7.4, 25 mM KCl, 5 mM MgCl2). Translation mix (25 μL) was added (300 pM saporin-L1 final) to the pre-incubated samples and were incubated at 37 °C for 1.5 hours. A 10 μL aliquot was sampled and luminescence was measured with the luciferase detection kit (luminescence) as described above. A control with the maximum inhibitor concentration without saporin-L1 established that the oligonucleotide itself did not affect luciferase expression. Percent translation relative to control (no saporin-L1) was plotted versus the log of inhibitor concentration and was fit to a dose-response curve for the calculation of EC50.
Results and Discussion
Saporin-L1 Catalysis
Initial rate kinetics were measured by coupling the adenine product to a luciferase–luciferin coupled assay with quantitation via luminescence (13). Saporin-L1 catalyzed deadenylation of A-10 gave a hyperbolic saturation curve with a kcat = 440 ± 16 min-1 and Km = 95 ± 7 μM at pH 7.7 (Table 1). MALDI-TOF analysis of the saporin-L1 reaction product showed that both adenosines in the GAGA tetraloop of A-10 were depurinated during prolonged incubations (Supporting information). Cyclic oxime RNA GAGA, a circular oligonucleotide substrate, was also depurinated by saporin-L1 with kinetics comparable to A-10 RNA with a kcat = 301 ± 27 min-1 and Km = 82 ± 15 μM (Table 1). The synthetic linker in circular oxime GAGA substrates folds the tetraloop for RIP recognition and is proposed to mimic the structure of stem-loop oligonucleotides (Figure 1) (16). Linear GAGA was also investigated as a saporin-L1 substrate and gave a kcat = 293 ± 29 min-1 and Km = 266 ± 39 μM (Table 1). The Km for linear GAGA is ∼3-fold higher than for A-10 or cyclic oxime RNA substrate while the catalytic turnover rate (kcat) is comparable. Linear GAGA is less structured in solution than stem-loop or cyclic oligonucleotides and requires higher concentrations for equivalent catalytic rates. To our knowledge, previous kinetic constants for saporin-L1 catalysis have only been reported for poly(A) RNA with a kcat = 61 ± 1 min-1 and Km = 639 ± 32 μM at pH 7.8 (Table 1) (5). A-10 RNA depurination by saporin-L1 is 10-fold faster (kcat) and 4.5-fold tighter (Km) than poly(A) RNA under comparable conditions, to give a 45-fold increased catalytic efficiency (kcat/Km).
Table 1. Kinetic parameters for saporin-L1.
| Substrate | kcat (min-1) | Km (μM) | kcat/Km (M-1s-1) |
|---|---|---|---|
| A-10 RNA | 440 ± 16 | 95 ± 7 | 7.7 × 104 |
| cyclic oxime GAGA | 301 ± 27 | 82 ± 15 | 6.1 × 104 |
| linear GAGA | 293 ± 29 | 266 ± 39 | 1.8 × 104 |
| poly(A)* | 61 ± 1 | 639 ± 32 | 1.6 × 103 |
| A-10 RNA (saporin-S6) | 0.35 ± 0.04 | 360 ± 60 | 16 |
kinetic constants for poly(A) were previously reported in 20 mM Tris/HCl, pH 7.8, 100 mM NH4Cl, 10 mM magnesium acetate.[5]
Saporin-S6 catalysis
Although saporin-S6 is more highly characterized than saporin-L1, it was resistant to inhibition by the transition state analogues. We measured A-10 catalysis by saporin-S6, an RIP from Saponaria officinalis seeds. Saporin-S6 was a commercial preparation and was further purified to remove contaminating saporin-L1 (Experimental Procedures). Saporin-S6 and saporin-L1 co-eluted on carboxymethyl resin but were separated by heparin chromatography (Supporting information). Saporin-S6 initial rate catalysis of A-10 RNA as substrate gave a kcat of 0.35 ± 0.04 min-1 and Km of 360 ± 60 μM at pH 7.7 (Table 1). Saporin-L1 catalyzes (kcat) A-10 RNA approximately 1,500 times faster and has a 4-fold lower Km than saporin-S6. Thus, saporin-L1 is 4,800-fold more efficient (kcat/Km) at catalyzing A-10 RNA depurination than saporin-S6 (Table 1). Ricin A-chain is incapable of a single turnover of A-10 above pH 6.5 but at pH 4.0 gives a kcat of ∼4 min-1 on this substrate (18). Saporin-S6 was previously reported to catalyze the depurination of a 35-mer synthetic SRL mimic with GAGA tetraloop at pH 7.6 with a kcat of 0.4 min-1 and Km of 9 μM, while RIPs trichosanthin, gelonin, cinnamomin A-chain, and ricin A-chain had no detectable activity at physiological pH values (19).
Saporin-L1 Action on 80S Ribosomes
Saporin-L1 action on 80S rabbit reticulocyte ribosomes (40 nM) showed multiple adenines released with a rate of 50 min-1 which is 250-fold faster than adenine release from A-10 RNA at an equivalent concentration. We observed a continuous, linear rate for the formation of adenine from 80S rabbit ribosomes extending well past one adenine/ribosome. Thus, saporin-L1 lacks sarcin-ricin loop specificity as the primary ribosomal depurination target. Previous reports indicate that saporin-L1 depurinates up to 36 adenines/mol from 80S rat ribosomes while saporin-S6 releases 1 to 2.5 adenine/mol (20). Moreover, saporin-L1 was reported to release ∼6 adenines from 80S rat ribosome before 50% inhibition of protein synthesis was observed in in vitro translation assays with a poly(U) transcript (5). Thus, adenines other than those at the sarcin-ricin loop are removed preferentially. Most other ribosome inactivating proteins are highly specific in releasing one mole of adenine per mole ribosome from the eukaryotic sarcin-ricin loop (1). Thus, both ricin A-chain and saporin-S6 release only 1 mole adenine per mole 80S rabbit ribosome in our assay conditions. Ribosomal proteins surrounding the sarcin-ricin loop are known to influence the ribotoxic mechanism of RIP substrate recognition (21, 22).
Saporin-L1 Inhibitors
DADMeA (9-DA in Figures) is a non-hydrolysable methylene bridged 9-deazaadenine 1-aza sugar with features of the transition state for depurination of A-10 constructs by ricin A-chain. The replacement of N for C at C1′ of the ribosyl group mimics the carbocation of the dissociated transition state (8, 10). Replacing adenine with 9-deazaadenine caused an elevated pKa at N7, another feature of the transition state. The methylene linker between the 9-deazaadenine and the hydroxypyrrolidine places the base analogue and ribocation at approximately the same distance as found at the transition state. The omission of the 2′-hydroxyl found in RNA is required for the chemical stability of 9-DA. Replacing the scissile adenosine with 9-DA within the 14-mer stem-loop [A-14 (9-DA) RNA] competitively inhibited saporin-L1 with A-10 RNA as substrate with a Ki*of of 3.7 ± 0.7 nM at pH 7.7 (Table 2). The slow onset inhibition (Ki*) observed for saporin-L1 binding of inhibitors such as A-14 (9-DA) RNA was common to all inhibitor constructs listed in Table 1 excluding s3(9-DA)s3 and 9-DA (Figure 2). Inhibitor A-14 (9-DA) DNA oligonucleotide inhibited saporin-L1 with a Ki* of 3.1 ± 0.5 nM, similar to A-14 (9-DA) RNA (Table 2). 2′-OMe A-14 (9-DA), a nuclease stable 14-mer oligonucleotide is also similar to the RNA/DNA versions with a Ki* value of 5.6 ± 0.8 nM (Table 2). Thus, high binding affinity is observed between A-14 (9-DA) constructs in RNA, DNA, and 2′-OMe structural motifs. A 10-mer inhibitor A-10 (9-DA) 2′OMe had a Ki*of of 4.2 ± 1.3 nM, similar to 2′-OMe A-14 (9-DA) (Table 2). This observation is reminiscent of previous reports with ricin A-chain in showing similar Km values for small constructs of RNA, DNA, and 2′-OMe modified substrates (23). Limited quantitative kinetic and substrate specificity data is available for other RIPs.
Table 2.
Inhibition constants for saporin-L1 inhibitorsA
| Inhibitor | Ki* (nM) |
|---|---|
| A-14 (9-DA) 2′-OMe | 5.6 ± 0.8 |
| A-14 (9-DA) RNA | 3.7 ± 0.7 |
| A-14 (9-DA) DNA | 3.1 ± 0.5 |
| A-10 (9-DA) 2′-OMe | 4.2 ± 1.3 |
| cyclic oxime (9-DA) 2′-OMe | 3.9 ± 0.5 |
| cyclic oxime (9-DA) DNA | 2.3 ± 0.1 |
| G(9-DA)GA 2′-OMe | 8.7 ± 2.3 |
| G(9-DA)Gs3 2′-OMe | 7.5 ± 1.6 |
| s3(9-DA)Gs3 2′- OMe | 6.4 ± 1.7 |
| s3(9-DA)s3 | 690 ± 100 B |
| 9-DA | > 0.5 × 106 B |
See Figure 1 for structures of inhibitors.
Values are Ki with no slow onset observed.
Figure 2.
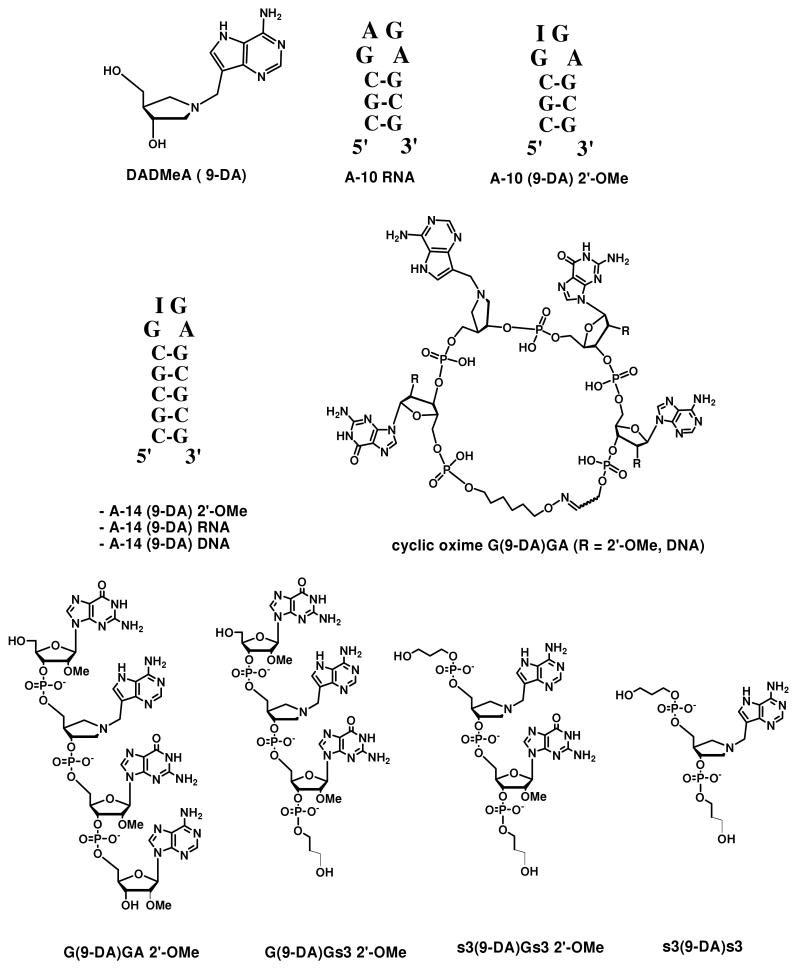
Slow-onset inhibition of saporin-L1 by transition state mimics. A) Plot of lumens (RLU) versus time for saporin-L1 catalysis of A-10 with increasing inhibitor [A14 (9-DA) 2′-OMe] concentrations. B) Competitive inhibition curve fit of rate versus increasing concentrations of tetramer cyclic oxime G(9-DA)GA DNA inhibitor. Kinetics were measured after a 10 minute enzyme-inhibitor pre-incubation equilibration to achieve slow onset binding (Ki*).
Circular Inhibitors
Cyclic DNA and cyclic 2′-OMe modified G(9-DA)GA oligonucleotides inhibited saporin-L1 with Ki* values of 2.3 ± 0.1 nM and 3.9 ± 0.5 nM respectively (Table 2, Figure 2B). Thus, 9-DA cyclic tetramers inhibit saporin-L1 ∼ 1.4 fold tighter than the larger A-14 stem-loop counterparts. This similarity in Ki* supports a primary role for the stem in sarcin-ricin stem-loop mimics to fold the tetraloop for RIP recognition (16). Circular oxime (9-DA) DNA binds saporin-L1 ∼40,000-fold tighter than A-10 RNA or cyclic oxime RNA substrate, a gain of ∼6 kcal/mole in binding energy.
Linear Inhibitors
We also investigated monomer, linear dimer, trimer and tetramer inhibitor scaffolds containing 9-DA in the RTA-scissile adenosine position (Figure 1). Linear tetramer G(9-DA)GA 2′-OMe bound saporin-L1 only ∼2-fold less tightly to than its cyclic oxime counterpart and gave a Ki* of 8.7 ± 2.3 nM (Table 2). Linear trimer [G(9-DA)Gs3] and dimer [s3(9-DA)Gs3] 2′-OMe inhibitors bound comparably with Ki* values of 7.5 ± 1.6 nM and 6.4 ± 1.7 nM respectively (Table 1, Figure 1). The linear dimer s3(9-DA)Gs3 phosphodiester also binds with affinity similar to stem-loop and cyclic oxime 2′-OMe inhibitors (Table 2). A monomer phosphodiester (s3(9-DA)s3) inhibited saporin-L1 with a Ki* value of 690 ± 100 nM while 9-DA alone was not an inhibitor (Table 2). Dimer (s3(9-DA)Gs3) is the tightest linear inhibitor to saporin-L1, binding ∼15,000-fold tighter than A-10 RNA, a gain of ∼5.6 kcal/mole in binding energy. Removal of the 3′- Gs3 moiety of dimer inhibitor to give monomer (s3(9-DA)s3) reduced binding affinity to saporin-L1 10-fold and therefore binding energy by ∼ 2-fold (Table 2). Thus, the 3′- Gs3 moiety of dimer s3(9-DA)Gs3 contributes half the binding energy for inhibitor association with saporin-L1. NMR structural determinations of GAGA tetraloops and the reported crystal structure of a 29-mer RNA SRL mimic show the G1A2G3A4 tretraloop conformation with A2 and G3 available for direct hydrogen bonding while the G1 and A4 form a sheared base pair (24-25). Surprisingly, none of the inhibitors for saporin-L1 (Table 2) were effective against saporin-S6 at concentrations up to 10 μM. Saporin-S6 also catalyzes A-10 deadenylation but at a much slower rate and higher Km (Table 1).
Saporin-L1 Translation Inhibition
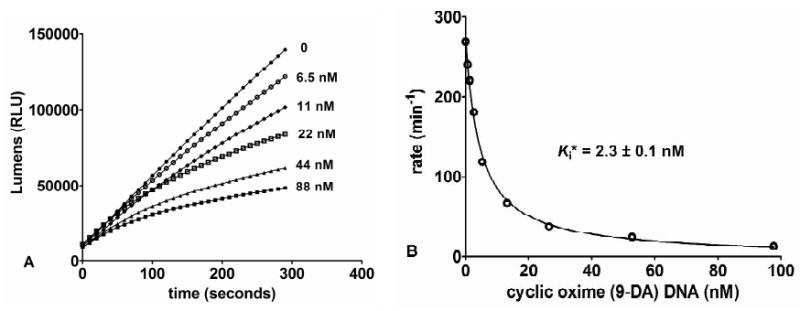
Saporin-L1 inhibited the translation of luciferase mRNA by rabbit reticulocyte lysate in a cell-free translation assay with an IC50 of 45 pM (Figure 3A). Previous reports of saporin-L1 inhibition (IC50) in ribosome translational assays are comparable and vary from 250 pM to 11 nM with poly (U) mRNA for poly-phenylalanine expression (5, 6, 7). Saporin-L1 has been reported to release more than 6 moles of adenine per ribosome to cause the arrest of translation (5). Although the sarcin-ricin loop is not the first adenylate targeted by saporin-L1 in the 80S ribosome, the robust depurination activity is highly toxic to ribosomes and protein translation. Ricin A-chain and saporin-S6 specifically depurinate the sarcin-ricin loop and have IC50 values of 30 pM and 8 pM respectively in rabbit reticulocyte lysate translation assays, similar to that of 45 pM for saporin-L1 (Figure 3A) (26).
Figure 3.
Inhibition of translation by saporin-L1 and protection by saporin-L-1 inhibitors. A) Saporin-L1 inhibition of protein translation (% relative to control) in rabbit reticulocyte lysate assays. The mean and SEM error of triplicate data points were fit to a dose-response curve. B) Protein translation rescue from 300 pM of saporin-L1 (% relative to no saporin-L1) with increasing concentrations [log(nM)] of dimer s3(9-DA)Gs3 inhibitor in a ribosome reticulocyte lysate assay. The mean and SEM error of triplicate data points were fit to a dose-response curve. C) Saporin-L1 (300 pM) rate of adenine release from 40 nM 80S ribosome versus concentrations of s3(9-DA)Gs3 inhibitor. D) Plot of Figure 3C as a dose-response curve fit for percentage of 80S catalysis [% relative to no s3(9-DA)Gs3)] versus increasing concentrations [log(nM)] of dimer s3(9-DA)Gs3 inhibitor. The zero inhibitor value shown in C is not plotted in D.
Saporin-L1 Inhibitor Implications for Toxin Immunotherapy
Saporin-L1 immunotoxin conjugates have been shown to be equally cytotoxic to cells as saporin-S6 conjugates (27). The catalytic A-chains of saporin-S6 and ricin have been investigated as anticancer agents in phase I/II clinical trials using chimeric immunotoxin conjugates in the treatment of lymphomas (11, 28, 29, 30). Therapeutic uses of immunotoxins have been limited by the vascular leak syndrome, a side effect resulting from inappropriate toxin targeting to the capillary bed of endothelial cells, causing edema and multi-organ failure (12, 31, 32, 33). Thus, cancer therapies that employ RIP-linked immunotoxins remain a challenge. Inhibitors to RIPs could provide a way to rescue normal cells following treatment of a targeted tissue with immunotoxins.
Saporin-L1 translation inhibition rescue by TS inhibitor
Protein translation, its inhibition by saporin-L1, and its rescue by inhibitors was studied in rabbit reticulocyte preparations. The minimal saporin-L1 inhibitor dinucleotide s3(9-DA)Gs3 2′-OMe was used (Figure 1). Saporin-L1 toxicity to cell-free translation caused ∼90% inhibition at 300 pM saporin-L1 and was rescued by increasing concentrations of s3(9-DA)Gs3 with an EC50 of 36 ± 2 nM (Figure 3B). The EC50 to Ki* ratio for s3(9-DA)Gs3 comparing translation assays and inhibition kinetics was ∼6, supporting strong saporin-L1 inhibition in complex assays at physiological pH (Figure 3B, Table 2). Dinucleotide s3(9-DA)Gs3 also inhibits saporin-L1 adenine release from purified 80S rabbit ribosomes and gave an IC50 of 7.8 ± 1.1 nM (Figure 2C, 2D). The calculation of a Km value for 80S rabbit ribosomes was limited by practical concentrations of ribosomes from reticulocyte lysate and therefore precluded direct Ki* calculation. However, the IC50 value for s3(9-DA)Gs3 was comparable to the Ki* measured for s3(9-DA)Gs3 in A-10 competition assays (Figure 2D, Table 2). Thus, TS inhibitors of saporin-L1 are equally effective in preventing ribosome depurination and depurination of small nucleic acid substrates.
Summary and Conclusions
Saporin-L1 is highly active on mammalian ribosomes and potently inhibited by the transition state mimic DADMeA (9-DA) when incorporated into the depurination site of stem-loop, circular and linear nucleic acid scaffolds appropriate for RIP recognition. This activity occurs at physiological conditions. The oligonucleotide context is essential as 9-DA alone is a poor inhibitor. Small oligonucleotide inhibitors featuring DADMeA bind up to 40,000 fold tighter than small stem-loop RNA substrates. Nine inhibitors exhibited slow onset binding to saporin-L1 with dissociation constants from 8.7 to 2.3 nM. Inhibitors constructed on RNA, 2′-OMe, and DNA scaffolds supported 9-DA binding with only small scaffold effects. Covalently closed circular inhibitors constructed on DNA or 2′-OMe RNA scaffolds were also excellent inhibitors. Dinucleotide s3(9-DA)Gs3 was the smallest tight binding inhibitor of saporin-L1. In ribosome translation rescue assays, s3(9-DA)Gs3 restored protein synthesis by inhibiting saporin-L1 depurination activities. Direct competition assays between 80S rabbit ribosomes and s3(9-DA)Gs3 established saporin-L1 inhibition with a dissociation constant of 7.8 nM.
Transition state inhibitor efficacy for RIP activity at physiologic pH has been previously limited by the pH 4.0 activity used for development of ricin A-chain transition state analogues. Ricin A-chain and other type I and II RIPs have a low pH catalytic optimum on nucleic acid substrates such as stem-loop RNA, poly(A) and/or hsDNA, while the natural ribosome substrate is depurinated optimally at physiologic pH (6, 18, 34) Saporin-S6 has a substrate specificity distinct from saporin –L1 and was not inhibited by saporin-L1 transition state mimics at neutral pH. Saporin-L1 rapidly catalyzes depurination of stem-loop, circular, and linear truncated mimics of the sarcin-ricin loop at neutral pH, a unique feature in the RIP family of N-glycohydrolases (Table 1) (35). Although saporin-L1 is reported to catalyze adenine release from poly(A), hsDNA, tRNA, E. coli rRNA, and globin mRNA at pH 7.8 (5, 6) the 80S ribosome is a preferred substrate.
Tight-binding inhibitors of saporin-L1 that prevent ribosome damage at physiological pH provide a breakthrough in developing immunotoxin cancer therapy. It should be possible to use saporin-L1 conjugates to target cancer cells. Following tumor lysis, rescue of the organism from vascular leak syndrome might be effected with the small, stable, and tight-binding inhibitors of saporin-L1 characterized here.
Supplementary Material
Footnotes
This work was supported by research grant CA072444 from the NIH.
Supporting Information Available: Supplementary information contains the details of saporin-L1 purification, mass spectral characteristics of chemically synthesized oligonucleotides and curve fits of kinetic data to obtain the kinetic and inhibition constants for saporin-L1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 2.Polito L, Bortolotti M, Farini V, Battelli MG, Barbieri L, Bolognesi A. Saporin induces multiple death pathways in lymphoma cells with different intensity and timing as compared to ricin. Inter Journ of Biochem and Cell Biol. 2009;41:1055–1061. doi: 10.1016/j.biocel.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: effect on DNA, RNA and poly(A) Nucleic Acids Res. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri L, Valbonesi P, Govoni M, Pession A, Stirpe F. Polynucleotide:adenosine glycosidase activity of saporin-L1: effects on various forms of mammalian DNA. Biochimica et Biophysica Acta. 2000;1480:258–266. doi: 10.1016/s0167-4838(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri L, Valbonesi P, Gorini P, Pession A, Stirpe F. Polynucleotide: adenosine glycosidase activity of saporin-L1 : effect on DNA, RNA and poly(A) Biochem J. 1996;319:507–513. doi: 10.1042/bj3190507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri L, Ciani M, Girbés T, Liu W, VanDamme EJM, Peumans WJ, Stirpe F. Enzymatic activity of toxic and non-toxic type 2 ribosome-inactivating proteins. FEBS Letters. 2004;563:219–222. doi: 10.1016/S0014-5793(04)00286-8. [DOI] [PubMed] [Google Scholar]
- 7.Ferreras JM, Barbieri L, Girbés T, Battelli MG, Rojo MA, Arias FJ, Rocher MA, Soriano F, Mendéz E, Stirpe F. Distribution and properties of major ribosome-inactivating proteins (28 S rRNA N-glycosidases) of the plant Saponaria officinalis L(Caryophyllaceae) Biochimica et Biophysica Acta. 1993;1216:31–42. doi: 10.1016/0167-4781(93)90034-b. [DOI] [PubMed] [Google Scholar]
- 8.Roday S, Amukele T, Evans GB, Tyler PC, Furneaux RH, Schramm VL. Inhibition of Ricin A-Chain with Pyrrolidine Mimics of the Oxacarbenium Ion Transition State. Biochemistry. 2004;43:4923–33. doi: 10.1021/bi0498499. [DOI] [PubMed] [Google Scholar]
- 9.Chen XY, Berti PJ, Schramm VL. Transition-State Analysis for Depurination of DNA by Ricin A-Chain. J Am Chem Soc. 2000;122:6527–6534. [Google Scholar]
- 10.Chen XY, Berti PJ, Schramm VL. Ricin A-Chain: Kinetic Isotope Effects and Transition State Structure with Stem-Loop RNA. J Am Chem Soc. 2000;122:1609–1617. [Google Scholar]
- 11.Schnell R, Borchmann P, Staak JO, Schindler J, Ghetie V, Vitetta ES, Engert A. Clinical evaluation of ricin A-chain immunotoxins in patients with Hodgkin's lymphoma. Annals of Oncology. 2003;14:729–736. doi: 10.1093/annonc/mdg209. [DOI] [PubMed] [Google Scholar]
- 12.Baluna R, Vitetta ES. Vascular leak syndrome: A side effect of immunotherapy. Immunopharmacology. 1997;37:117–132. doi: 10.1016/s0162-3109(97)00041-6. [DOI] [PubMed] [Google Scholar]
- 13.Sturm MB, Schramm VL. Detecting ricin: sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. Femtomolar transition state analogue inhibitors of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 15.Angelis FD, Tullio AD, Spanò L, Tucci A. Mass spectrometric study of different isoforms of the plant toxin saporin. Journal of Mass Spectrometry. 2001;36:1237–1239. doi: 10.1002/jms.239. [DOI] [PubMed] [Google Scholar]
- 16.Sturm MB, Roday S, Schramm VL. Circular DNA and DNA/RNA Hybrid Molecules as Scaffolds for Ricin Inhibitor Design. J Am Chem Soc. 2007;129:5544–5550. doi: 10.1021/ja068054h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–84. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen XY, Link TM, Schramm VL. Ricin A-Chain: Kinetics, Mechanism, and RNA Stem-Loop Inhibitors. Biochemistry. 1998;37:11605–11613. doi: 10.1021/bi980990p. [DOI] [PubMed] [Google Scholar]
- 19.Tang S, Hu R, Liu W, Ruan K. Non-Specific Depurination Activity of Saporin-S6, a Ribosome-Inactivating Protein, under Acidic Conditions. Biol Chem. 2000;381:769–772. doi: 10.1515/BC.2000.098. [DOI] [PubMed] [Google Scholar]
- 20.Barbieri L, Ferreras JM, Barraco A, Ricci P, Stirpe F. Some ribosome-inactivating proteins depurinate ribosomal RNA at multiple sites. Biochem J. 1992;286:1–4. doi: 10.1042/bj2860001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vater CA, Bartle LM, Leszyk JD, Lambert JM, Goldmacher VS. Ricin A chain can be chemically cross-linked to the mammalian ribosomal proteins L9 and L10e. J Biol Chem. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 22.Chan DS, Chu LO, Lee KM, Too PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amukele TK, Schramm VL. Ricin A-chain substrate specificity in RNA, DNA, and hybrid stem-loop structures. Biochemistry. 2004;43:4913–4922. doi: 10.1021/bi0498508. [DOI] [PubMed] [Google Scholar]
- 24.Correll CC, Munishkin A, Chan YL, Ren Z, Wool IG, Steitz TA. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc Natl Acad Sci USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jucker FM, Heus HA, Yip PF, Moors EH, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 26.Hale M. Microtiter-Based Assay for Evaluating the Biological Activity of Ribosome-Inactivating Proteins. Pharmacol Toxicol. 2001;88:255–260. [PubMed] [Google Scholar]
- 27.Barbieri L, Bolognesi A, Valbonesi P, Polito L, Olivieri F, Stirpe F. Polynucleotide: Adenosine Glycosidase Activity of Immunotoxins Containing Ribosome-Inactivating Proteins. Journal of Drug Targeting. 2000;8:281–288. doi: 10.3109/10611860008997906. [DOI] [PubMed] [Google Scholar]
- 28.Blakey DC, Skilleter DN, Price RJ, Watson GJ, Hart LI, Newell DR, Thorpe PE. Comparison of the Pharmacokinetics and Hepatotoxic Effects of Saporin and Ricin A-Chain Immunotoxins on Murine Liver Parenchymal Cells. Cancer Research. 1988;48:7072–7078. [PubMed] [Google Scholar]
- 29.Falini B, Bolognesi A, Flenghi L, Tazzari P, Broe M, Stein H, Dtirkop H, Aversa F, Corneli R, Pizzolo G, Barbabietola G, Sabattini E, Pileri S, Martelli M, Stirpe F. Response of refractory Hodgkin's disease to monoclonal anti-CD30 immunotoxin. Lancet. 1992;339:1195–1196. doi: 10.1016/0140-6736(92)91135-u. [DOI] [PubMed] [Google Scholar]
- 30.Polito L, Bolognesi A, Tazzari PL, Farini V, Lubelli C, Zinzani PL, Ricci F, Stirpe F. The conjugate Rituximab/saporin-S6 completely inhibits clonogenic growth of CD20-expressing cells and produces a synergistic toxic effect with Fludarabine. Leukemia. 2004;18:1215–1222. doi: 10.1038/sj.leu.2403378. [DOI] [PubMed] [Google Scholar]
- 31.Baluna R, Rizo J, Gordon BE, Ghetie V, Vitetta ES. Evidence for a structural motif in toxins and interleukin-2 that may be responsible for binding to endothelial cells and initiating vascular leak syndrome. Proc Natl Acad Sci USA. 1999;96:3957–3962. doi: 10.1073/pnas.96.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baluna R, Coleman E, Jones C, Ghetia V, Vitetta ES. The Effect of a Monoclonal Antibody Coupled to Ricin A Chain-Derived Peptides on Endothelial Cells in Vitro: Insights into Toxin-Mediated Vascular Damage Exp. Cell Res. 2000;258:417–424. doi: 10.1006/excr.2000.4954. [DOI] [PubMed] [Google Scholar]
- 33.Smallshaw JE, Ghetie V, Rizo J, Fulmer JR, Trahan LL, Ghetie MA, Vitetta ES. Genetic engineering of an immunotoxin to eliminate pulmonary vascular leak in mice. Nature Biotechnology. 2003;27:387–391. doi: 10.1038/nbt800. [DOI] [PubMed] [Google Scholar]
- 34.Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F. Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: effect on DNA, RNA and poly(A) Nucleic Acids Research. 1997;25:518–522. doi: 10.1093/nar/25.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbieri L, Gorini P, Valbonesi P, Castiglioni P, Stirpe F. Unexpected activity of saporins. Nature (London) 1994;372:624. doi: 10.1038/372624a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.