Summary
S. cerevisiae senses glucose and galactose differently. Glucose is detected through sensors that reside in the cellular plasma membrane. When activated, the sensors initiate a signal transduction cascade that ultimately inactivates the Rgt1 transcriptional repressor by causing degradation of its co-repressors Mth1 and Std1 [1, 2]. This results in expression of many HXT genes encoding glucose transporters [3]. The ensuing flood of glucose into the cell activates Mig1, a transcriptional repressor that mediates ‘glucose repression’ of many genes, including the GAL genes; hence, glucose sensing hinders galactose utilization [4-6]. Galactose is sensed in the cytoplasm via Gal3. Upon binding galactose (and ATP), Gal3 sequesters the Gal80 protein, thereby emancipating the Gal4 transcriptional activator of the GAL genes [7]. Gal4 also activates expression of MTH1 encoding a co-repressor critical for Rgt1 function [8]. Thus, galactose inhibits glucose assimilation by encouraging repression of HXT genes. C. albicans senses glucose similarly to S. cerevisiae, but does not sense galactose through Gal3/Gal80/Gal4 [9]. Its genome harbors no GAL80 orthologue, and the severely truncated CaGal4 does not regulate CaGAL genes [9, 10]. We present evidence that C. albicans senses galactose with its Hgt4 glucose sensor, a capability that is enabled by transcriptional ‘rewiring’ of its sugar-sensing signal transduction pathways (Fig. 1). We suggest that galactose sensing through Hgt4 is ancestral in fungi.
Results and Discussion
Hgt4 affects cell growth and filamentation on galactose
C. albicans Δhgt4 mutants cannot grow on glucose in the presence of the respiration inhibitor antimycin A [11], which forces cells to ferment glucose and demands a high rate of glucose influx. Because galactose and glucose are structurally similar, it seemed plausible that the Hgt4 glucose sensor might sense galactose. Indeed, Δhgt4 cells have a marked growth defect on galactose with antimycin A (Fig. S1A), suggesting that Hgt4 is required for galactose utilization (See Table S3 for strains used in this study). Galactose induces robust filamentation (yeast-to-hyphal morphogenesis) of C. albicans cells, and the Δhgt4 cells are also defective in this response (Fig. S1B). Thus, in the absence of Hgt4, C. albicans cells display growth and morphological defects in galactose.
Galactose and glucose induce expression of the same genes
Expression of 49 genes increased by 2-fold or greater (Table 1, Groups I-III) in response to 2% galactose (compared to glycerol). Most of these galactose-induced genes (40, or 82%) are also significantly induced by 2% glucose (Table 1, Group I). Six of the nine genes that were not induced by 2 % glucose are in fact induced by low glucose levels (<0.2%), but have been shown to be repressed in cells exposed to the high level of glucose used here (Table 1, Group II) [11-15]. Only three genes are modestly induced by galactose but not induced by glucose (Table 1, Group III). Therefore, 94% (46/49) of the characterized genes that are induced in response to galactose are also induced in response to low or high levels of glucose.
Table 1. Genes Induced in Response to Sugars.
| GROUP I | |||||
|---|---|---|---|---|---|
| (Genes induced in 2% galactose and in 2% glucose)* | |||||
| Galactose | Glucose | ||||
| Name | ORF ID | Fold Up | Fold Up | Annotation | |
| HGT7 | orf19.2023 | 32.2 | 30.8 | Putative glucose transporter | |
| QDR1 | orf19.508 | 18.6 | 56.1 | Antibiotic resistance transporter | |
| AOX2 | orf19.4773 | 13.7 | 8 | Alternative oxidase | |
| CRZ2 | orf19.2356 | 10.1 | 12.8 | Putative transcription factor | |
| FET99 | orf19.4212 | 10 | 19.1 | Multicopper oxidase family | |
| RHR2 | orf19.5437 | 9.2 | 25.1 | Putative glycerol 3-phosphatase | |
| RNR22 | orf19.1868 | 8.1 | 25.4 | Ribonucleoside di-Phosphate reductase | |
| TPO3 | orf19.4737 | 7.4 | 16.4 | Possible polyamine transporter | |
| PDC11 | orf19.2877 | 6.2 | 9.1 | Similar to pyruvate decarboxylase | |
| HGT6 | orf19.2020 | 5.3 | 3.2 | Putative glucose transporter | |
| MNN22 | orf19.3803 | 5.3 | 9.1 | Golgi alpha-1, 2-mannosyltxferase | |
| FMA1 | orf19.6837 | 5.2 | 9.9 | Membrane-assoc. protein | |
| HAK1 | orf19.6249 | 5.1 | 3.4 | Putative potassium transporter | |
| HXK2 | orf19.542 | 5 | 8 | Hexokinase II | |
| TYE7 | orf19.4941 | 4.4 | 5.7 | Putative bHLH transcription factor | |
| GDH3 | orf19.4716 | 4.3 | 20.2 | NADP-glutamate dehydrogenase | |
| CMK1 | orf19.5911 | 4.1 | 5 | Ca2+/Calmodulin-dependent kinase | |
| FET34 | orf19.4215 | 3.8 | 6.8 | Similar to multicopper ferroxidase | |
| STP4 | orf19.909 | 3.8 | 5.7 | Putative transcription factor | |
| AOX1 | orf19.4774 | 3.7 | 3.3 | Alternative oxidase | |
| EHT1 | orf19.3040 | 3.4 | 8.4 | Similar to Eht1p | |
| PFK1 | orf19.3967 | 3.2 | 7 | α-subunit of phosphofructokinase | |
| CRP1 | orf19.4784 | 3.2 | 6.9 | Copper transporter | |
| PFK2 | orf19.6540 | 3.1 | 5.7 | β-subunit of phosphofructokinase | |
| PHO15 | orf19.4444 | 3 | 5.8 | 4-nitrophenyl phosphatase | |
| UBC15 | orf19.5337 | 2.9 | 3 | Ub-conjugation, DNA repair | |
| MIG1 | orf19.4318 | 2.9 | 3.2 | Transcriptional repressor | |
| AHP1 | orf19.2762 | 2.9 | 6.3 | Putative alkyl hydroperoxide reductase | |
| ARG1 | orf19.7469 | 2.7 | 4 | Similar to argininosuccinate synthase | |
| PHO113 | orf19.2619 | 2.6 | 3.6 | Constitutive acid phosphatase | |
| NDE1 | orf19.339 | 2.5 | 2.7 | Putative NADH dehydrogenase | |
| GPX2 | orf19.85 | 2.5 | 2.25 | Similar to glutathione peroxidase | |
| ROD1 | orf19.1509 | 2.4 | 3.8 | Drug tolerance; Rgt1-repressed | |
| FCR1 | orf19.6817 | 2.3 | 2.8 | Put. Zn-cluster transcription factor | |
| OPT9 | orf19.2584 | 2.1 | 3.1 | Probable pseudogene | |
| EBP7 | orf19.5816 | 2 | 1.9 | Stress-induced via Cap1p | |
| ARG5 | orf19.4788 | 2 | 5 | Arginine biosynthetic enzyme | |
| ARG4 | orf19.6689 | 2 | 3.9 | Argininosuccinate lyase | |
| DOG1 | orf19.3392 | 1.9 | 2.6 | Put. 2-deoxygluc-6-phosphatase | |
| YIM1 | orf19.847 | 1.9 | 1.8 | Similar to mitochondrial protease | |
| GROUP II | |||||
| (Genes induced in 2% galactose, but repressed in 2% glucose) | |||||
| Galactose | Glucose | ||||
| Name | ORF ID | Fold Up | Fold Up | Annotation | |
| HGT12 | orf19.7094 | 6.74 | 0.07 | Glucose, fructose, mannose transporter | |
| HXT10 | orf19.4384 | 2.6 | 0.21 | Sugar transporter | |
| HGT2 | orf19.3668 | 2 | 0.02 | Putative glucose transporter | |
| GAL1 | orf19.3670 | 6.1 | 0.8 | Galactokinase | |
| GAL10 | orf19.3672 | 6 | 0.7 | UDP-glucose 4-epimerase | |
| GAL7 | orf19.3675 | 5.9 | 0.9 | UDP-hexose-1-P uridylyltransferase | |
| GROUP III | |||||
| (Genes induced in 2% galactose, NOT induced in 2% glucose) | |||||
| Galactose | Glucose | ||||
| Name | ORF ID | Fold Up | Fold Up | Annotation | |
| PGA37 | orf19.3923 | 3.2 | 1.1 | Putative GPI-anchored protein | |
| HSP30 | orf19.4526 | 2.9 | 0.48 | Similar To heat shock protein | |
| STB3 | orf19.203 | 2.3 | 1.4 | Predicted Sin3 Binding protein | |
Fold induction indicates gene expression in each sugar relative to its expression in glycerol cultures. For all genes listed, p<0.05 (student's t-test). Uncharacterized open reading frames induced in galactose and glucose are shown in Table S4.
Hgt4 affects the transcriptional response to galactose
Expression of five of the top genes listed in Table 1 (Group I) was re-examined by RT-PCR analysis. In cells grown on glycerol, these genes are either not expressed (HGT7, QDR1, AOX2) or expressed at low levels (CMK1, HXK2), and all five are induced in response to galactose in an Hgt4-dependent manner (Fig. 2). HGT12, encoding a glucose transporter related to Hgt4 [11, 16], does not affect the expression of these genes. Induction of GAL1 expression by galactose is significantly diminished in the Δhgt4 mutant (Fig. S2), consistent with the previous observation that the Hgt4 signal increases GAL1 and GAL7 expression two-fold [11, 12]. Galactose still induces GAL1 expression in Δhgt4 cells, indicating another signaling pathway contributes to GAL1 expression, possibly by acting upon Cph1 (a C. albicans homologue of S. cerevisiae Ste12) [9].
Figure 2. Hgt4 regulates galactose-induced genes.
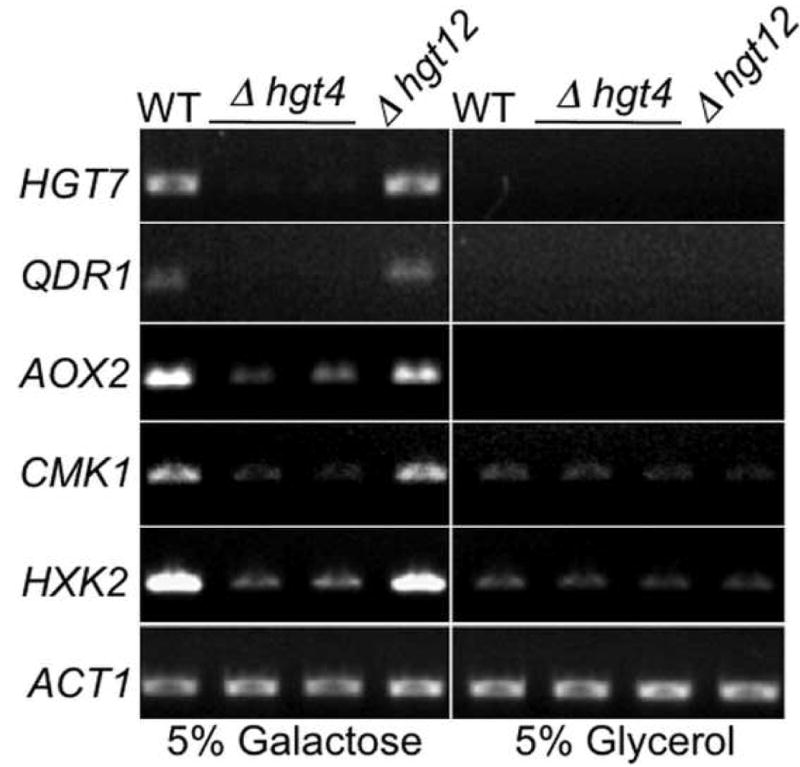
Log phase cultures of WT (BWP17), Δhgt4 (CM9 and CM10), or Δhgt12 (CM64) cells were split and incubated in fresh media containing 5% galactose or 5% glycerol at 30°C for 2 hours. Total RNA was reverse transcribed and PCR amplified with primers for HGT7 (orf19.2023), QDR1 (orf19.508), AOX2 (orf19.4773), CMK1 (orf19.5911), HXK2 (orf19.542), and ACT1 (orf19.5007). Control reactions lacking reverse transcriptase yielded no products (not shown).
The CaHGT7 gene, encoding a hexose transporter, is highly induced – over 30-fold – by both galactose and glucose (Table 1). HGT7 expression is activated by low levels (0.04%) of glucose, fructose, or mannose (Fig. 3A, top), and by a high level (1.6%) of galactose (Fig. 3A, bottom). HGT7 expression in response to sugars is entirely dependent on HGT4 (Fig. 3B), and Hgt4 mediates the dose-dependent galactose induction of HGT7 expression at concentrations as low as 0.6% (Fig. S3).
Figure 3. HGT7 is induced in response to galactose.
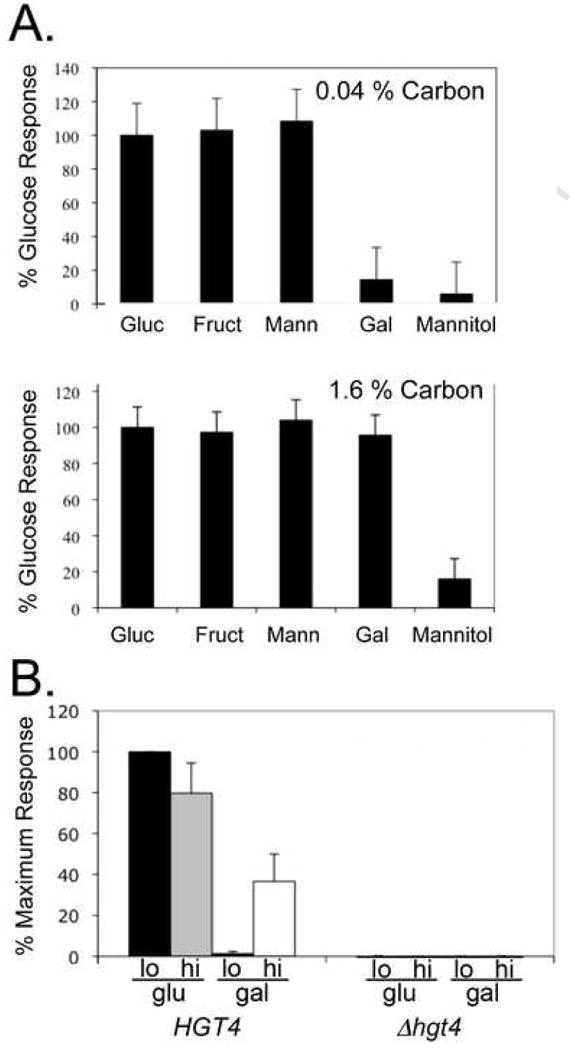
(A) The HGT7 promoter was fused to Streptococcus thermophilus lacZ gene, and this construct was integrated into the C. albicans genome at the native HGT7 locus. Cells with HGT7::HGT7-lacZ (CM79 and CM80) were grown in glycerol media, split, and incubated at 30°C for 2 hours in fresh media containing glycerol or 0.04% (top panel) or 1.6% (bottom panel) of the sugars indicated. Cells were lysed, assayed for β-galactosidase activity (in quadruplicate (top) or in triplicate (bottom)), and the results were normalized to the lacZ activity in the glycerol media. Data are presented as the mean +/- one standard deviation. (B) Cells with HIS1::HGT7-lacZ (HGT4; CM230 and CM 231), (Δhgt4; CM232 and CM233) were grown in media with glycerol as carbon source, split, and incubated at 30°C for 2 hours in fresh media lacking histidine but containing glycerol or the sugar indicated, (n=10 for HGT4; n=10 for Δhgt4). Black bar: 0.04% glucose; grey bar: 1.6% glucose; striped bar: 0.04% galactose; white bar: 1.6% galactose. All values were normalized to activity in glycerol, and expressed as the percent of the maximum response in 0.04% glucose. Data are the mean +/- one standard deviation.
Galactose-induced genes have Rgt1-binding sites
Of the 50 genes most highly induced by galactose, 34 of them (68%) contain at least one consensus Rgt1 consensus DNA-binding motif (5′-CGGANNA-3′) within 1 kilobase upstream of the translational start codon (Table S1). This is a significant enrichment (p<10-3) — only 46% of promoters genome-wide harbor an Rgt1 motif — that is similar to the enrichment of consensus Rgt1-binding sites upstream of genes regulated by glucose via Hgt4 and CaRgt1 (66%, p<10-5) (See Experimental Procedures in Supplementary Material for statistical methods). The CaCph1 transcription factor has also been implicated in the expression of the CaGAL genes in response to galactose [9], but its binding-site is not enriched in other galactose-induced genes (Table S1). The promoters of the GAL1-10 and GAL7 genes (encoding the enzymes for galactose metabolism) each contain both a perfect Cph1 response element (5′-TGTAACGTTACA-3′) [9] and two Rgt1 recognition sequence motifs, consistent with the idea that Rgt1 and Cph1 coordinately regulate these genes in response to galactose.
Hgt4 senses galactose in S. cerevisiae
In S. cerevisiae, HXT genes are induced by glucose, fructose, and mannose, but not by galactose, ostensibly because Snf3 and Rgt2 do not bind galactose [17]. If Hgt4 binds galactose, then expressing it in S. cerevisiae should cause galactose-induction of HXT genes. The HGT4 sugar-binding domain (codon optimized) was expressed in S. cerevisiae from the RGT2 promoter (see Experimental Procedures in Supplementary Material). Because the C-terminal cytoplasmic tails of the glucose sensors have diverged almost completely in the ∼200 million years since the C. albicans and S. cerevisiae lineages diverged, the Hgt4 sugar-binding domain was fused to the Rgt2 tail to enable coupling of the sensor to the S. cerevisiae signal transduction pathway (Fig. S4) [11]. Exchanging the intracellular signaling tails of glucose sensors does not affect their response to glucose (V. Brown unpublished data; V. Brachet, unpublished data), so we are confident that the Hgt4-Rgt2 chimera retains the sugar-sensing specificity of Hgt4. In S. cerevisiae cells expressing the Hgt4 chimera, HXT1 is not induced by galactose (Fig. 4A, black bars; Fig. S5, third row). However, galactose induces MTH1 expression in S. cerevisiae via Gal4 [8], and the resulting increase in Mth1 levels would be expected to reinforce Rgt1-mediated repression of HXT1, effectively masking any galactose signal generated by Hgt4 in S. cerevisiae. Deleting ScGAL4 eliminates this control element, and reveals robust activation of the HXT1-lacZ reporter in response to galactose in cells expressing the Hgt4 chimera (Fig. 4A, blue bars; Fig. S5, bottom row) [18]. In contrast, neither Rgt2 nor Snf3 (which are present in these strains) respond to galactose (indicated by cells with the vector control, Fig. 4A, grey bars; Fig. S5, first column). Thus, expression of the Hgt4 sugar-binding domain in S. cerevisiae confers a novel galactose response upon baker's yeast.
Figure 4. C. albicans HGT4 confers a novel galactose-response upon S. cerevisiae.

(A) S. cerevisiae strains were grown in media containing glycerol, cell densities were normalized, and the culture was split and incubated overnight at 30°C in fresh media containing 5% glycerol or 5% galactose, then lysed and β-galactosidase activity was assayed (Experimental Procedures in Supplementary Material). Data are the average of biological duplicates. White bars: wild-type cells + pRS316 vector (YM7642); black bars: wild-type cells +Hgt4-Chimera (YM7643); grey bars: Δgal4 cells + pRS316 vector (YM7644); blue bars: Δgal4 cells + Hgt4-Chimera (YM7645). (B) MTH1 orthologues are galactose-induced in diverse fungi. A phylogenetic tree showing the relationship of yeasts spanning ∼200 million years of evolution is shown [42-45]. Characteristics of the galactose-sensing pathways in these species are described in Table S2. The black circle represents a whole genome duplication event, the white circle represents the proposed appearance of the Gal4-Gal80 gene regulatory mechanism; asterisks indicate the species analyzed in (C). (C) Each species was grown overnight in glycerol media, and incubated in fresh media containing 5% glycerol or 5% galactose at 30°C for 3 hours. RT-PCR was performed on total RNA using species-specific primers for either ACT1 or the MTH1/STD1 orthologue (Fungal strains are described in Experimental Procedures Supplementary Material). First strand cDNAs served as templates for quantitative PCR. Each MTH1/STD1 signal was normalized to the ACT1 signal in that sample, and the ΔΔCt values are expressed as ‘Fold Induction’ of expression in galactose relative to expression in glycerol (2ΔΔCt). Data are the average of duplicates. Separate experiments were performed using semi-quantitative PCR to confirm the results (see Fig. S6).
Galactose induces MTH1 expression in diverse fungi
C. albicans did not undergo a whole genome duplication, so it has one homologue of the S. cerevisiae MTH1 and STD1 paralogues (CaSTD1). CaStd1 (orf19.6173), is 27% identical to both the S. cerevisiae Std1 (43% similar) and Mth1 (41% similar), and harbors a conserved motif (SxSxxSSIFS, residues 62-71) that is critical for glucose-induced phosphorylation of ScStd1 and ScMth1 (which leads to their degradation) [19]. We surmised that since Hgt4 functions as a galactose sensor in C. albicans, CaSTD1 expression must not be induced by galactose. Indeed, CaSTD1 expression is unaffected by galactose (Table 1 and data not shown), a result confirmed by RT-PCR and RT-qPCR analyses (Figs. S6 and 4C respectively). To assess the evolutionary conservation of this galactose response, we measured expression of MTH1 orthologues in a diverse sampling of fungi spanning ∼200 million years of evolution (Fig. 4B). In all species tested except C. albicans, expression of MTH1 is induced in response to galactose (Fig. 4C and S6). Induction occurs even in C. glabrata, which has lost the GAL4 gene, as well as in K. lactis, which lacks canonical Gal4 binding sites in the promoter of its MTH1 orthologue (Table S2). Galactose-induced activation of ScMTH1 expression by ScGal4 in S. cerevisiae [8] appears to antagonize the galactose signal generated by Hgt4, and such antagonism is likely in the four fungi in the S. cerevisiae to K. lactis clade that we analyzed.
These data illuminate the evolution of galactose sensing in fungi. Sensing galactose through both the Gal4 and the Hgt4/Snf3/Rgt2 pathways seems imprudent because it would lead to cross-repression of genes in both pathways (see Summary and Fig. 1). Within the Ascomycetes, Candida glabrata, Kluyveromyces waltii, and Ashbya gossypii have no canonical galactose sensor because GAL4, or GAL80, or both are absent, but they have also lost all galactose utilization pathway enzymes (GAL1, GAL7, and GAL10), and thus cannot utilize galactose in any case [10, 20, 21]. The Gal4-mediated galactose-sensing pathway is intact in a few yeasts that diverged before the duplication, such as K. lactis and S. kluyveri [22-24]. Debaromyces hansenii and Pichia stipitus have GAL4 homologues, but no obvious GAL80 homologues. In contrast, all the Candida species we surveyed (except C. glabrata), as well as Yarrowia lipolytica, and Lodderomyces elongisporus, harbor genes encoding the enzymes for galactose metabolism, but their GAL4 genes are more similar to CaGAL4 (than ScGAL4), and they all lack a GAL80 functional homologue. The implication is that the Ascomycetes that can metabolize galactose, but have no Gal4 or Gal80 regulators, utilize an Hgt4-like sensing pathway to control galactose-response genes. This supports the notion that the Gal4-Gal80 control circuit arose prior to the origin of the S. cerevisiae – K. lactis clade, but after this clade and Candida species diverged from their common ancestor (Fig. 4B, white dot), and suggests that Hgt4 represents an ancestral sensor of galactose. In C. albicans, the altered specificity of the Hgt4 glucose sensor in combination with the absence a canonical Gal4 pathway has enabled this fungus to sense galactose through Hgt4.
Figure 1. Sugar sensing pathways in C. albicans and S. cerevisiae.

Glucose signaling begins at the cell surface with the sensors (CaHgt4, or ScSnf3 and ScRgt2), and ends in the nucleus with deactivation of the Rgt1 transcriptional repressor [1, 11]. The keystone proteins are the transcriptional co-repressors (CaStd1, or ScStd1 and ScMth1), which associate with both the sensor and the transcriptional repressor, and it is the levels of these proteins that translate the environmental signal into gene expression changes. Sugar binding to a sensor activates yeast casein kinase (Yck), which then phosphorylates Std1 and Mth1, thereby marking them for ubiquitylation by the SCFGrr1 complex, and dooming them to destruction by the proteasome. Depletion of the co-repressors renders Rgt1 impotent, which results in transcriptional derepression of downstream genes. In S. cerevisiae, galactose enters the cell, is phosphorylated and binds (with ATP) to the Gal3 protein. This complex binds and sequesters Gal80, and relieves the inhibition of the Gal4 transcriptional activator. In C. albicans, CaGal4 does not regulate the GAL genes. Instead, galactose is sensed by the Hgt4 glucose sensor, and likely also through Cph1 (a homologue of the S. cerevisiae Ste12).
C. albicans Std1 functions in the sugar sensing pathway
Since the Gal4 signaling pathway is structured differently in C. albicans, it was possible that the Hgt4 pathway had also changed. If sugar sensing by C. albicans is analogous to S. cerevisiae glucose-sensing, the CaStd1 co-repressor should be a key protein in the pathway (Fig. 1). Indeed it is, because homozygous Δstd1 null mutant cells have the same hyper-filamented morphology as Δrgt1 mutant cells (Fig. S7A), and as cells carrying the constitutively signaling HGT4-1 mutation (Fig. S7B), and this phenotype is reversed by reintroducing one wild-type allele into these cells (Fig. S7B). This result supports previous observations that implicated the Hgt4 pathway in C. albicans filamentation [11, 12]. Further, HGT7 expression is constitutive in the Δstd1 mutant (just like in the HGT4-1 mutant), and reintroducing one copy of CaSTD1 into this mutant reverses this phenotype (Fig. 5A). Thus, although the galactose-sensing pathways are completely different between C. albicans and S. cerevisiae, the glucose-sensing pathway remains the same (Hgt4-CaStd1-CaRgt1).
Figure 5. CaSTD1 and ScSTD1 function similarly.
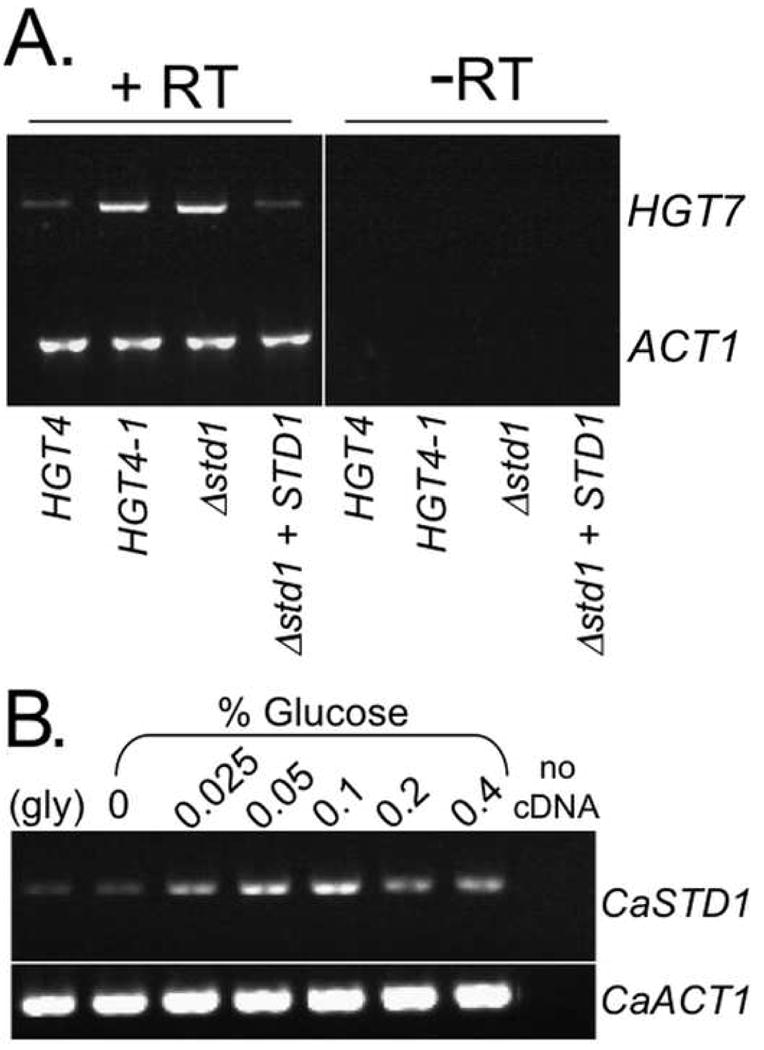
(A) CaSTD1 plays a role in the HGT4 pathway. Isogenic strains [HGT4 (CM87) vs. HGT4-1 (CM36) and Δstd1 (CM222) vs. STD1 (CM224)] were grown at 30°C to log phase in media containing glycerol. Cells were harvested, snap frozen, and total RNA was purified for RT-PCR analysis of HGT7 (orf19.2023) or ACT1 (orf19.5007). (B) CaSTD1 is glucose-induced. C. albicans cells (SC5314) were grown to log-phase in media containing glycerol, then incubated at 30°C for 2 hours in fresh media with glycerol (gly), or with the indicated concentrations of glucose (0 indicates no carbon source). Cells were harvested, snap frozen, and total RNA was purified for RT-PCR analysis using primers for CaSTD1 (orf19.6173) or ACT1 (orf19.5007).
Examining CaStd1 function in C. albicans sheds light on the separate functions of the S. cerevisiae paralogues. CaSTD1 expression resembles that of ScSTD1, not ScMTH1: it is induced by glucose but not by galactose (Figs. 5B and 4C respectively) [25]. This implies that ScStd1 has a more ancestral role, and ScMth1 a more derived role, in this signal transduction pathway. The Hgt4/Snf3/Rgt2 sugar sensing pathway may be universally involved in fungal morphology: disrupting ScMTH1 represses filamentous growth in baker's yeast in the Σ1278b pseudohyphal strain [26]. Further studies on pre- and post-duplication yeast species will be necessary to determine whether Mth1 and Std1 function redundantly, cooperatively, or in opposition to each other, and whether they affect fungal morphogenesis throughout this kingdom.
It seems clear that the glucose and galactose sensing systems in fungi work together as a network to regulate transcription of genes such as GAL1 in C. albicans and HXT1 in S. cerevisiae. In fact, transcriptional regulation of the HXT genes in S. cerevisiae is the result of at least seven interconnected signal transduction cascades: (I.) glucose-sensing through Snf3/Rgt2 [27], (II.) sugar sensing through the Gpr1 G-protein coupled receptor [28], (III.) osmo-sensing through the Hog1 MAP kinase pathway [29], (IV.) glucose repression mediated by Mig1 and Mig2 [6], (V.) the TOR1 protein kinase pathway [30], (VI.) oxygen availability [31], and finally (VII.) galactose-sensing through Gal4. These signal transduction pathways provide a malleable framework for responding to extracellular nutrients.
Supplementary Material
Acknowledgments
We thank Chris Todd Hittinger and Chandra Tucker for critical reading of this manuscript, and Jim Dover and Christine Carle for technical assistance. This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (K01DK077878 to V.B.), and by grants from the National Institute of General Medical Sciences (F32GM076967 to J.S., and R01GM32540 to M.J.), and by funds provided to M.J. by the James S. McDonnell Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnston M, Kim JH. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:247–252. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- 2.Santangelo GM. Glucose Signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston M, Flick JS, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Vit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 9.Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun BR, van Het Hoog M, d'Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, et al. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1:36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown V, Sexton JA, Johnston M. A glucose sensor in Candida albicans. Eukaryot Cell. 2006;5:1726–1737. doi: 10.1128/EC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sexton JA, Brown V, Johnston M. Regulation of sugar transport and metabolism by the Candida albicans Rgt1 transcriptional repressor. Yeast. 2007;24:847–860. doi: 10.1002/yea.1514. [DOI] [PubMed] [Google Scholar]
- 13.Murad AM, d'Enfert C, Gaillardin C, Tournu H, Tekaia F, Talibi D, Marechal D, Marchais V, Cottin J, Brown AJ. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol Microbiol. 2001;42:981–993. doi: 10.1046/j.1365-2958.2001.02713.x. [DOI] [PubMed] [Google Scholar]
- 14.Singh V, Satheesh SV, Raghavendra ML, Sadhale PP. The key enzyme in galactose metabolism, UDP-galactose-4-epimerase, affects cell-wall integrity and morphology in Candida albicans even in the absence of galactose. Fungal Genet Biol. 2007;44:563–574. doi: 10.1016/j.fgb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Fan J, Chaturvedi V, Shen SH. Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J Mol Evol. 2002;55:336–346. doi: 10.1007/s00239-002-2330-4. [DOI] [PubMed] [Google Scholar]
- 16.Luo L, Tong X, Farley PC. The Candida albicans gene HGT12 (orf19.7094) encodes a hexose transporter. FEMS Immunol Med Microbiol. 2007;51:14–17. doi: 10.1111/j.1574-695X.2007.00274.x. [DOI] [PubMed] [Google Scholar]
- 17.Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly DE, Lamb DC, Kelly SL. Genome-wide generation of yeast gene deletion strains. Comp Funct Genomics. 2001;2:236–242. doi: 10.1002/cfg.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci U S A. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hittinger CT, Rokas A, Carroll SB. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc Natl Acad Sci U S A. 2004;101:14144–14149. doi: 10.1073/pnas.0404319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cliften PF, Fulton RS, Wilson RK, Johnston M. After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics. 2006;172:863–872. doi: 10.1534/genetics.105.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 24.Langkjaer RB, Cliften PF, Johnston M, Piskur J. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature. 2003;421:848–852. doi: 10.1038/nature01419. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Brachet V, Moriya H, Johnston M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:167–173. doi: 10.1128/EC.5.1.167-173.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki C, Hori Y, Kashiwagi Y. Screening and characterization of transposon-insertion mutants in a pseudohyphal strain of Saccharomyces cerevisiae. Yeast. 2003;20:407–415. doi: 10.1002/yea.970. [DOI] [PubMed] [Google Scholar]
- 27.Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem. 2006;281:26144–26149. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- 29.Tomas-Cobos L, Casadome L, Mas G, Sanz P, Posas F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J Biol Chem. 2004;279:22010–22019. doi: 10.1074/jbc.M400609200. [DOI] [PubMed] [Google Scholar]
- 30.Tomas-Cobos L, Viana R, Sanz P. TOR kinase pathway and 14-3-3 proteins regulate glucose-induced expression of HXT1, a yeast low-affinity glucose transporter. Yeast. 2005;22:471–479. doi: 10.1002/yea.1224. [DOI] [PubMed] [Google Scholar]
- 31.Rintala E, Wiebe MG, Tamminen A, Ruohonen L, Penttila M. Transcription of hexose transporters of Saccharomyces cerevisiae is affected by change in oxygen provision. BMC Microbiol. 2008;8:53. doi: 10.1186/1471-2180-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 35.Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spreghini E, Davis DA, Subaran R, Kim M, Mitchell AP. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot Cell. 2003;2:746–755. doi: 10.1128/EC.2.4.746-755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorch Y, Kornberg RD. A region flanking the GAL7 gene and a binding site for GAL4 protein as upstream activating sequences in yeast. J Mol Biol. 1985;186:821–824. doi: 10.1016/0022-2836(85)90400-0. [DOI] [PubMed] [Google Scholar]
- 39.Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 40.Uhl MA, Johnson AD. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology. 2001;147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- 41.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 42.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 44.Scannell DR, Frank AC, Conant GC, Byrne KP, Woolfit M, Wolfe KH. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci U S A. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


