Abstract
MyD88 participates in signal transduction by binding to the cytoplasmic Toll/IL-1 receptor (TIR) domains of activated Toll-like receptors (TLR). Yeast two-hybrid experiments reveal that the TIR domains of human TLR differ in their ability to associate with MyD88: The TIR of TLR2 binds to MyD88 but the TIR of the closely related TLR1, 6, or 10 do not. Using chimeric TIR domains, we define the critical region responsible for differential MyD88 binding, and use a computational analysis of the critical region to reveal the amino acids that differ between MyD88 binders and non-binders. Remarkably, a single missense mutation created in TLR1 (N672D) confers on it the ability to bind MyD88, without affecting its association with other proteins. Mutations identified as critical for MyD88 binding also affect signaling of TLR pairs in mammalian cells. To investigate the difference between MyD88 binders and non-binders,we identify novel interacting proteins for each cytoplasmic domain of TLR1, 2, 6, and 10. For example, heat shock protein (HSP) 60 binds to TLR1 but not to TLR2, and HSP60 and MyD88 appear to bind the same region of the TIR domain. In summary, interactions between the TLR, MyD88, and novel associated proteins have been characterized.
Keywords: Gain-of-function, Protein interaction, Toll-like receptor
Introduction
Innate immune cells express Toll-like receptors (TLR) that detect microbial antigens and other 'danger' signals and activate a conserved innate immune response [1]. Ten TLR are encoded in the human genome, and each recognizes unique ligands that bind to TLR extracellular domains. The resulting oligomerization of receptors activates intracellular signaling [2].
For all TLR except TLR3, the initial event in signal transduction is recruitment of the MyD88 to the activated receptor. Signaling by a TLR culminates in activation of genes involved in the inflammatory response, but the transcriptional responses to the different activated TLR differ. TLR3, 4, 5, 7, 8, and 9 can each signal as homo-oligomers, but TLR1, 2, 6 (and, it is presumed, the closely related TLR10) cannot [3, 4]. Tethering of TLR intracellular domains to other proteins that dimerize, such as the extracellular domains of the cluster of differentiation antigen 4 (CD4) or the integrin receptor, has allowed signal transduction to be assessed without the variable of efficiency of ligand recognition [4]. In such experiments, homodimerized cytoplasmic domains of TLR1, 2, and 6 do not activate nuclear factor kappa B (NF-κB), but TLR2 in combination with either TLR1 or 6 efficiently signals through NF-κB. Thus, TLR2 appears to require the assistance of either TLR1 or 6 to signal [4–6]. These observations suggest that close juxtaposition of particular Toll/IL-1 receptor (TIR) domains is sufficient to recruit MyD88 [7].
The intracellular domains of TIR have similar sequences, but are not functionally identical [8]. TIR domains all share common sequence motifs, termed Box 1, Box 2, and Box 3. Boxes 1 and 2 are critical for MyD88-dependent signaling. The crystal structures of the TIR domains from TLR1 and 2 reveal that a conserved proline critical for MyD88 signaling (TIR2 P681) lies in the apparently flexible 'BB' loop of Box 2 [9]. MyD88 binds to activated receptors [10, 11] and recruits the IRAK Ser/Thr protein kinase, the TRAF6 ubiquitin-protein ligase and the TGFβ-activated protein kinase to the activated TLR complex. A subsequent series of phosphorylation and ubiquitination steps culminates in activation of the NF-κB, p38 and AP-1 transcription factors, which results in transcription of genes involved in the inflammatory response [12]. Other proteins that associate with TLR intracellular domains, such as the MyD88 adapter-like/TIR domain-containing adapter protein (MAL/TIRAP), likely confer an additional level of specificity to the innate immune response [13, 14]. TLR loss-of-function mutations have been identified that affect many different sites in the TIR domain [15], but the determinants of the TLR-MyD88 interaction are far from clear. All TLR (except TLR3), as well as IL receptors 1 and 18, require MyD88 for function. MyD88 knockout mice predictably suffer from increased susceptibility to bacterial and viral infections but, surprisingly, have a reduced capacity for adjuvant-mediated tumor regression, and also show a decreased level of atherosclerosis in response to elevated serum lipids [16, 17]. Because of its central role in inflammation, MyD88 has been suggested as a 'drugable' target for modulating inflammation in human disease [18]. An ultimate goal would be the specific modulation of certain MyD88-dependent signaling pathways without destroying (or triggering) the entire Toll and IL receptor signal network. Before this goal can be achieved, the specific protein-protein interactions involving MyD88 must be fully understood.
Saccharomyces cerevisiae provides an excellent platform for studying interactions of proteins with TLR because it possesses no homologues of the TLR or their adaptors. Using the yeast two-hybrid method, we show that only a few TLR cytoplasmic domains bind MyD88, and we define amino acids that are responsible for the differential binding of TLR TIR domains to MyD88.
Results
TLR TIR2 directly binds MyD88
We fused the yeast Gal4 DNA-binding domain (BD) to the complete cytoplasmic domain of each of the ten human TLR. Because the Gal4 BD binds to DNA as a dimer, the BD-TLR fusions are forced homodimers. Although the spatial arrangement of the TLR domains in the Gal4-DNA complex is unknown, we presume that dimerization of the Gal4 BD closely juxtaposes the TLR cytoplasmic domains. Such close association is sufficient for signaling in mammalian cells, as observed when the extracellular domain of CD4 unites the TLR cytoplasmic domains [4]. In contrast, the Gal4 transcriptional activation domain (AD) does not force the dimerization of AD-TLR fusion proteins.
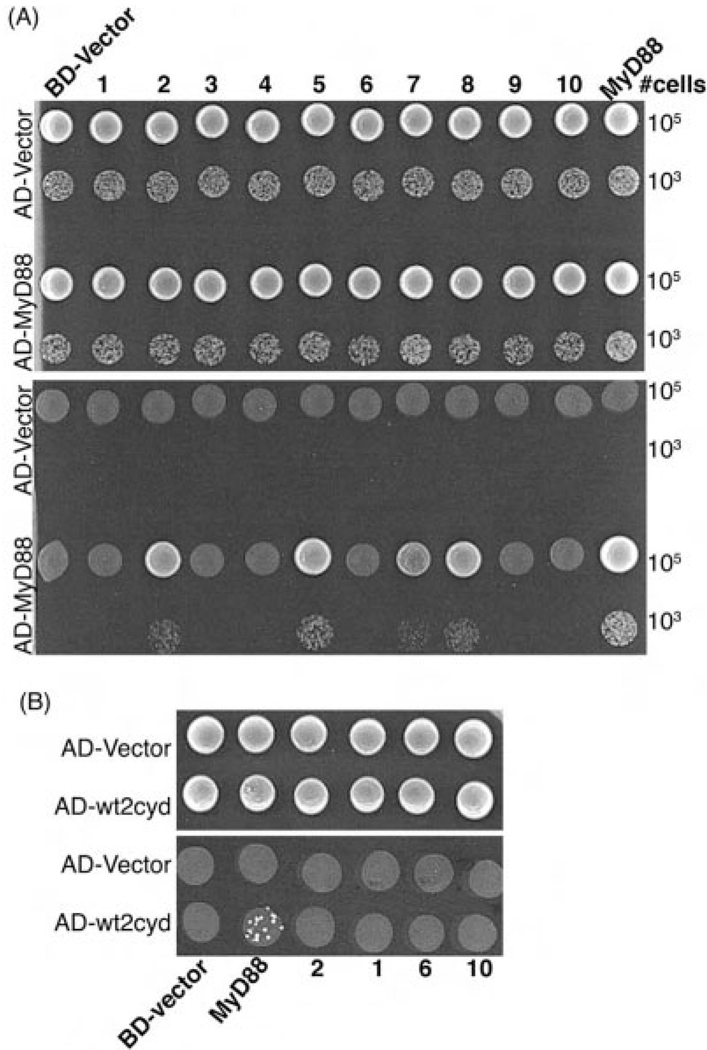
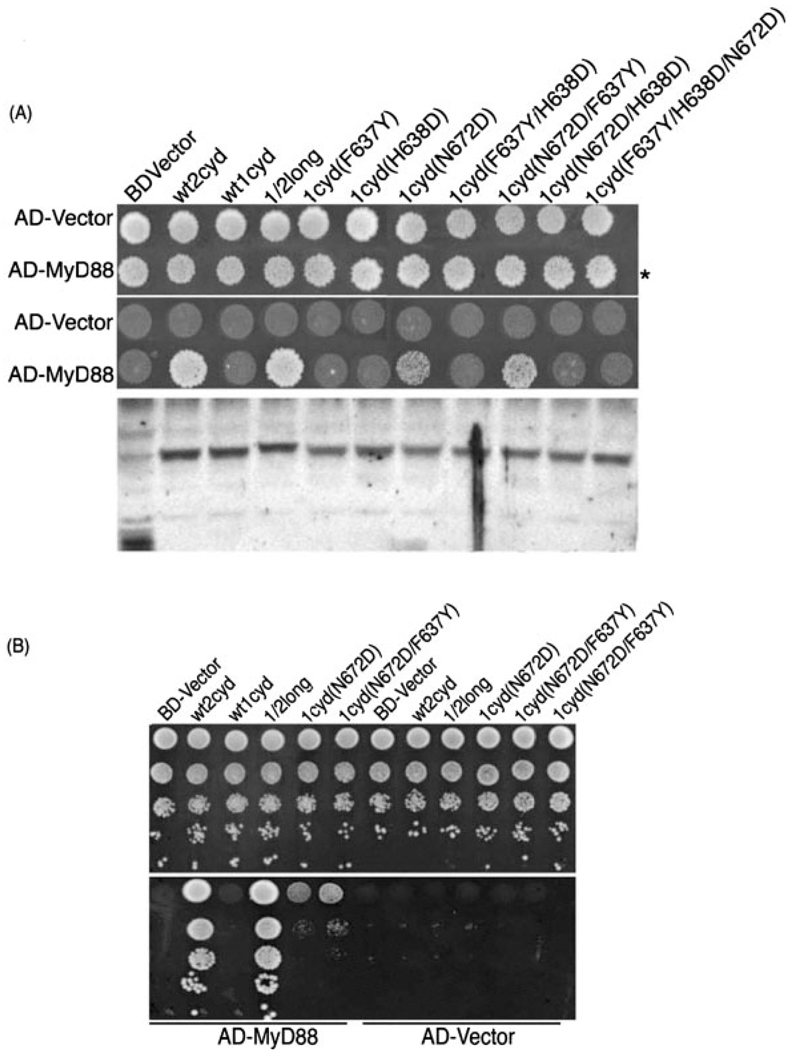
The BD-TLR were tested for binding to full-length human MyD88 protein (AD-MyD88) (Fig. 1A). MyD88 homodimerizes [7] and served as the positive control: BD-MyD88 stimulated robust growth of yeast cells that also express AD-MyD88. The BD-TLR homodimers differentially associated with MyD88; TLR2, 5, 7, and 8 bound, but TLR1, 6, and 10 did not (TLR3 does not signal through MyD88, and its cytoplasmic domain did not bind MyD88 in the two-hybrid assay). The cytoplasmic domains of TLR4 and 9 did not bind MyD88, presumably due to either a low level of expression of the fusions or perhaps they require accessory proteins for the interaction. In contrast to what is expected based on signaling data in mammalian cells, the BD-TLR2 cytoplasmic domain bound to AD-MyD88 in the absence of TLR1 or 6. TLR2-MyD88 interaction also occurred in the reciprocal format, when the AD-TLR2 cytoplasmic domain was not a 'forced' dimer. This result could reflect weak dimerization of the TLR2 cytoplasmic domain, allowing it to bind MyD88 (although no signal for TLR2 dimerization in the assay was observed), or it could reflect a weak interaction between the monomeric AD-TLR2 cytoplasmic domain and MyD88 (Fig. 1B). In any case, the results show that, regardless of signaling capacity, the dimerized TLR2 cytoplasmic domain is capable of directly binding MyD88.
Figure 1.
(A) MyD88 binds to the TLR cytoplasmic domains 2, 5, 7, and 8. Yeast were transformed with the Gal4 BD fusions indicated (along top) and the Gal4 AD fusions indicated (at left). Transformants were selected on synthetic media lacking tryptophan and leucine (−TL, top panel), and protein interactions were identified by plating transformants on synthetic media lacking tryptophan, leucine, and histidine, and supplemented with 10 mM 3AT media (−TLH/+10 mM 3AT, bottom panel). (B) The reciprocal TLR2-MyD88 interaction. Cells expressed the BD fusions indicated (along bottom) and the AD fusions indicated (at left). Transformants were selected on −TL media (top panel) and on −TLH/+10 mM 3AT (bottom panel).
A conserved proline is necessary but not sufficient for MyD88 binding
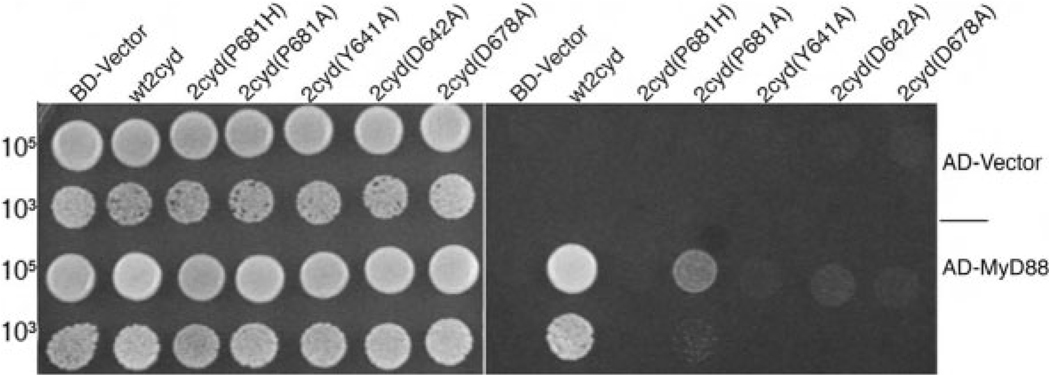
The MyD88 non-binders TLR1, 6, and 10 are closely related to the MyD88 binder TLR2 [19] (Fig. 2A). This observation prompted us to attempt to identify the specific amino acid residues that account for the difference in MyD88 binding. The conserved proline in Box 2 of the TIR domain (TLR1 P675 or TLR2 P681; Fig. 2B) is necessary for signaling: The P712H missense in TLR4 renders lpsd mice hyporesponsive to endotoxin, and the analogous P681H mutation in TLR2 is a dominant-negative mutation that abolishes wild-type TLR2 signaling in mammalian cells [20]. This mutation is thought to render the TIR domains unable to recruit MyD88 [21], and this was recapitulated in the yeast two-hybrid assay: A P681 mutation abrogated the TLR2 cytoplasmic domain-MyD88 two-hybrid interaction, as did mutation of other highly conserved residues in the TIR domain (Fig. 3). However, the TLR cytoplasmic domains 1, 6, and 10 have retained this conserved proline, and thus other amino acid differences in this subset of TLR must account for the differential MyD88 binding activity.
Figure 2.
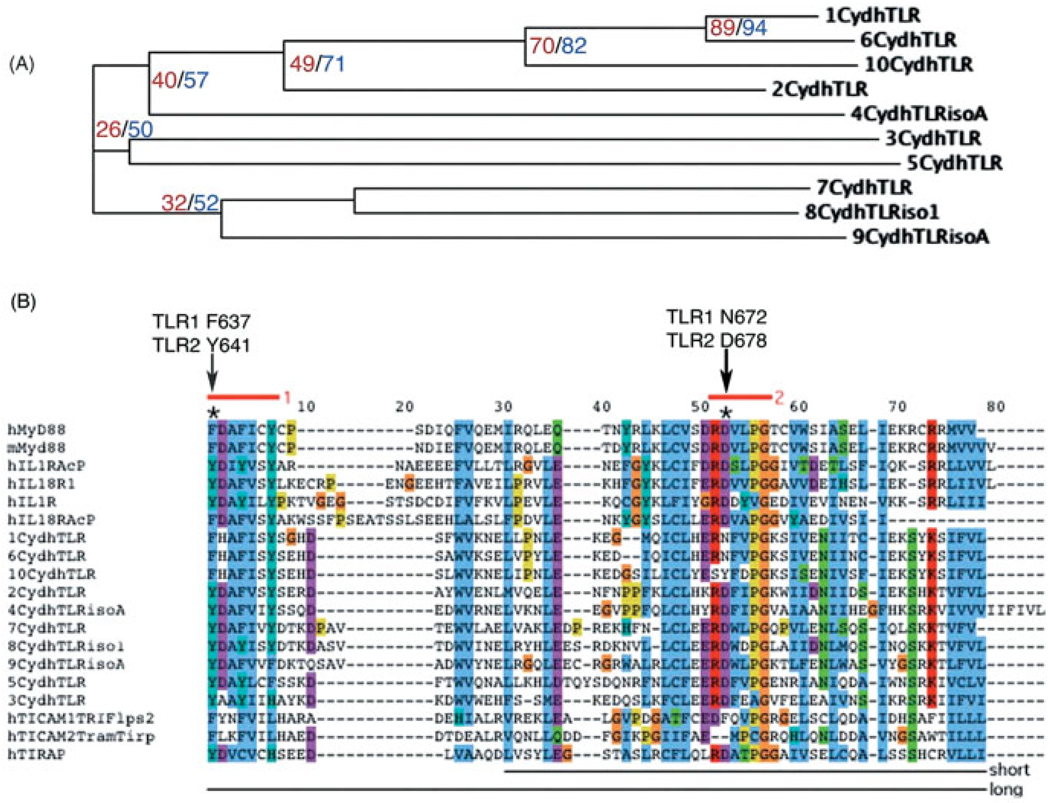
(A) ClustalW alignment of the human TLR cytoplasmic domains (Cyd) showing the closely related MyD88 non-binders 1, 6, and 10, and the next nearest relative, TLR2, which binds MyD88. Numbers in red indicate percent identity, and in blue indicate percent similarity between the TLR1 Cyd and the other TLR cytoplasmic domains. (B) Protein alignment of the TIR region that converts the cytoplasmic domains of TLR1 and 6 into MyD88-binding proteins. Boxes 1 and 2 (red bars) are highly conserved hallmarks of the TIR domain and are critical for receptor signaling. 'Short' or 'long' bars indicate regions of TLR2 or 5 swapped into TLR1, 6, and 10 to generate cytoplasmic domain chimeras. Asterisks at the top indicate positions of gain-of-function point mutations in TLR1, and analogous residues in TLR2.
Figure 3.
Point mutations in the TLR2 cytoplasmic domain abrogate MyD88 binding. Yeast co-transformed with the AD fusions indicated (at right) and the BD fusions indicated (along top) were selected on −TL media (left panel) and on −TLH/+3 mM3AT media (right panel).
Chimeric TIR domains of TLR1 and TLR2 bind MyD88
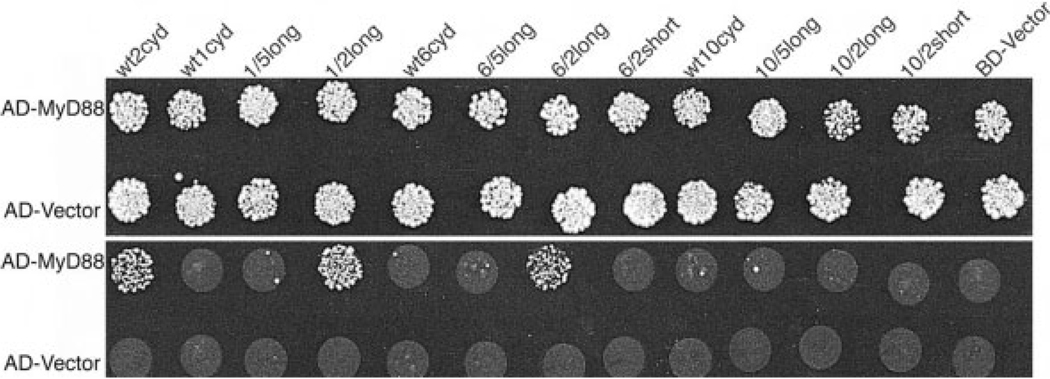
Because TLR1, 6, and 10 are homologous to TLR2, a chimera that incorporates a portion of TLR2 into any one of these TLR should not drastically perturb the overall structure of the TIR domain. We constructed BD fusion chimeras with either a 'long' 60-residue segment or a 'short' 48-residue segment of the TLR2 TIR domain replacing the equivalent region in TLR1, 6, or 10, and tested them for MyD88 binding (Fig. 2B and 4). The 60-residue segment of the TLR2 TIR domain conferred MyD88 binding activity to TLR1 and 6 (but not to TLR10); the corresponding segment of the more distantly related TLR5 did not impart this gain-of-function (Fig. 4). Therefore, the 60-amino acid segment of TLR2 contains all the necessary determinants to bind MyD88 when placed in the context of the TLR1 or 6 cytoplasmic domains.
Figure 4.
MyD88 binds chimeric cytoplasmic domains of TLR1 and 6. Yeast were co-transformed with the BD fusions indicated (along top) and the AD fusions indicated (at right). The transformation reaction was split in half and spotted onto −TL media (top panel) and onto −TLH/+3 mM3AT media (bottom panel). The "wt" indicates wild-type cytoplasmic domains, "long" indicates TLR2 amino acids 641–700, or TLR5 amino acids 693–758, swapped into the cytoplasmic domains of TLR1, 6, or 10, replacing the endogenous region, and "short" indicates TLR2 amino acids 659–700 swapped into the TLR cytoplasmic domains.
Amino acid co-variation in TIR domains
Previous studies indicated that the conserved Boxes 1 and 2 of the TIR domain are required for signaling (Fig. 2B; red bars) [22]. Inspection of the TIR domains indicated that three amino acids in Boxes 1 and 2 are strictly conserved in the MyD88 binders but not in the non-binders (Fig. 2). TLR1 phenylalanine 637, histidine 638 and asparagine 672 (also present in TLR6) are diverged from the analogous residues in all other TLR that signal by MyD88, which harbor tyrosine, aspartic acid, and another aspartic acid residue, respectively, in these positions (Fig. 2B; asterisks). The dichotomy between MyD88 binders (e.g. TLR2) and non-binders (e.g. TLR1 and 6) at these three residues implies functional significance regarding the MyD88 binding activity of the receptors.
If these amino acids are functionally linked, then their relationship should be detected by a statistical coupling analysis [23]. This method mines information in a multiple-sequence alignment to identify amino acids that appear to interact statistically, i.e. a perturbation of one amino acid is coupled to a change at a second-site residue (also known as co-variation). The 60-residue core MyD88-binding region of human TLR2 (Fig. 2B) was used to curate sequences for TLR1, 2, 6, 10 and MyD88 from Danio rerio to Homo sapiens in the NCBI database, and these were aligned using ClustalW (supplemental Fig. S1). 'Coupling energy' analyses (29) were performed focusing on TLR1 F637, H638, and N672. If F637 was designated as the invariant residue, then only two other positions, H638 and N672, were identified that appear to be linked to F637. If the H638 was designated as invariant, no other residues were predicted to harbor any particular amino acid. When N672 was designated as invariant, linkage to F637 and H638 was again predicted, and weaker linkages to H646, I666, Y691, S693, and I694 were uncovered (supplemental Table S1). The visual analysis was supported by the computational method and predicted the strongest co-variation of the TLR TIR1 residues F637 and H638 with N672, suggesting a functional link between these amino acids.
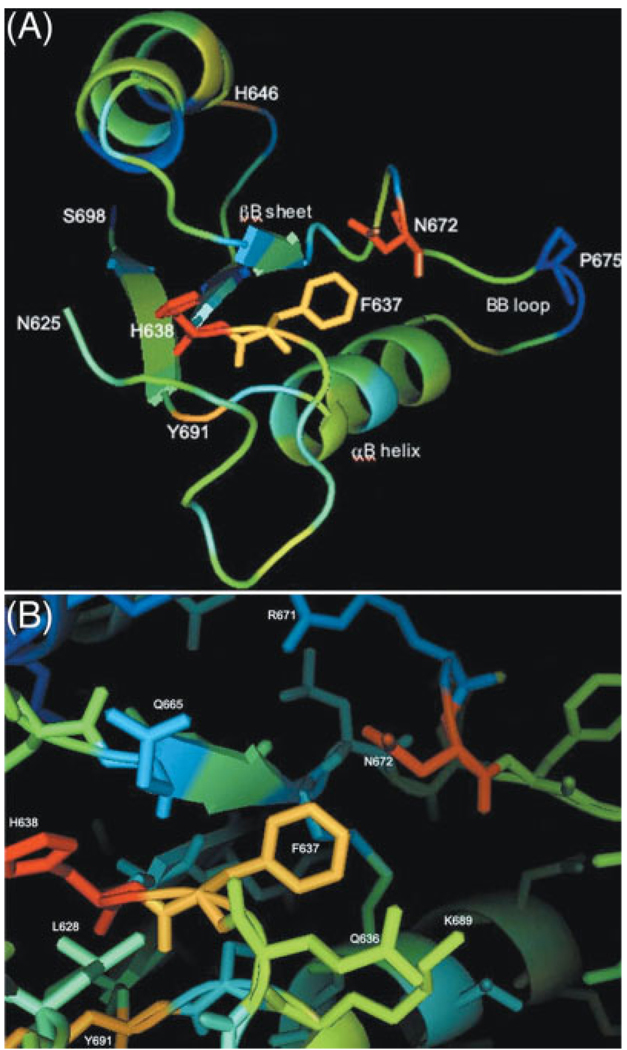
Co-varied residues are juxtaposed in TIR domain structure
A relationship had emerged between TLR1 residues F637, H638, and N672, but these residues are scattered throughout the linear protein sequence. Close arrangement of these residues in the TIR fold might support the plausibility of a functional interaction. Using PyMol (see Materials and methods), we plotted the values from the N672 coupling analysis onto the crystal structure of the TLR1 TIR domain. PyMol transformed the values into a 'heat map' showing relatively strong (red, highest values), moderate (yellow), or weak (blue, lowest values) relationships to the designated N672 residue (Fig. 5A) [9]. The predicted critical MyD88-binding residues, also statistically linked by computational analysis, appear to be in close proximity in the TLR1 TIR domain; indeed, the side chains of TLR1 residues F637 and N672 appear to be directly juxtaposed (Fig. 5B). Similar results were obtained with the TLR2 TIR domain structure (data not shown). The side chains of the H646, I666, Y691, S693, and I694 residues, weakly linked to N672 by statistical analysis, did not appear to lie in close proximity to F637, H638, or N672 (Fig. 5; see Supplemental methods). Subsequent experiments focused on the residues with strongest statistical linkage and in close proximity to each other: TLR1 F637, H638, and N672.
Figure 5.
(A) Predicted co-variation in the TLR1 TIR domain. A coupling energy analysis was run designating the TLR1 N672 amino acid as the invariant position (red). The statistical coupling analysis provided a vectorized numerical value, , for each residue, which reflects the statistical coupling of the residues to N672. The values for each amino acid were linked to the crystal structure of the TLR1 TIR domain and imported into the PyMol program. The image was colorized in PyMol, using 'b-factors', and a heat map was generated of the domain. Orange indicates relatively strong co-variation with the invariant residue, yellow indicates weaker correlation, and green or blue indicates no evidence of co-variation between the N672 and a given residue. See Supplemental data for the multiple sequence alignment and values. (B) A closer view of the positions of the TLR1 N672 and F637 side chains, as predicted by the TIR crystal structure. Other amino acids with side chains that appear spatially oriented near F637, H638, and N672 in the crystal structure are also labeled.
A single point mutation confers gain-of-function MyD88 binding ability
To test the relationship between the TLR1 F637, H638, and N672, we performed site-directed mutagenesis on the TLR TIR domain, changing native amino acids to the analogous TLR2 residues. All the possible combinations of single, double, and triple mutations at these three positions were created, and Western blots of yeast lysates showed comparable expression of wild-type and mutant proteins (Fig. 6A).
Figure 6.
(A) Gain-of-function point mutations in TIR1 confer MyD88 binding activity. Yeast were transformed with the BD fusions indicated (along top) and the AD fusions indicated (at left). Transformants were selected on −TL media (top panel) and on −TLH/+10 mM 3AT media (middle panel). The bottom panel shows comparable expression of the BD-TLR cytoplasmic domain fusions. Lanes on the Western blot correspond to labeled columns in (A). Cultures were inoculated from the −TL spots in (A) (asterisk indicates row of spots used for inoculation) and grown to log phase in media lacking tryptophan to select for the BD fusions. Protein lysates were prepared and 5 × 106 cell equivalents each were immunoblotted for the Gal4 BD. (B) Serial dilutions of transformants showing a two-hybrid signal for MyD88 binding. The experiment in (A) was reproduced with independent clones bearing the samemutations.AD fusions are indicated along the top and BD fusions are indicated along the bottom. Transformants were grown in −TL liquid media, diluted to OD600 = 0.45, and a series of tenfold dilutions were made for each culture. Of each serial dilution, 10 µL was spotted on −TL (top panel) and −TLH/+10 mM 3AT media (bottom panel).
Remarkably, a single missense in TLR1 (N672D) conferred the ability to bind MyD88, and the N672D/F637Y double mutation conferred an even stronger gainof- function (Fig. 6). Neither the F637Y single mutation nor the H638D single mutation produced any MyD88 binding ability. The N672D/H638D double mutant and the N672D/H638D/F637Y triple mutant did not bind MyD88, implying a negative effect by the H638D change. Although the F637Y and H638D single mutations did not create effects on their own, they clearly modulated the activity of the N672D mutation, indicating that the residues are functionally linked, critical determinants of MyD88 binding. Supporting this conclusion, the reciprocal TLR2 mutations (D678N, Y641A and D642A) eliminated MyD88 binding by TLR2, as did the previously characterized P681H mutation in TLR2 (Fig. 3). Replacing the critical P681 of TLR2 with an alanine instead of a histidine did not completely abrogate the two-hybrid signal for TLR2-MyD88 interaction (Fig. 3). These data indicate that the highly conserved TLR2 P681 proline is necessary but not sufficient for MyD88 binding, and that other residues (TLR2 D678, Y641 and D642) function concertedly to determine the differential MyD88 binding activity of the TLR.
Critical residues affect TLR signaling in mammalian cells
We assessed whether the mutations characterized in the yeast two-hybrid experiments affect TLR signaling in mammalian cells. Similar to experiments that characterized signaling by wild-type TLR cytoplasmic domains [4], the extracellular domain of the constitutively dimerizing CD4 antigen was fused to the transmembrane-intracellular domains of the wild-type (wt) TLR1, the wt TLR2, the 'gain-of-function' double mutant (dm) TLR1 (F637Y/N672D) and the 'loss-of-function' dm TLR2 (Y641F/D678N) (Fig. 7). As expected, the TLR2 alone (T2wt or T2dm) or TLR1 alone (T1wt or T1dm) did not signal, but the combination of wt TLR2 and 1 together (2wt/1wt) signaled through NF-κB. Replacing either wt TLR with a dm partner (2wt/1dm or 2dm/1wt) diminished signaling, indicating that the two amino acid residues we identified as critical for MyD88 binding indeed impact signaling by the TLR1 + 2 pair. Importantly, the double mutant pair (2dm/1dm) signaled just as well as the wild-type pair (2wt/1wt), suggesting that not only are these two residues critical for signaling, but that the TLR complex requires a precise arrangement of these residues in order to bind MyD88 and produce a signal: One partner must have the F-N configuration, and the other partner must have the Y-D configuration.
Figure 7.
A precise combination of residues is required for TLR signaling. The CD4-TLR chimeras harbored the transmembrane and cytoplasmic domains of wild-type TLR2 (2wt), double mutant TLR2 Y641F/D678N (T2dm), wild-type TLR1 (1wt), or double mutant TLR1 F637Y/N672D (T1dm). The constructs were transfected into CHO cells alone or in the indicated pairs, and signal transduction was assessed by activation of the ELAM-1 promoter driving a firefly luciferase gene. Activation was measured in relative luciferase units (RLU). See Materials and methods.
Novel protein interactions with specific TLR cytoplasmic domains
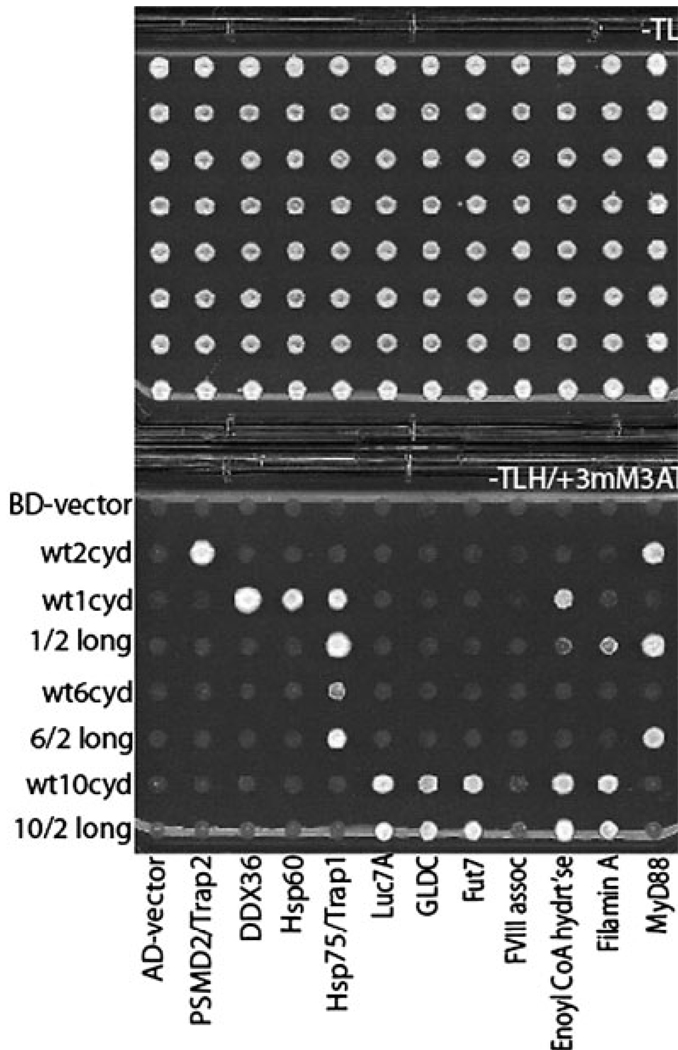
To investigate the specificity of TIR domain interactions, we used the cytoplasmic domains of TLR1, 2, 6, and 10 to screen a human B cell cDNA library of activation domain hybrids in search of proteins that bind specifically to each TIR. We identified at least one protein that specifically interacted with only a subset of the ten TLR cytoplasmic domains (Table 1). Heat shock protein (HSP) 60 specifically bound toTLR1, and HSP75, or tumor necrosis factor receptor-associated protein (TRAP) 1, bound specifically to TLR1 and 6. HSP have been recently described as TLR signal modulators [24–26]. The DDX36 protein, an RNA helicase of unknown function, bound only TLR1, and PSMD2, or TRAP2, which is a regulatory subunit of the proteasome, bound specifically to TLR2. The activity of proteasomes has recently be shown to be affected by TLR2 stimulation [27], and the ubiquitin ligase TRIAD3A associates with the cytoplasmic domains of TLR3, 4, and 9 [28]. The TLR10 screen yielded six specific interactions: LUC7A, a protein of unknown function with homology to the RNA splicing machinery [29]; GLDC, a glycine decarboxylase [30]; FUT7, a fucosyl transferase involved in generating sialyl Lewis-X carbohydrate moieties [31]; F8A, a coagulation factor VIII-associated transcript [32]; enoyl CoA hydratase, a peroxisomal protein involved in lipid oxidation [33]; and Filamin A, a protein that links cytoskeletal organization with many cell surface receptors [34–36]. When retested against the panel of ten TLR cytoplasmic domains, which all have related TIR domains, most of the newly identified protein partners interacted with only a subset of TLR. This result suggests that the interactions are specific.
Table 1.
Novel interactions with TLR cytoplasmic domains
| BD fusiona) | AD fusion | Bounde) |
|---|---|---|
| TLR1 cydb) | p36/DDX36, DNA/RNA helicase | Cyd 1 |
| Hsp75/TRAP1, heat shock protein | Cyd 1, 6 | |
| Hsp60/HSPD1, heat shock protein | Cyd 1 | |
| TLR2 cydc) | Psmd2/TRAP2, 26S proteasome subunit | Cyd 2 |
| TLR6 cydc) | Hsp75/TRAP1, heat shock protein | Cyd 1, 6 |
| Ptpn6/SHP-1, tyrosine phosphatase | Cyd 10 | |
| TLR10 cydd) | Luc7A, cisplatin resistance associated protein | Cyd 10 |
| Gldc, glycine dehydrogenase | Cyd 10 | |
| Fut7, fucosyl transferase | Cyd 10 | |
| Enoyl CoA hydratase | Cyd 10 | |
| Filamin A, actin-associated protein | Cyd 1, 10 | |
| Catalase, antioxidant enzyme | Cyd 10 | |
| IL enhancer BP | not retested | |
| Bicaudal D, dynein recruiting protein | not retested |
Each BD fusion was co-expressed with a human B cell cDNA library, and two-hybrid interactions were identified; Cyd indicates cytoplasmic domain.
The screen was performed in the presence of 10 mM 3AT.
The screen was performed in the presence of 3 mM 3AT.
The screen was performed in the presence of 25 mM 3AT.
AD fusion plasmids were retransformed into yeast to determine binding specificity for the panel of TLR1 through 10.
Since the TLR1/2 chimera changed the MyD88 binding specificity of TLR1, we determined whether this mutant had altered binding specificity toward the newly identified protein partners of TLR1 (Fig. 8). For the TLR1/2 chimera, gain of MyD88 binding capacity was concomitant with the loss of the ability to bind DDX36 or HSP60, but did not affect binding to HSP75/TRAP1. This result implies that MyD88, DDX36, and HSP60 all bind to the same general region of the TIR domain. The TLR1 N672D/F637Y double mutant bound MyD88, but did not show altered specificity toward any other proteins (data not shown).
Figure 8.
Binding of TLR and chimeras to newly identified proteins. Yeast expressing the BD fusions indicated (at left) and the AD fusion partners indicated (along bottom, see Table 1) were grown on −TL media (top panel), then replica-plated onto −TLH/+3 mM 3AT media (bottom panel).
Discussion
The TLR are assumed to directly bind MyD88, yet evidence for direct unassisted MyD88 interaction is sparse. Our experiments using the yeast two-hybrid assay tested direct protein-protein interactions between MyD88 and the ten human TLR, avoiding confounding factors such as accessory adapter proteins, coreceptors, or differential affinity for ligands. We showed that (1) MyD88 directly binds to homodimers of TLR2, 5, 7, and 8; (2) a 60-amino acid region from TLR2, but not the analogous region from TLR5, can confer MyD88 binding capacity toTLR1 and 6; (3) specific amino acids in the critical MyD88-binding region of the TLR show covariance, and the co-varying amino acids F637, H638, and N672 in TLR1 are functionally linked with respect to MyD88 binding; (4) a single mutation in TLR1 (N672D) confers gain-of-function MyD88 binding capacity; (5) the amino acids identified by the computational analysis and characterized by the two-hybrid system are critical for signaling in mammalian cells, and the TLR complex requires a precise arrangement of these residues in order to signal; and (6) newly identified protein partners specifically associate with TLR1, 2, 6, and 10.
The TLR2-MyD88 two-hybrid interaction was unexpected, because TLR2 cannot signal by itself in mammalian cells, even as a forced homodimer [4]. It is possible that TLR2 might bind MyD88 in mammalian cells, but that binding is not sufficient for signaling. Other proteins brought to the activated receptor complex by TLR1 (or 6) likely enable signaling through TLR2.
The high sequence homology and nearly identical crystal structure between the TIR domains of TLR1 and 2 provided a unique platformfor searching for gain-of-function mutations affecting the MyD88 association [9]. The N672D mutation was sufficient for TLR1-MyD88 interaction and the F637Y or H638D mutations modulated the MyD88 binding ability of N672D. None of the point mutants produced the robust two-hybrid signal for MyD88 binding displayed by the wild-type TLR2 or the TLR1/2 chimera, and the triple mutant TLR1 (F637Y/H638D/N672D), which has the same amino acid configuration as TLR2, did not bind MyD88. Together, these results suggest that other residues, likely those implicated in the statistical coupling analysis, are involved in strong MyD88 binding. The yeast two-hybrid experiments showed that the TLR1 F637Y/N672D double mutant bound MyD88, and in mammalian cells, a precise arrangement of these residues in each partner was required in order to signal: One partner must have the F-N (TLR1) configuration, and the other partner must have the Y-D (TLR2) configuration (Fig. 7). However, other regions of the TLR cytoplasmic domains must be important for MyD88 signaling because the 1wt/1dm pair (which harbors the F-N + Y-D configuration) could not signal. These data could be interpreted in two ways: First, MyD88 might not bind by the 1wt/1dm complex. If so, then additional amino acid changes may be needed to convert a mutant TLR1 into a strong MyD88 binder in mammalian cells. Alternatively, MyD88 may bind the 1wt/1dm complex, but other proteins (recruited specifically by the other regions of the TLR2 cytoplasmic domain) are required to signal, again suggesting that MyD88 binding is necessary but not sufficient to enable signaling.
The TLR2 P681 residue is found in the 'flexible' BB loop between sheet βB and helix αB, for which little positional information is gained from the crystal structure. A P681H mutation does not affect the overall TIR domain fold, but destroys signaling through MyD88 [9]. This proline may be required for TLR dimerization or for MyD88 contact. Our results imply that its role is in contacting MyD88, because the TLR2 P681H abrogated MyD88 association in the context of a 'forced' dimer; however, some evidence suggests that this proline does not directly contact MyD88 [37]. The TLR2 P681 might be indirectly involved in MyD88 contact by determining the topology of the flexible loop that harbors the critical D678; thus, the TLR2 P681H mutation might disturb a critical spatial arrangement of contact residues. Supporting this contention, the TLR2 P681A mutation, which presumably puts little spatial constraint on the flexible loop, did not completely destroy MyD88 binding in the two-hybrid assay (Fig. 3). Absolute affinity of the interaction cannot be determined in this type of experiment, but clearly, the P681A mutation does not produce the same effect as the P681H mutation in TLR2.
We have also uncovered novel protein interactions that lend insight into TLR signaling. For example, in Drosophila melanogaster, the dToll-dTube-dMyD88 complex, which signals for dorsal-ventral patterning as well as anti-fungal immune responses in the fly, is associated with Filamin [36, 38, 39]. In addition to Filamin, we found at least five other proteins that directly bound specifically to the cytoplasmic domain of TLR10 in the absence of MyD88.
Proteins that specifically associate with each TLR impart signal specificity to the Toll pathway [40]. TLR1 or 6 may recruit MAL/TIRAP or other, as yet unidentified, proteins to TLR2 in order to assemble a competent signaling complex. Thus, the novel interactions of HSP60 and HSP75 with TLR1 and 6 found here are worthy of future analysis. HSP60 has been shown to activate TLR4 signaling, although the mode of action is contentious [24, 25], and activated TLR4 associates with intracellular HSP70 and HSP90 [26].
Since this is the first report of proteins specifically associated only with TLR1 or 6, it will be of interest to determine whether the newly identified protein interactions described in this work affect signaling by TLR2 hetero-complexes in mammalian cells.
Materials and methods
Reagents
Templates for PCR amplification of the TLR cytoplasmic domains and MyD88 were provided by Dr. Alan Aderem (Institute for Systems Biology, Seattle, WA) and Dr. Christopher Wilson (University of Washington, Seattle, WA). Two-hybrid vectors were pOBD2 and pOAD [41], the host yeast was the strain PJ69-4α [42], and the human B cell cDNA λACT library was from ATCC (cat. no. 87003).
Constructs
Transmembrane domains were identified using TMPred (www.ch.embnet.org/software/TMPRED_form.html) and the Simple Modular Architecture Research Tool database [43]. The Gal4 BD or AD was fused to the N termini of the human TLR cytoplasmic domains or human MyD88. Interaction domains were PCR-amplified and cloned into pOBD2 or pOAD by gap repair in the yeast PJ69-4α.
Primers
All forward PCR primers harbored a 5′ recombination sequence to direct gap repair: 5′-GAATTCCAGCTGACCACCATG- followed by the domain-specific (annealing) sequence: TLR1 -GATCTGCCCTGGTAT CTCAGGATGG-3′; TLR2 -GCCCGTTTCCATGGCCTGTGGT ATATGAA-3′; TLR3 -GAGGGCTGGAGG ATAATCTTTTTATTGG-3′; TLR4 -GCCC ACCTGATGCTTCTTGCTGGCTGCATAAAG- 3′; TLR5 -GCCTCTTTTATCTGTTATAA GACAGCCCA-3′; TLR6 -GATCTGCCC TGGTATCTCAGGATC- 3′; TLR7 -ACAGC AAGTCACCTCTATTTCTGG-3′; TLR8 - AAAGGCTACAGGTCTCTTTCCACATCCC-3′; TLR9 -TGCTTCCACCTGTGCCTGGCC TGGCTTCCC-3′; TLR10 -GCCCACTTTGAT CTGCCCTGGTATCTCAG-3′; MyD88 -GCTGCAGGAGGTCCCGGCGCGGGGTCTGCGG- 3′. All reverse primers harbored a different 5′ recombination sequence to direct gap repair: 5′-GATCCCCGGGAATTGCCATGG- followed by the domain-specific sequence: TLR1 -CTATTT CTTTGCTTGCTCTGTCAGCTTA- 3′; TLR2 -CTAGGGACTTTATCGCAGCTCTCAGAT- 3′; TLR3 -TTAATGTACAGAGTTTTTGGAT CCAAGTGC- 3′; TLR4 -TCACTAAGATGTTG CTTCCTGCCAATTGCA-3′; TLR5 -TTAGG AGATGGTTGCTACAGTTTGCAA-3′; TLR6 - TTAAGATTTCACATCATTGTTTTCAGTGA-3′; TLR7 -CTAGACCGTTTCTTGAACACCTG ACT-3′; TLR8 -TTAGTATTGCTTAATGG AATCGACATAC-3′; TLR9 -CTATTCGGCCG TGGGTCCCTGGCAGAAG- 3′; TLR10 -TTATA GACAATCTGTTCTCATCAGAGAG- 3′, MyD88 -TCAGGGCAGGGACAAAGCCTTGGCA-3′.
Yeast two-hybrid screen
Protein interactions were selected for in synthetic complete media lacking tryptophan, leucine, and histidine and supplemented with 3-aminotriazole (3AT), an inhibitor of the His3 enzyme.
Protein alignments
TLR cytoplasmic domain sequences were aligned via ClustalW http://www.ebi.ac.uk/clustalw/, using default parameters.
Statistical coupling analysis
The software and other tools for the statistical coupling analyses were provided by Dr. Rama Ranganathan (University of Texas, Southwestern Medical Center, Dallas, TX). Coupling energy analysis was performed on a multiple-sequence alignment, designating one amino acid as invariant (F637, H638, or N672 of TLR1). Vectorized data for each position in the sequence alignment were imported into the PyMol program (http://pymol.sourceforge.net/) and mapped onto the crystal structure of the TLR1 TIR domain using the 'b-factors' for color designation. Only the 60-amino acid region that confers MyD88 binding activity is shown.
Site-directed mutagenesis
Mutations were generated using the Stratagene Quickchange Site-Directed Mutagenesis Kit as per the manufacturer's protocol.
Western blots
Yeast expressing the BD fusion proteins were grown to log phase in synthetic media lacking tryptophan. Three OD600 equivalents of cells were pelleted, resuspended in 50 µL 2.5 mM NaOH and placed on ice for 10 min. Trichloroacetic acid was added to 25% final concentration, and samples were incubated for 10 min on ice. Precipitates were pelleted for 10 min at 14 000 × g at 4°C. Pellets were resuspended in 50 µL 8 M urea buffer and neutralized with 20 µL 1 M Tris-Cl, pH 8.0. Samples were boiled for 5 min, pelleted, and 15 µL of the supernatants were separated through a 7.5% SDS-PAGE gel. Blots were probed with anti-Gal4 BD primary antibody (Upstate, cat. no. 06-262) and a secondary antibody conjugated to horseradish peroxidase (Amersham).
CD4-TLR signaling assays
The CD4-TLR chimeras were overexpressed in CHO cells using the pEF6 expression vector (Invitrogen). The signal reporter construct consisted of the NF-κB-dependent endothelial cell-leukocyte adhesion molecule-1 (ELAM-1) promoter driving the firefly luciferase gene, and activation was measured in luciferase units relative to the constitutively expressed Renilla luciferase gene (the transfection control reporter); the activities were measured by the Dual-Luciferase Reporter Assay System (Promega). The empty expression vector provided the negative control, and TLR4, which signals strongly as a homodimer, served as the positive control for reporter activation.
Library screens
Cells expressing the BD-human TLR cytoplasmic domain construct were grown overnight in synthetic medium lacking tryptophan and supplemented with 2 × adenine. Cultures were diluted to OD600 = 0.5 in YPD medium [44] and grown for 4 h. Transformations were carried out as described previously [45], using 0.1, 0.5, 1.0, and 2.0 µg of the human B cell cDNA AD fusion library. Transformants were selected in media lacking tryptophan, leucine, and histidine, supplemented with 3–25 mM 3AT, and grown for 7 days at 30°C. His+ colonies were re-streaked onto selective plates, AD fusion constructs were rescued through leuB− E. coli, and the inserts were sequenced. Constructs were co-transformed into the PJ69-4α strain along with the panel of BD-TLR cytoplasmic domains and tested for binding specificity.
Supplementary Material
Acknowledgements
The authors thank Adeline Hajjar for insightful discussions, and Mark Johnston for critical review of the manuscript. This work was supported by fellowship DRG 1744-02 from the Damon Runyon Cancer Research Foundation, and grant CFFT MILLER00V0 from the Cystic Fibrosis Foundation. S.F. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- 3AT
3-aminotriazole
- AD
Gal4 transcriptional activation domain
- BD
Gal4 DNA-binding domain
- ELAM-1
endothelial cell-leukocyte adhesion molecule-1
- MAL/TIRAP
MyD88 adapter-like/TIR domain-containing adapter protein
- TIR
Toll/IL-1 receptor domain
- −TL
lacking tryptophan and leucine
- −TLH
lacking tryptophan, leucine, and histidine
- TRAP
tumor necrosis factor receptor-associated protein
Footnotes
Supporting information for this article is available at http://www.wiley-vch.de/contents/jc_2040/2006/35158_s.pdf
References
- 1.Takeda K, Akira S. TLR signaling pathways. Semin. Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Armant MA, Fenton MJ. Toll-like receptors: A family of pattern-recognition receptors in mammals. Genome Biol. 2002;3:3011.1–3011.6. doi: 10.1186/gb-2002-3-8-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishiya T, DeFranco AL. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J. Biol. Chem. 2004;279:19008–19017. doi: 10.1074/jbc.M311618200. [DOI] [PubMed] [Google Scholar]
- 4.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proc. Natl. Acad. Sci. USA. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hajjar AM, O'Mahony DS, Ozinsky A, Underhill DM, Aderem A, Klebanoff SJ, Wilson CB. Cutting Edge: Functional interactions between Toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J. Immunol. 2001;166:15–19. doi: 10.4049/jimmunol.166.1.15. [DOI] [PubMed] [Google Scholar]
- 6.Sandor F, Latz E, Re F, Mandell L, Repik G, Golenbock DT, Espevik T, et al. Importance of extra- and intracellular domains of TLR1 and TLR2 in NFkappa B signaling. J. Cell Biol. 2003;162:1099–1110. doi: 10.1083/jcb.200304093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns K, Martinon F, Esslinger C, Pahl H, Schneider P, Bodmer JL, Di Marco F, et al. MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]
- 8.Athman R, Philpott D. Innate immunity via Toll-like receptors and Nod proteins. Curr. Opin. Microbiol. 2004;7:25–32. doi: 10.1016/j.mib.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 10.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 11.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: An adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 12.Martin MU, Wesche H. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochim. Biophys. Acta. 2002;1592:265–280. doi: 10.1016/s0167-4889(02)00320-8. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 15.Ronni T, Agarwal V, Haykinson M, Haberland ME, Cheng G, Smale ST. Common interaction surfaces of the Toll-like receptor 4 cytoplasmic domain stimulate multiple nuclear targets. Mol. Cell. Biol. 2003;23:2543–2555. doi: 10.1128/MCB.23.7.2543-2555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akazawa T, Masuda H, Saeki Y, Matsumoto M, Takeda K, Tsujimura K, Kuzushima K, et al. Adjuvant-mediated tumor regression and tumor-specific cytotoxic response are impaired in MyD88-deficient mice. Cancer Res. 2004;64:757–764. doi: 10.1158/0008-5472.can-03-1518. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkbacka H, Kunjathoor VV, Moore KJ, Koehn S, Ordija CM, Lee MA, Means T, et al. Reduced atherosclerosis in MyD88-null mice links elevated serum cholesterol levels to activation of innate immunity signaling pathways. Nat. Med. 2004;10:416–421. doi: 10.1038/nm1008. [DOI] [PubMed] [Google Scholar]
- 18.Bartfai T, Behrens MM, Gaidarova S, Pemberton J, Shivanyuk A, Rebek J., Jr A low molecular weight mimic of the Toll/IL-1 receptor/resistance domain inhibits IL-1 receptor-mediated responses. Proc. Natl. Acad. Sci. USA. 2003;100:7971–7976. doi: 10.1073/pnas.0932746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beutler B, Rehli M. Evolution of the TIR, Tolls and TLRs: Functional inferences from computational biology. Curr. Top. Microbiol. Immunol. 2002;270:1–21. doi: 10.1007/978-3-642-59430-4_1. [DOI] [PubMed] [Google Scholar]
- 20.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 21.Lee HK, Dunzendorfer S, Tobias PS. Cytoplasmic domain-mediated dimerizations of Toll-like receptor 4 observed by beta-lactamase enzyme fragment complementation. J. Biol. Chem. 2004;279:10564–10574. doi: 10.1074/jbc.M311564200. [DOI] [PubMed] [Google Scholar]
- 22.Slack JL, Schooley K, Bonnert TP, Mitcham JL, Qwarnstrom EE, Sims JE, Dower SK. Identification of two major sites in the type I interleukin-1 receptor cytoplasmic region responsible for coupling to pro-inflammatory signaling pathways. J. Biol. Chem. 2000;275:4670–4678. doi: 10.1074/jbc.275.7.4670. [DOI] [PubMed] [Google Scholar]
- 23.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 24.Vabulas RM, Wagner H, Schild H. Heat shock proteins as ligands of Toll-like receptors. Curr. Top. Microbiol. Immunol. 2002;270:169–184. doi: 10.1007/978-3-642-59430-4_11. [DOI] [PubMed] [Google Scholar]
- 25.Gao B, Tsan MF. Recombinant human heat shock protein 60 does not induce the release of tumor necrosis factor alpha from murine macrophages. J. Biol. Chem. 2003;278:22523–22529. doi: 10.1074/jbc.M303161200. [DOI] [PubMed] [Google Scholar]
- 26.Triantafilou M, Triantafilou K. Heat-shock protein 70 and heatshock protein 90 associate with Toll-like receptor 4 in response to bacterial lipopolysaccharide. Biochem. Soc. Trans. 2004;32:636–639. doi: 10.1042/BST0320636. [DOI] [PubMed] [Google Scholar]
- 27.Link C, Gavioli R, Ebensen T, Canella A, Reinhard E, Guzman CA. The Toll-like receptor ligand MALP-2 stimulates dendritic cell maturation and modulates proteasome composition and activity. Eur. J. Immunol. 2004;34:899–907. doi: 10.1002/eji.200324511. [DOI] [PubMed] [Google Scholar]
- 28.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat. Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 29.Nishii Y, Morishima M, Kakehi Y, Umehara K, Kioka N, Terano Y, Amachi T, Ueda K. CROP/Luc7A, a novel serine/arginine-rich nuclear protein, isolated from cisplatin-resistant cell line. FEBS Lett. 2000;465:153–156. doi: 10.1016/s0014-5793(99)01744-5. [DOI] [PubMed] [Google Scholar]
- 30.Sellner L, Edkins E, Greed L, Lewis B. Detection of mutations in the glycine decarboxylase gene in patients with nonketotic hyperglycinaemia. Mol. Genet. MeTable. 2005;84:167–171. doi: 10.1016/j.ymgme.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Bengtson P, Lundblad A, Larson G, Pahlsson P. Polymorphonuclear leukocytes from individuals carrying the G329A mutation in the alpha 1,3-fucosyltransferase VII gene (FUT7) roll on E- and P-selectins. J. Immunol. 2002;169:3940–3946. doi: 10.4049/jimmunol.169.7.3940. [DOI] [PubMed] [Google Scholar]
- 32.Peters MF, Ross CA. Isolation of a 40-kDa Huntingtin-associated protein. J. Biol. Chem. 2001;276:3188–3194. doi: 10.1074/jbc.M008099200. [DOI] [PubMed] [Google Scholar]
- 33.Koski KM, Haapalainen AM, Hiltunen JK, Glumoff T. Crystal structure of 2-enoyl-CoA hydratase 2 from human peroxisomal multi-functional enzyme type 2. J. Mol. Biol. 2005;345:1157–1169. doi: 10.1016/j.jmb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, et al. Localized mutations in the gene encoding the cytoskeletal protein Filamin A cause diverse malformations in humans. Nat. Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 35.Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U. Physical and functional interaction of Filamin (actin-binding protein-280) and tumor necrosis factor receptor-associated factor 2. J. Biol. Chem. 2000;275:271–278. doi: 10.1074/jbc.275.1.271. [DOI] [PubMed] [Google Scholar]
- 36.Edwards DN, Towb P, Wasserman SA. An activity-dependent network of interactions links the Rel protein Dorsal with its cytoplasmic regulators. Development. 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- 37.Dunne A, Ejdeback M, Ludidi PL, O'Neill LA, Gay NJ. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J. Biol. Chem. 2003;278:41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 38.Horng T, Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl. Acad. Sci. USA. 2001;98:12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. USA. 2002;99:12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of TLR7 signaling. J. Biol. Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 41.Hudson JR, Dawson EP, Rushing KL, Jackson CH, Lockshon D, Conover D, Lanciault C, et al. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 1997;7:1169–1173. doi: 10.1101/gr.7.12.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherman F, Fink GR, Hicks JB, editors. Methods in yeast genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 45.Bartel PL, Fields S. The yeast two-hybrid system. New York: Oxford University Press; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.