Abstract
Although the correlative evidence relating the presence of amyloid fibrils and certain disease states (e.g. Alzheimer’s disease and Type 2 Diabetes) is overwhelming, a direct causative role for amyloid has proved harder to establish. Current thinking links a narrow region of the aggregate protein size distribution, the so called ‘early aggregate’ domain to cellular toxicity. A troubling feature of this theory however is that the nucleated reaction mechanism by which amyloid formation is believed to occur results in a very low number concentration of early aggregates which are rapidly extended to form amyloid fibrils. This situation means that the concentration of early aggregates is very low at the time when they are supposedly at their most toxic. Here we adopt a novel explicit simulation strategy to examine a kinetic regime involving nucleated growth combined with fibril fragmentation under which this situation can be reversed so as to produce a high number concentration of small on-pathway toxic aggregates. Dependent upon the rate of fragmentation, the time scale for generation of toxic early aggregates may be coupled, uncoupled or disassociated from the time scale for the appearance of amyloid fibrils. Furthermore the model presents itself as a biochemical ‘switch’ transitioning between modes of amyloid induced cell death dependent upon either specific amyloid toxicity or non-specific solid mass induced tissue damage.
Introduction
In modern day structural biology the term amyloid is used to describe a certain class of linear protein fibers that are highly enriched in β-sheet structural elements and are capable of binding specific intercalating ligands (1,2). In humans there are more than twenty-five different proteins for which conversion of the protein into amyloid is strongly coupled with disease onset (3–6). Two such amyloid linked disease processes of particular note (due to their prevalence and societal impact) are Alzheimer’s disease and Type 2 Diabetes which involve the formation of amyloid fibers from the A-β (7–9) and amylin (10–12) peptides respectively. However despite a strong correlative relationship between the occurrence of amyloid fibrils and a number of these disease states a detailed causative mechanistic role for amyloid in disease progression is currently lacking in most cases (1,3–6,13). What is clear however, is that nearly all amyloid related disease involve, at some stage in their etiology, tissue-specific apoptotic (13,14) or necrotic (2,15,16) processes causing harm or death to the afflicted individual.
Despite the fact that amyloid fibrils can be formed from a variety of different protein precursors1- (6,17–19) all amyloid fibrils share a high degree of structural similarity at the mesoscopic level. This structural similarity is borne out by the fact that all amyloid fibrils specifically bind to a common set of ligands and their proto-fibril width distribution lies within a narrow range of about 3 to 10 nm (1,2). Due to these common structural characteristics that are irrespective of the particular monomeric protein building blocks, a common underlying mechanism explaining amyloid’s role in disease has been sought (1–6,17–19). Towards this direction a number of theories have been put forward which posit different structural roles for amyloid’s action as a harmful agent to the cell. These roles include (i.) a physical disruptor of the cell or tissue due to incompatible dimensions or the physical amount of the amyloid produced (2,15,16), (ii.) a biochemical disruptor of enzymes involved in essential cellular processes such as proteasome(19,20,21), chaperone (14,22–24), redox control pathways (14,25,26) and (iii.) a pore forming membrane disruptor leading to loss of cellular homeostasis by formation of unwanted pores in both the cell membrane and intracellular compartment membranes (14,27,28).
We previously described a ‘two hit’ model of amyloid’s role in disease progression (29) that supposed that (i.) the production of a certain critical mass of amyloid was related to disease initiation, and (ii.) that the production of this critical mass of amyloid could be caused through a combination of both genetic and environmental factors. Our model was therefore similar in nature to the loss of heterozygosity (LOH) model used to explain the banding patterns seen in the age dependent development of retinal blastoma (30)1.
Although our previous model was capable of providing a rational explanation for a number of the vagaries associated with both the spontaneous development of disease and the existence of a species barriers in the transmission of infectious forms of amyloid (known as prions(31–33)), it was not able to shed light on the possible mechanistic role played by amyloid in any disease initiation/ progression process. In the current work we seek to correct this deficiency by developing a new model that can incorporate additional experimental information, the two major strands of which are (i.) findings from numerous experimental studies that suggest that not all types of amyloid fibrils are equally toxic, but rather a certain range of small amyloid oligomers possess the majority of the cytotoxic properties(12,34–37) (ii.) results from autopsies of patients who have expired as a result of Diabetes or Alzheimer’s which indicate that extensive tissue damage is not always correlated with amyloid fibril load (38–41). Our new model of amyloid’s relationship to disease development is more complex in nature than our previous ‘two hit’ model. However the increase in complexity carries with it a range of fascinating new insights into the possible kinetic linkages existing between amyloid formation and the onset of disease.
Theory and Methods
Simulation Model
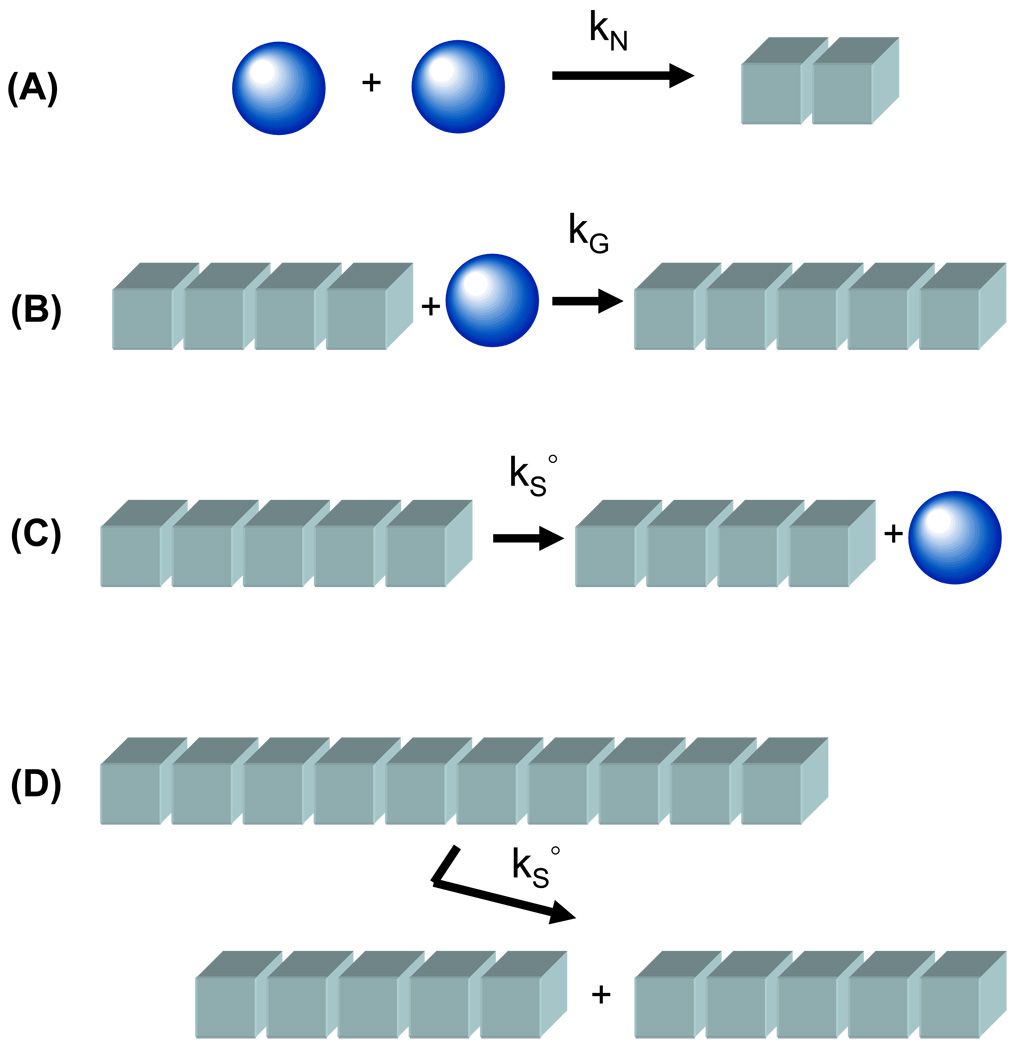
To model amyloid formation from a starting pool of monomer we explicitly treated the many thousands of rate equations specified by our kinetic model (Appendix 1). Despite the many excellent previous modelling works on the subject the arguments made within, and insight provided by this paper were only possible because of the explicit simulation approach adopted. As such the results outlined within the current work have not been previously explored. The conceptual basics of our model are highlighted in Fig. 1. Amyloid fibril formation is considered to be initiated via a slow nucleation step, governed by a rate constant kN and a critical nucleus size n, which is followed by a number of faster growth steps, governed by identical rate constants kG. Amyloid formation is considered to occur by monomer addition in a uni-directional manner. Amyloid fragmentation may occur by monomer fracture between any two monomers in the amyloid fibre at a rate constant ks°. If monomer fracture results in the production of a fragmentation species of the size of monomer then this species is considered to rejoin the monomer pool. Fragmentation species of size greater than monomer are considered to be short amyloid fragments.
Fig. 1.
Schematic description of the kinetic rate model used for the explicit solution of amyloid formation by a nucleated reaction mechanism that is subject to fibril scission/fragmentation. (A) Critical nucleus formation (n = 2) from monomeric proteins is governed by a bi-molecular rate constant kN. (B) Uni-directional fibril growth occurs by monomer addition to a fibril end, the process is governed by a bi-molecular growth rate constant kG. (C) Scission at the end of the fibril results in release of a monomer that rejoins the native monomer pool. Scission from either end of the fibril is regulated by a uni-molecular rate constant kS°. (D) In a fibril consisting of j sub-units, fibril fragmentation is technically possible at any of the j-3 internal sites at a characteristic intrinsic scission rate kS°. In the numerical solution of our model we confine breakage to the central position with this breakage event governed by a rate constant set to (j-3) kS° (see Hall and Edskes, 2004 – ref 29).
Model Postulates
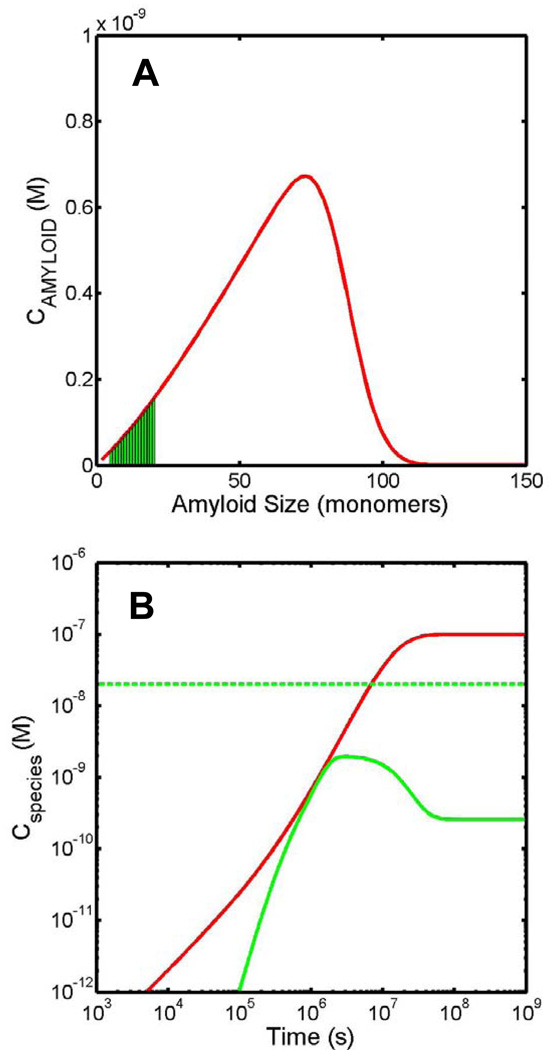
In keeping with the concept of the toxic early oligomer hypothesis our starting postulate involves designating an arbitrary region of the amyloid size distribution with these toxic attributes (Fig. 2a). We next postulate that disease initiation/phenotype display corresponds to the production of a threshold concentration of toxic oligomer production (Fig. 2b) and not the total mass of protein existing in amyloid. In the simulation exercises to come we will plot both the total amount of toxic oligomer and the total mass of protein existing in all amyloid forms.
Fig. 2.
Diagram showing the two fundamental postulates in our modelling approach. (A) A certain section of the amyloid size distribution, known as the toxic oligomer region has cytotoxic properties. In this paper we consider the toxic oligomer region to extend from aggregates containing 5 to 20 protein monomer units. Here we see a representative amyloid size distribution (red line) with the toxic oligomer region highlighted (green shaded region). (B) The onset of disease corresponds to the exceeding of a threshold level of toxic oligomers. Here the threshold level is set at 20 nM and is shown by the dotted green line. The solid green line describes the time dependent formation of the summed total of all toxic oligomers, and the solid red line denotes the time dependent formation of the summed total of all amyloid species. We note that for the current example the critical threshold value is not reached.
Parameter Values
We recycle the parameter values for n, kN, kG, and ks° from our previous work29 noting that the values are not meant to, in any way, reflect values characteristic of any living system but rather are chosen to effect the smooth transition from one type of limiting case behaviour to another. In this fashion we have used the following parameter set, n = 2, kN = 0.01M−1s−1, kG = 100 M−1s−1. The parameter ks° was allowed to vary from 0 s−1 to 1×10−6 s−1. Two disparate cases of monomer concentration were chosen to reflect the two extremes of protein monomer regulation in the cell. The first case which we shall designate as Case A corresponded to a situation in which the regulation of monomer production was much faster than the time scale of the amyloid formation reaction such that the concentration of free monomer was kept at a constant level at all times (MFREE = 100nM). The second case, designated as Case B, corresponded to a situation whereby the rate of monomer production/regulation was very slow in relation to the time scale of the amyloid formation reaction. For this case we treated the total amount of protein in the system as a constant defined by MTOT such that any formation of amyloid resulted in a decrease in the free monomer concentration (MTOT = 100nM).
Effecting a Solution of the Model
At the core of this work lies the solution of a very large set of equations describing all possible chemical reactions specified by a nucleated growth mechanism of amyloid fibril formation featuring fibril fragmentation. This explicit simulation approach has provided us with a full temporal description of the amyloid fibril distribution. Subsequent grouping and analysis of relevant sections of that distribution has allowed us to make the conceptual discoveries presented within this paper. The chemical rate equations outlined in Eqn. A1, A6 and A7 were solved using a self written numerical integration procedure programmed in MATLAB R2008b on a standard desktop personal computer. The numerical integration procedure employed was based on a fourth order Runge-Kutta procedure and has been previously described (29).
Results
Here we present the findings of our modelling approach first for limiting Case A and then for limiting Case B. In both simulation sets we use a fixed set of nucleation and growth rate constants in conjunction with a fixed nucleus size and we vary the intrinsic fracture rate constant ks° over three orders of magnitude such that the scission rate constant took on a value of either ks° = 1×10−6 s−1 (relatively fast scission) or ks° = 1×10−9 s−1 (reflecting relatively slow scission). The results are contrasted against a classical model of nucleated growth in which fibril growth occurs by endwise addition of monomer only (with no associated fibril fragmentation or endwise depolymerization such that ks° = 0 s−1).
Case A: Fixed free concentration of monomer
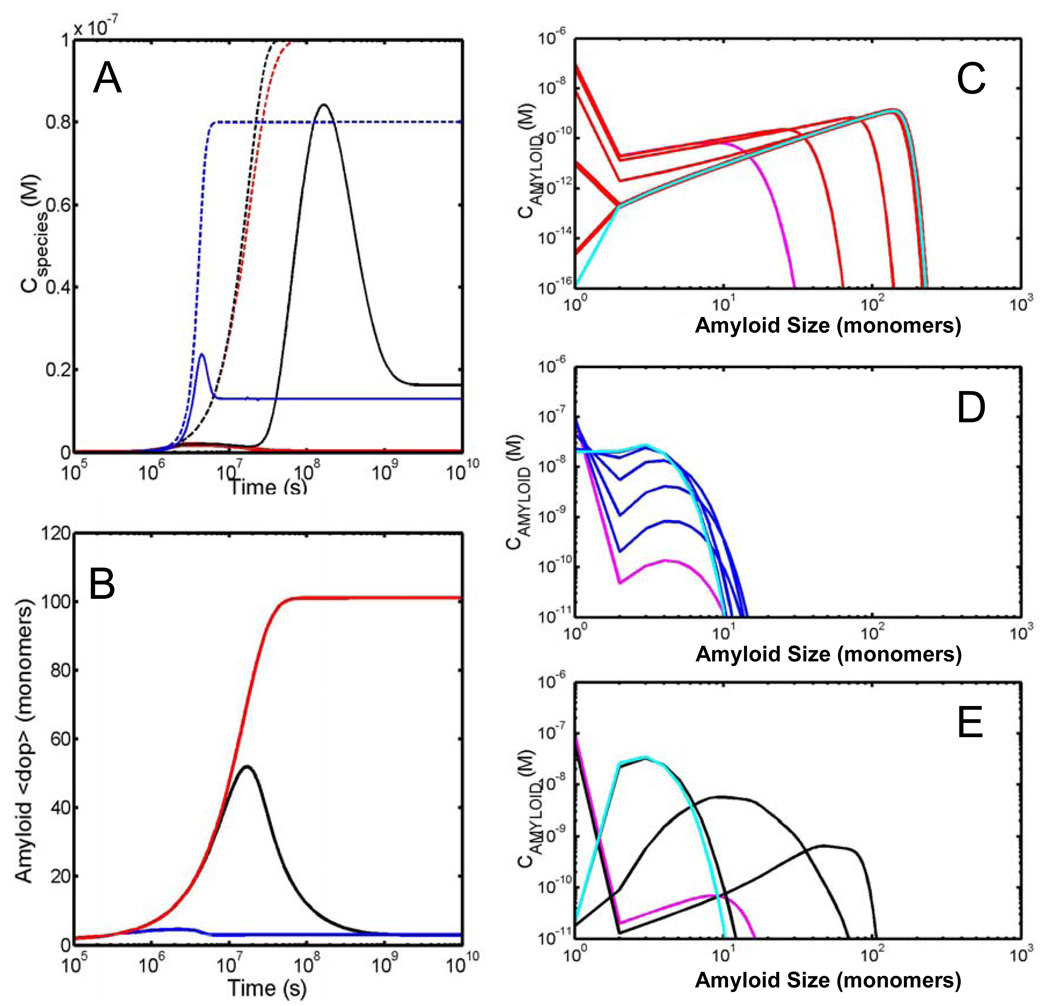
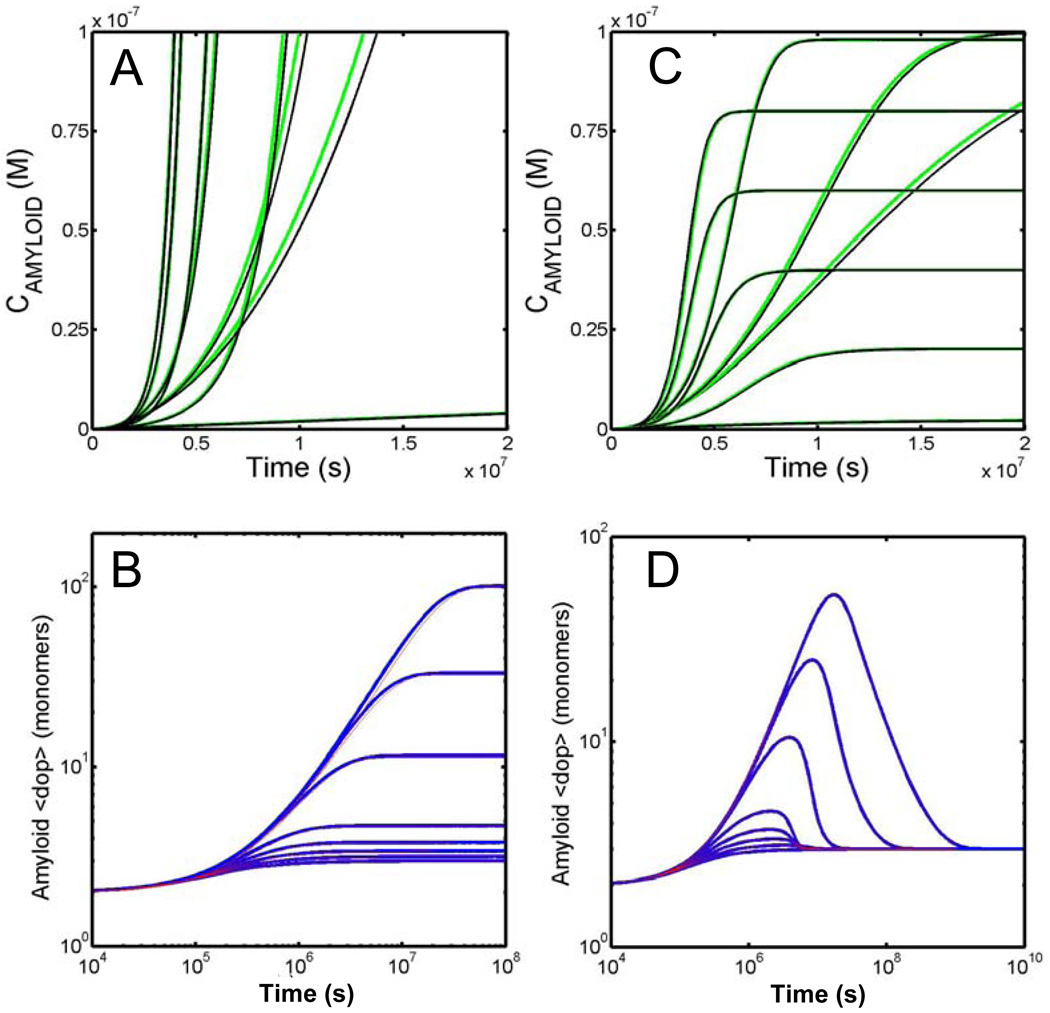
For the case of a constant concentration of free monomer (Fig. 3a) we note that the relationship between the time-scales relating protein in all amyloid forms and that specifically in the toxic early oligomer form can be widely disparate depending on the rate of fibril fracture/scission. For the case of no scission, ks° = 0 s−1, the two time scales are virtually identical with both appearing around the 1×106 second mark (total amyloid - dashed red line; toxic oligmer – solid red line). However for this kinetic regime the total extent of toxic oligomer species never becomes appreciable as its rate of production and rate of loss (via its incorporation into higher molecular weight amyloid species) reaches a steady state level of approximately 2.5 nM. For this irreversible case the average degree of polymerization of all amyloid species continues to grow in an unbounded fashion approximately reaching 5000 monomers by the end of the simulation (Fig. 3b). The shape of the distribution constituting the amyloid product at a number of different stages along the time course is described in Fig. 3c. The distribution is asymmetric, the degree of asymmetry increasing with increasing extents of amyloid formation. Thus for this kinetic regime characterised by no scission/fragmentation (ks° = 0 s−1) we conclude that the time scale for the appearance of toxic oligomers is highly coupled to the time scale for the production of long amyloid fibrils although as the total extent of amyloid fibres increases the ratio of the mass of toxic oligomers to the total mass of amyloid drops substantially.
Fig. 3.
Model development for the situation of a fixed free concentration of monomer (Case A). (A) Time dependent formation of different amyloid species: the summed total all amyloid species (dotted line) and summed total of all toxic oligomer species (solid line) for three different values of the scission/fragmentation rate – red (kS°= 0s−1), blue (kS°= 1×10−6s−1) and black (kS°= 1×10−9s−1). (B) Time dependent evolution of the amyloid average degree of polymerization for the three different scission/fragmentation rates – red (kS°= 0s−1), blue (kS°= 1×10−6s−1) and black (kS°= 1×10−9s−1). (C–E) Snapshots of the kinetic evolution of the amyloid size distribution for the three different scission/fragmentation rates (C) kS°= 0s−1, (D) kS°= 1×10−6s−1 and (E) kS°= 1×10−9s−1 with magenta representing the earliest time and cyan representing the latest time. The displayed snapshots correspond to the following time windows (described in seconds) (C) 1×106, 3.2×106, 1×107, 3.2×107, 1×108. (D) 1×106, 2×106, 3.2×106, 4×106, 5×106. (E) 1×106, 3.2×106, 1×107, 3.2×107, 1×108.
For the case of relatively fast scission, ks° = 1×10−6 s−1, the relationship between the time scale of production of toxic early oligomers and all amyloid forms (Fig. 3a, total amyloid - dashed blue line; toxic oligomer – solid blue line) is essentially identical to the time scale of toxic oligomer production due to the fact that the majority of polymerized protein exists in the toxic oligomer form. Indeed it can be seen from the evolution of the average degree of polymerization (Fig. 3b) that the average size never varies significantly from approximately 4 monomers. The range of species making up the amyloid distribution at a series of different times is shown in Fig. 3d. As can be noted the extent of toxic oligomer production quickly passes the critical level associated with disease without ever being associated with a significant build up of long amyloid fibres. So for this kinetic regime characterised by a relatively fast fragmentation/scission rate (ks° = 1×10−6 s−1) we observe that the production of toxic oligomer species is disassociated from the appearance of amyloid fibrils.
For the situation in which the scission rate is relatively slow, ks° = 1×10−9 s−1, there is a significant separation of time scales between the appearance of long amyloid fibres (Fig. 3a, dashed black line - around 3×106 seconds) and toxic early oligomers (Fig. 3a, solid black line - around 1×108 seconds). This timing difference is slightly preceded by a roll-over in the average degree of polymerization (Fig. 3b) which occurs at/from about 2×107 seconds. Unlike the previous ‘fast fragmentation’ case, in this relatively slow fragmentation/scission rate regime (ks° = 1×10−9s−1) the formation of toxic oligomer to a level associated with disease is associated with the appearance of long amyloid fibres however the two characteristic time scales of production are strongly decoupled. This is demonstrated very clearly by a series of time snapshots of the growing polymer distribution (Fig. 3e). The broad range of aggregates centred around an eventually uniformly shaped distribution with an average size of approximately 100 monomers means that the total mass of protein in amyloid form must become large before the amount of ‘toxic oligomers’ existing at the outer edge of the distribution can become significant.
Case B: Fixed total concentration of monomer
In this case we examine the effect of changes in the fragmentation/scission rate constant on the characteristic time scales describing the appearance of protein existing in all amyloid forms and the protein specifically in ‘toxic oligomer’ form for the situation of a total fixed amount of protein existing in the system.
Perhaps the most striking difference between this case and the preceding one is the observation (Fig. 4a) of either one or two time dependent maxima, in the time course of the toxic oligomer species for all cases of the varied scission rate (solid red line − ks° = 0s−1; solid black line − ks° = 1×10−9s−1; solid green line − ks° = = 1×10−6s−1). As can be noted from Fig. 4b this temporary production of a relatively large amount of toxic oligomer species is due to the decay of the average degree of polymerization of the amyloid, which dependent upon the operative rate of scission may occur on a time scale vastly slower than the original polymerization reaction. With reference to the particular amyloid distribution profiles recorded at different times (Figs. 4C – E) and in a fashion similar to the preceding case, the appearance of toxic oligomers may be either coupled to the appearance of amyloid fibrils (Fig. 4A dotted red line −ks°=0s−1), uncoupled from the appearance of long amyloid fibrils (Fig. 4A dotted black line − 1×10−9s−1) or alternatively not associated with the appearance of amyloid fibrils (Fig. 4A dotted blue line − 1×10−6s−1).
Fig. 4.
Model development for the situation of a fixed total concentration of monomer present in the system (Case B). (A) Time dependent formation of different amyloid species: the summed total all amyloid species (dotted line) and summed total of all toxic oligomer species (solid line) for three different values of the scission/fragmentation rate – red (kS°= 0s−1), blue (kS°= 1×10−6s−1) and black (kS°= 1×10−9s−1). (B) Time dependent evolution of the amyloid average degree of polymerization for the three different scission/fragmentation rates – red (kS°= 0s−1), blue (kS°= 1×10−6s−1) and black (kS°= 1×10−9s−1). (C–E) Snapshots of the kinetic evolution of the amyloid size distribution for the three different scission/fragmentation rates (C) kS°= 0s−1, (D) kS°= 1×10−6s−1 and (E) kS°= 1×10−9s−1 with magenta representing the earliest time and cyan representing the latest time. The displayed snapshots correspond to the following time windows (described in seconds) (C) 1×106, 3.2×106, 1×107, 3.2×107, 1×108, 1×109, 1×1010. (D) 1×106, 2×106, 3×106, 4×106, 5×106, 6×106, 7×106. (E) 1×106, 1×107, 1×108, 1×109, 1×1010.
Discussion
This work differs significantly from our previous modelling study (29) at the level of the starting postulate describing a causative link between the production of amyloid and the onset of disease progression (1–6). In the current work we have adopted a point of view based on experimental findings (34–37), that the toxic component of amyloid is associated with a narrow region of its size distribution encompassing a range of small on pathway aggregates2. In the previous work we considered disease onset/phenotype display to be associated with the total mass of protein in amyloid form (29). This subtle distinction has opened a new window from which we can view the link between the production of amyloid fibrils and the onset of disease.
Using a kinetic model of nucleated polymerization that incorporates internal fragmentation in combination with the starting assumption that a certain defined region of the amyloid distribution has toxic properties we have shown that, dependent upon the rate of fibril scission/fragmentation the characteristic time scale of production of significant levels of toxic oligomer may be either coupled, uncoupled or not associated with the appearance of amyloid fibrils. Such a finding is of particular interest when discussed in relation to (i.) Cases where no fibrils are seen but disease is present (35–37,39,42) (ii.) Cases where fibrils are extensively present but no disease is manifested (3,40) until disease results from a substantial physical mass of amyloid leading to tissue rupture and necrosis (3,15,16).
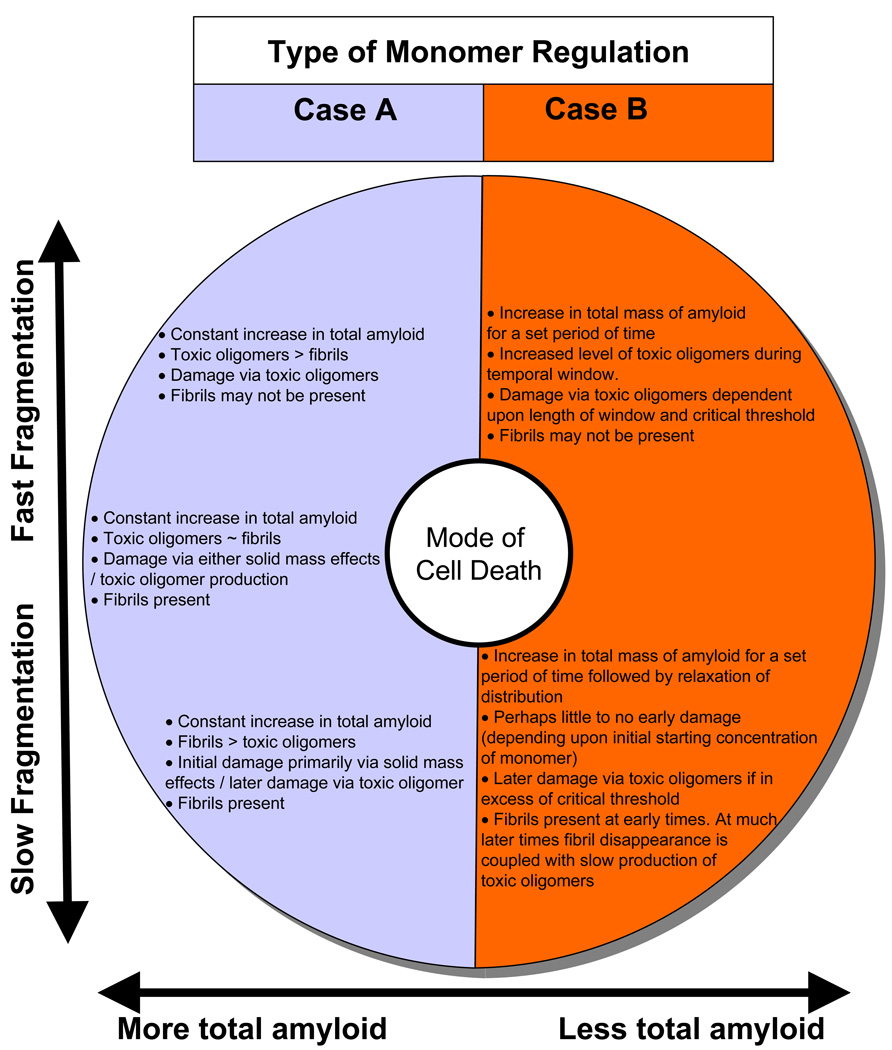
We explore the ramifications of such possible coupling/uncoupling between the production of toxic oligomers and the generation of longer amyloid fibres in Fig. 5. We note that our model provides an operative switch between the given modes of cell death and tissue injury posited in the introduction. These different modes ranged from the specific malefaction of toxic oligomers to general deleterious physical effects related to the growth of solid masses. For instance our model indicates that a fast scission case A regime would effect cell death as a result of a constant increase in the concentration of toxic oligomer. Alternatively cell death from a slow scission case A regime may occur initially from the significant mass of amyloid produced. If the surrounding cells/tissue did not succumb to this initial challenge then it may be defeated at longer time scales when cell death would result from the production of significant amounts of toxic oligomer produced via fragmentation. As indicated in Fig. 5 for the case B monomer regulation regime the total mass of amyloid produced will always be less than for the case A type system (given identical rate parameters and the same initial free concentration of monomer). Therefore for fast scission case B type systems, cell death would most likely result from the production of toxic oligomers (if occurring at a level above the critical threshold and for a long enough time during the temporal window of toxicity). Such toxic oligomer production would likely proceed with few, if any, long amyloid fibrils being present. For a slow scission case B regime little cell death/tissue damage would be expected over a long period of time despite the appearance of long amyloid fibres. However cell death due to toxic oligomer production may proceed at a much later time due to the fragmentation of these fibres if the level of toxic oligomers produced exceeded the required critical threshold for a sufficient period of time.
Fig. 5.
The model outlined in this paper provides a convenient switch between the different causative modes of amyloid related cell death due to (i.)toxic oligomer effects or (ii.) solid mass effects. Comments within the coloured circle make reference to the type of monomer regulation i.e. case A or case B type systems.
In this work we have explored the two extremes of cellular/physiological regulation of monomer concentration in the hope that the truth may lie somewhere between these two limiting cases3. In the first case we considered the situation in which the free monomer concentration was maintained at a fixed level irrespective of the degree of amyloid formation. Such a case might represent disease states for which the monomer has a required biological function such as is the case in systemic amyloidosis involving immunoglobulin light chain (3,43) or familial amyloidosis involving transthyretin (44). In the second case we considered the situation where the system was closed and the total amount of monomer in the system was a constant irrespective of the structural form (amyloid or free monomer) in which the protein existed. This second case might be descriptive of amyloid prions in yeast, the phenotype of which is only displayed upon near complete conversion of the monomer to the amyloid form (45,46). Importantly, for both limiting cases of monomer regulation we found the same general behaviour with regards to association between the variation of the fibril fragmentation rate and the variation in the degree of coupling (or association) of the time scales for production of toxic oligomers and formation of long amyloid fibrils. One possibly important difference that was observed between the two cases of monomer regulation was that for situations more reflective of a fixed total concentration of total monomer (Case B) a time dependent maximum in the production of toxic oligomer was seen to occur. The existence of such a time dependent maximum may have important therapeutic implications if the pertinent cell or tissue cannot withstand exposure to a certain concentration of toxic oligomer beyond a set critical window of time. If such a situation was indeed the case we would have to introduce a kinetic consideration to the starting of an anti-amyloid forming drug regimen (47).
Another important finding that pertains to both cases of monomer regulation is that for parameter regimes reflective of the situation in which polymer fragmentation is slow in relation to polymer formation there will exist two vastly different time scales governing (i.) the formation of amyloid, (ii.) the relaxation of the amyloid size distribution. The existence of two such disparate time scales means that the polymer distribution event may occur over a time scale that is many orders of magnitude greater than the initial polymerization event. Indeed this second relaxation process may occur so slowly as to be apparently invisible to researchers conducting experiments over realistically accessible time periods. Also it may be difficult to discern from comparison of theoretical amyloid size distributions garnered by numerical integration of rate equations over a non-logarithmic time scale4.
Although our explicit rate model captures many realistic features of the amyloid polymerization event one aspect of the reaction that it is incapable of accounting for is the possible growth of amyloid fibrils by the annealing of smaller fibrils. Such a process has been considered by McCammon and others using a closed rate model for the polymerization of G-actin (48). A similar averaged rate model has recently been applied to the description of amyloid formation by Schuck et. al. (49). Another model also inclusive of fibril annealing was developed by Pallito and Murphy (50) for the description of amyloid formation kinetics exhibited by the A-β peptide. If such annealing is shown to be a common property of medically important amyloid forming systems then annealing may provide an alternative mechanism for decoupling the production of toxic species from the appearance of long amyloid fibrils. In the future we hope to extend our explicit rate model to incorporate such possible annealing to determine to what extent this additional factor will complicate matters.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Diabetes Digestive and Kidney Diseases. This research was also supported by the Japanese Science and Technology Agency (JST) and the University of Tsukuba under the special coordinated scheme ‘Funds in Aid for the Promotion of Young Scientists’ Independent Research.’
Appendix 1: Justification of mathematical approximation for breakage
Here we describe the basic components of our explicit rate model based on nucleated growth. Explicit solution of the distribution at all time points and for all sizes for the model described in our paper would rely upon numerical integration of the complete set of equations described by Eqn. A1.
| [A1a] |
| [A1b] |
The simplest approximation for the scission rate constant describing the separation of a species of any size z into two species of size any size x (where x < z) and z-x is ks(x,z-x) = ks°. Although computationally possible the solution becomes quite slow for large distribution sizes due to the fact that the number of calculations at each time point will increase as ~Nmax2 where Nmax is the maximum number of discrete species in solution. In order to make calculation more rapid we have adopted a step function type approximation in which the fragmentation pathway of any species of size z is considered as shown by Eqn. A2.
| [A2a] |
where,
| [A2b] |
The terms rl and rh respectively denoting the low and high rounding to the nearest integer. During a time interval Δt, a species of size z and molar concentration CZ will suffer a decrease in number concentration due to its own fragmentation, resulting in an incremental molar decrease of that species concentration of ΔCZ (Eqn. A3a). Due to the flat probability distribution associated with an assumed fragmentation rate constant that is independent of position, each particular species resulting from the fragmentation event will be produced at statistically equal concentrations, denoted ΔC0 (Eqn. A3b).
| [A3a] |
| [A3b] |
It is noted that the step approximation described in Eqn. A2 satisfies the colligative conservation criteria in a manner similar to the exact equation (Eqn. A4).
| [A4a] |
| [A4b] |
We additionally note that the step approximation satisfies the required mass conservation criteria (Eqn. A5).
| [A5a] |
| [A5b] |
By explicit accounting for the rate of formation and loss of all species by fibril fragmentation within the confines of the step approximation we develop an approximate alternative to the exact equation which similarly provides full distribution time evolution information and whose computing time scales5 with ~ Nmax
| [A6a] |
| [A6b] |
For the simulation of amyloid formation we have adopted a parameter convention for nucleated growth whereby a critical nucleus oligomer size, n, is assigned. All association steps involving oligomers of degree lower than n are considered to be governed by a rate constant kN. Conversely all steps involving oligomers of degree greater than or equal to n are considered to be governed by a rate constant kG.
Appendix 2: Justification of the basic correctness of the solution method
We compare certain averaged properties of our explicit rate model (total mass of amyloid as a function of time and average degree of polymerization as a function of time) against an alternative method of solutionA1 based on the summation (with subsequent collapse and cancellation of terms) of the all the rate equations involved in an explicit description of a nucleated polymerization reaction featuring internal fragmentation. Although containing less information than our explicit rate model, in that it is only capable of providing averaged properties and not full distribution information, this alternative approach does not rely upon the step function approximation adopted in our solution method for coping with the multitudinous potential pathways of the fragmentation event and therefore provides a good check of our model’s basic correctness. The rate equations resulting from such an approach for the description of uni-directional amyloid growth, and bi-directional dissociation are as follows
| [A7a] |
| [A7b] |
Here CN refers to the number concentration of all amyloid species, CA the equivalent concentration of monomers existing in all amyloid forms and C1 the operative concentration of free monomer. As can be seen (App. Fig. 1) in the present case the level of agreement between the predictions of our explicit rate model and the limited averaged parameter model is excellent at both the level of total amyloid mass and the average degree of polymerization for both A and B limiting cases of monomer regulation6.
App. Fig. 1.
Cross validation of the method used in this paper to explicitly simulate the amyloid distribution information in a system undergoing nucleated polymerization in combination with fibril fragmentation/scission against a limited alternative procedure that is capable of only predicting averaged properties of the growing amyloid system. With reference to Case A monomer conditions the left hand column displays the solutions provided by the two alternative procedures for calculating the time dependence of (A) the summed total of all amyloid forms (green – explicit model, black – limited model), (B) the amyloid’s average degree of polymerization (blue – explicit model, red – limited model). The right hand column describes the similar information for the Case B monomer conditions. The scission rates considered were as follows (units s−1) 5×10−6, 4×10−6, 3×10−6, 2×10−6, 1×10−6, 1×10−7, 1×10−8, 1×10−9.
Appendix 3: Model findings in the light of results from molecular simulations
A general feature of non-specific protein aggregation reactions is the potential lack of a unique mechanism – meaning that any particular aggregate of a chosen size may be constituted as any one of a number of possible structural isomers. Dependent upon the interplay between solution conditions and the protein sequence, certain reaction pathways may be disfavoured thereby channelling the aggregation reaction into a more defined pathway. With the decrease in pathway complexity there is a concomitant increase in the degree of structural order and a lessening of the number of potential isomeric states associated with each particularly sized aggregate. Amyloid formation is an interesting protein aggregation reaction from this perspective as the specific amyloid reaction may proceed amidst other competing reactions or it may proceed as a conjunction of non-ordered and ordered processesA2. With regard to this point we note that a molecular level simulation procedure such as Monte Carlo (MC) or Molecular Dynamics (MD) may provide an alternative information stream on which to assess the basis of our argument. We deal with this point by first pointing out the practical necessity of approaching the problem in the manner which we did. Finally we finish with a limited review of the molecular level amyloid simulation literature highlighting some key findings which relate to our argument.
Practical necessity of a rate equation based description
In this study we were interested in examining the likely time dependence of the build up of small amyloid oligomers stemming from fibril fragmentation occurring concurrently with fibril formation. We examined three limiting case behaviours, zero fragmentation rate, fragmentation rates that were relatively fast in relation to the time course of aggregation and fragmentation rates slow in relation to the time course of aggregation. A full-scale molecular level representation of the problem would require sufficient protein/peptide monomeric units to have a statistical representation (> 10) of each length of fibril. This would require a sufficient starting concentration of monomers to allow the generation of multiple nucleation centres. A quick examination of the average degree of polymerizations reached in this study reveals that the computational burden of such a computer experiment would be too great for current technology to carry from beginning to end. However one possible route for incorporating smaller particle number and smaller time period MD simulation runs into a traditional rate equation model was recently suggestedA3. Such a combination approach in which MD simulation is used to both help determine mechanismA4 and provide estimates of the molecular rate constantsA3,A5 may indeed prove useful in the future.
Some key findings from recent molecular level amyloid simulation studies
Where molecular level simulation has been particular insightful has been in shining light on the factors that affect peptide/protein amyloidogenicity. From simple particle models to more complex MD based calculations, consideration of the particle nature of the aggregating peptide has offered clues as to the effect of polypeptide chain lengthA3, amyloidogenic peptide position within a larger polypeptide sequenceA3, salt bridgesA4, amino acid mutationA6 and added solution components such as trifluoroethanolA7. Particle level simulation has also helped to postulate a range of possible early oligomeric structuresA8,A9 and to identify which of these structures may have a toxic roleA10,A11.
Appendix 4: Justification of mechanistic assumptions present within the model
The central mechanistic assumption of the current work which is that fibrils undergo internal breakage. With regard to this point a range of amyloid and amyloid like fibres have been shown to undergo breakage both in vitroA1, A12–A15 and in vivoA16,A17. These studies and others have also shown that subjecting amyloid fibres to either sonication or shearing forces resulting from rapid stirring can stimulate fibre fracture rates and enhance the rate of amyloid growth. Although these findings do not necessarily carry the weight of an inductive proof they do suggest that internal breakage events may not be an uncommon property of amyloid fibres. Furthermore it is known that there exists chaperone proteins within the cell that have ‘dis-aggregase’ properties, i.e. they are capable of accelerating the rate of fibre breakage in an energy dependent mannerA18,A19. We note that within the bounds of the simple model presented (and its even simpler representation by Eqn. A7) we may isolate the conditions (in terms of rate constant and monomer concentration values) for significant amyloid production for Case B monomer production by first setting dCA/dt = 0 in Eqn. 7b and solving for CN and then substituting this result into Eqn. 7a after setting dCN/dt = 0 and then solving for CA. The effect of variation of the scission rate constant can be noted in Appendix Fig. 1.
A second mechanistic assumption present within our modelling approach is that of a site breakage rate that is independent of position for an assumed linear fibre having identical internal bonding patterns. Stated more simply this assumption means that the probability distribution for breakage as a function of position is flat. Whether or not there exists a more complicated position dependence to rate of breakage depends upon the structure of the amyloid fiberA20 (for instance the bonding arrangements in helical fibres will exhibit pronounced end effects due to different number of bonding contacts formed between end located monomers and internally located monomers) and the designated mode of fibril breakage (for instance breakage occurring due to increased temperature would be expected to be governed by a different probability distribution than would breakage effected by a shear fieldA13). Simulation of the amyloid polymerization kinetics with a different position dependence would certainly yield different kinetic results in the particular – however the central implication of this study, that amyloid fibrils once formed may act as a reservoir of toxic oligomer production (or toxic oligomer precursors), would remain unchanged.
The third mechanistic assumption in our model is that the toxic oligomer region is an on-pathway species. Although we did stipulate in the text that we felt that this assumption was not critical we elaborate on the robustness of our model findings in light of the possibility that toxic oligomers are off pathway species. Generally speaking if an off pathway oligomer was the toxic form then it would be separated from the on pathway oligomeric forms via either a kinetic barrier (Eqn. A8a) or an equilibrium barrier (Eqn. A8b).
| [A8a] |
| [A8b] |
For either case we might surmise that the concentration of toxic off pathway species would be related to the concentration of the relevant precursor on pathway species by a transformation rate or equilibrium constant (Eqn. A9) thereby extending the arguments made in this paper specifically for the on pathway toxic oligomer form to off pathway toxic oligomer forms.
| [A9a] |
| [A9b] |
A special case of off pathway toxicity with specific reference to Alzheimer’s disease has been suggestedA21 that relates to an alternative mechanism of possible toxicity due to the non A-β peptide fragment of the A-β pro-polypeptide that is liberated upon enzymatic cleavage of the propolypeptide by surface bound γ-secretase. As this cleavage step is both essentially irreversible and occurs prior to amyloid formation it would not be influenced by any subsequent change in the aggregation state of the amyloid and therefore is not at all explainable by this model which deals only with possible toxicity of oligomers generated during the general process of amyloid formation.
The final mechanistic assumption within our model is the description of two limiting cases of monomer regulation, one corresponding to a case of amyloid formation that is slow on the time scale of monomer regulation (Case A) and one corresponding to a case of amyloid formation that is fast on the time scale of monomer regulation. We might more stringently codify these definitions using the following (massively oversimplified) scheme (Eqn. A10) to depict the general features of regulation of monomer (M) concentration in a system capable of amyloid formation from monomer M (governed by a first order rate constant kA (s−1)) that is occurring in conjunction with monomer formation from some constant precursor (α) (regulated by a first order rate constant kF (s−1)) and monomer breakdown to species β (governed by a first order rate constant kB (s−1)).
| [A10] |
The rate of monomer formation and loss from the precursor (at constant concentration Cα) can be written as Eqn. A11a and the integrated solution is present as Eqn. A11b.
| [A11a] |
| [A11b] |
If the relative rates of the system are such that kACM(t=0) << kBCM(t=0) and kACM(t=0) << kFCα the time dependent concentration of monomer is approximately constant and is given by CM(t) ~ CM(t=0). If alternatively the rates are such that kACM(t=0) >> kBCM(t=0) and kACM(t=0) >> kFCα then the time dependent concentration of monomer is approximately given by CM(t)~CM(t=0)exp(−kAt). Appendix Fig. A2 makes this point clear with these two extreme cases along with some intermediate examples where kACM(t=0) ~ kBCM(t=0) ~ kFCα. It was our hope in this paper to encapsulate the widest range of possible behaviors by explicitly simulating the two extremum. In likening an experimental system to either case we are relying upon a limit argument i.e. real experimental systems will more closely resemble one of the two extreme forms thus displaying features reflective of that particular case. In Fig. A2 we try to demonstrate as much by simulating the two extreme case behaviors along with a number of intermediate cases. So although we cited the transthyretin and immunoglobulin light chain amyloid diseases and the yeast Sup 35 and Ure2 protein based prion systems as respective examples of case A and B they will not fully conform to these limiting case requirements. Interestingly, an anonymous reviewer of this manuscript has suggested that neuronal loss as a result of Alzheimer’s disease (A22) might constitute an example of a system having some similarity to a Case B system. The reasoning behind this suggestion is that neuronal cell death would remove the source of A-β production. However this notion may be complicated by the fact that other nearby healthy cells will continue with, and may even up-regulate, their A-β production. Furthermore we have no firm idea of the differential rate of monomer and fibril breakdown as nearby cells die and let loose their complement of proteases and degrading enzymes etc. into the intercellular space. As of the present time we are unable to reach agreement on the basic correctness of the reviewer’s hypothesis but due to its potential importance in stimulating discussion on the topic of amyloid formation kinetics in Alzheimer’s disease we have decided to include it in this appendix.
App. Fig. 2.
Graphical representation of monomer regulation as codified by Eqn A10 and A11. We see the relative decay of monomer from its starting value for the cases where kA* = 0, kF* = kB* = 1 (blue line); kA* = 100, kF* = kB* = 0 (red line); kA* = 0.1, kF* = kB* = 1 (green dotted line); kA* = 1, kF* = kB* = 1 (green solid line) and kA* = 100, kF* = kB* = 1 (green dashed line). The two extreme cases considered in this paper are represented by the red line (Case B) and the blue line (Case A). Rate constants and time have been scaled by arbitrary characteristic values kc and tc such that kA* = kA/kc, kB* = kB/kc, kF* = kF/kc and t* = t/tc. The relative starting concentration was set to kF*Cαα/kB* with kF* = kB* = 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Our finding (29) that phenotypic markers of amyloid could result from an induced change affecting amyloid’s structural/replicative properties had a number of important consequences. Chief amongst these was that amyloid related disease could develop from modification of an existing phenotypically silent amyloid formation process rather than because of the emergence of a previously non-existent amyloid pathway. A year after our paper was published this conceptual linkage was demonstrated experimentally by Tanaka et. al. (51).
Although we have limited our discussion to on-pathway intermediates it is interesting to note that the argument is applicable to off pathway intermediates, (such as spherulites or circularized loops) if they are themselves the products of small on pathway oligomers
By choosing these extremes we encompass relevant situations of protein synthesis and degradation (see Appendix 4).
In reference 29 we concluded that for a constant total concentration of protein for the case of a slow scission/fragmentation rate (ks° = 1×10−9s−1) the amyloid size distribution would establish an equilibrium around an average dop of ≈ 50 monomers. In this work we see that this distribution will relax back to an average dop of ≈ 3 monomers over a time scale approximately 100 times longer than the initial polymerization event. For the alternative monomer regulation situation in which the free concentration of monomer is a constant, a large stable size distribution will be attained and maintained
For very large systems of equations a second step approximation may be desirable although as seen in Appendix 2 such an additional approximation was not required in this work.
There is a uniform multiplicative factor of approximately 2 between the time scales reported here and our earlier work reported in (29).
Contributor Information
Damien Hall, Email: damienhall@md.tsukuba.ac.jp.
Herman Edskes, Email: edskes@helix.nih.gov.
References
- 1.Baxa U. Structural Basis of Infectious and Non-Infectious Amyloids. Current Alzheimer Research. 2008;5:308–318. doi: 10.2174/156720508784533367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 3.Pepys MB. Amyloidosis. Annu. Rev. Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 4.Meredith SC. Protein Denaturation and Aggregation: Cellular Responses to Denatured and Aggregated Proteins. Ann. N.Y. Acad. Sci. 2005;1066:181–221. doi: 10.1196/annals.1363.030. [DOI] [PubMed] [Google Scholar]
- 5.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 6.Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch. Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 7.Glenner GG, Wong CW. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Com. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 8.Selkoe DJ. Amyloid β-peptide is produced by cultured cells during normal metabolism: a reprise. J. Alzheim. Dis. 2006;9:163–168. doi: 10.3233/jad-2006-9s319. [DOI] [PubMed] [Google Scholar]
- 9.Chiang PK, Lam MA, Luo Y. The many faces of amyloid-β in Alzheimer’s disease. Curr. Mol. Med. 2008;8:580–584. doi: 10.2174/156652408785747951. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich JC, Ratner IM. Amyloidosis of the islets of the Langerhans. A restudy of islet hyaline in diabetic and non-diabetic individuals. Am. J. Pathol. 1961;38:49–59. [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien TD, Butler PC, Westermark P, Johnson KH. Islet amyloid polypeptide: a review of its biology and potential roles in the pathogenesis of diabetes mellitus. Vet. Pathol. 1993;30:317–332. doi: 10.1177/030098589303000401. [DOI] [PubMed] [Google Scholar]
- 12.Haatja L, Gurlo T, Huang CJ, Butler PC. Islet Amyloid in Type 2 Diabetes, and the Toxic Oligomer Hypothesis. Endocrine Rev. 2008;29:303–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winderickx J, Delay C, De Vos A, Klinger H, Pellens K, Vanhelmont T, Van Leuven F, Zabrocki P. Protein folding diseases and neurodegeneration: Lessons learned from yeast. Biochim. Biophys. Acta. 2008;1783:1381–1395. doi: 10.1016/j.bbamcr.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Resende R, Ferreiro E, Pereira C, Resende de Oliveira C. Neurotoxic effect of oligomeric and fibrillar species of amyloid-β peptide 1–42: involvement of endoplasmic reticulum calcium release in oligomer induced cell death. Neuroscience. 2008;155:725–737. doi: 10.1016/j.neuroscience.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 15.Sueyoshi E, Sakamoto I, Okimoto T, Hayashi K, Tanaka K, Toda G. Cardiac Amyloidosis: Typical Imaging Findings and Diffuse Myocardial Damage Demonstrated by Delayed Contrast-Enhanced MRI. Cardiovasc. Intervent. Radiol. 2006;29:710–712. doi: 10.1007/s00270-004-0162-x. [DOI] [PubMed] [Google Scholar]
- 16.Shiels SA, Hasan SI, Darowski A. Collapse in a 79-year-old: a rare case of amyloid of the pelvis. Age and Aging. 2005;34:648–650. doi: 10.1093/ageing/afi161. [DOI] [PubMed] [Google Scholar]
- 17.Dobson CM. The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glabe CG. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol. Aging. 2006;27:570–575. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 19.Rahimi F, Shanmugam A, Bitan G. Structure – Function Relationships of Pre-Fibrillar Protein Assemblies in Alzheimer’s Disease and Related Disorders. Curr. Alzheim. Res. 2008;5:319–341. doi: 10.2174/156720508784533358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregori L, Fuchs C, Figueiredo-Pereira ME, Van Nostrand WE, Goldgaber D. Amyloid-β protein inhibits ubiquitin-dependent protein degradation in vitro. J. Biol. Chem. 1995;270:19702–19708. doi: 10.1074/jbc.270.34.19702. [DOI] [PubMed] [Google Scholar]
- 21.Cecarini V, Bonfili L, Amici M, Angeletti M, Keller JN, Eleuteri AM. Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Res. 2008;1209:8–18. doi: 10.1016/j.brainres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Csermely P. Chaperone overload is a possible contributor to civilization diseases. Trends Genet. 2001;17:701–704. doi: 10.1016/s0168-9525(01)02495-7. [DOI] [PubMed] [Google Scholar]
- 23.Söti C, Csermely P. Protein stress and stress proteins: implications in aging and disease. J. Biosci. 2007;32:511–515. doi: 10.1007/s12038-007-0050-z. [DOI] [PubMed] [Google Scholar]
- 24.Wilson MR, Yerbury JJ, Poon S. Potential roles of abundant extracellular chaperones in the control of amyloid formation and toxicity. Mol. BiolSyst. 2008;4:42–52. doi: 10.1039/b712728f. [DOI] [PubMed] [Google Scholar]
- 25.Smith DG, Cappai R, Barnham KJ. The redox chemistry of the Alzheimer's disease amyloid-β peptide. Biochim Biophys Acta. 2007;1768:1976–1990. doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Mattson MP, Mattson EP. Amyloid peptide enhances nail rusting: novel insight into mechanisms of aging and Alzheimer's disease. Ageing Res. Rev. 2002;1:327–330. doi: 10.1016/s1568-1637(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 27.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 28.Lashuel HA, Lansbury PT., Jr Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 29.Hall D, Edskes H. Silent prions lying in wait: a two-hit model of prion/amyloid formation and infection. J. Mol. Biol. 2004;336:775–786. doi: 10.1016/j.jmb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U.S.A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prusiner SB. In: Nobel Lectures, Physiology or Medicine 1996–2000. Jörnvall Hans., editor. Singapore: World Scientific Publishing Co.; 2003. pp. 74–129. [Google Scholar]
- 32.Wickner RB, Shewmaker F, Kryndushkin D, Edskes HK. Protein inheritance (prions) based on parallel in-register beta-sheet amyloid structures. Bioessays. 2008;30:955–964. doi: 10.1002/bies.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baskakov IV. Branched chain mechanism of polymerization and ultrastructure of prion protein amyloid fibrils. FEBS Journal. 2007;274:3756–3765. doi: 10.1111/j.1742-4658.2007.05916.x. [DOI] [PubMed] [Google Scholar]
- 34.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. (U.S.A.) 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartley DM, Walsh DM, Ye CP, Diehl T, Vasquez S, Vassilev PM, Teplow DB, Selkoe DJ. Protofibrillar intermediates of amyloid beta-protein induce acute electrophysiological changes and progressive neurotoxicity in cortical neurons. J Neurosci. 1999;19:8876–8884. doi: 10.1523/JNEUROSCI.19-20-08876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 37.Malisauskas M, Ostman J, Darinskas A, Zamotin V, Liutkevicius E, Lundgren E, Morozova-Roche LA. Does the cytotoxic effect of transient amyloid oligomers from common equine lysozyme in vitro imply innate amyloid toxicity? J. Biol. Chem. 280:6269–6275. doi: 10.1074/jbc.M407273200. [DOI] [PubMed] [Google Scholar]
- 38.Katzman R, Terry R, DeTeresa R, Brown T, Davies P, Fuld P, Renbing X, Peck A. Clinical, pathological, and neurochemical changes in dementia: a subgroup with preserved mental status and numerous neocortical plaques. Ann. Neurol. 1988;23:138–144. doi: 10.1002/ana.410230206. [DOI] [PubMed] [Google Scholar]
- 39.Berg L, McKeel DW, Jr, Miller JP, Storandt M, Rubin EH, Morris JC, Baty J, Coats M, Norton J, Goate AM, Price JL, Gearing M, Mirra SS, Saunders AM. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998;55:326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 40.Westermark P. The nature of amyloid in islets of Langerhans in old age. New York: Academic Press; 1976. [Google Scholar]
- 41.Westermark P. Fine structure of islets of Langerhans in insular amyloidosis. Virchows. Arch. A. Pathol. Anat. 1973;359:1–8. doi: 10.1007/BF00549079. [DOI] [PubMed] [Google Scholar]
- 42.Oddo S, Caccamo A, Tran L, Lambert MP, Glabe CG, Klein WL, LaFerla FM. Temporal profile of amyloid-beta (Aβ) oligomerization in an in vivo model of Alzheimer disease. A link between Aβ and tau pathology. J Biol Chem. 2006;281:1599–1604. doi: 10.1074/jbc.M507892200. [DOI] [PubMed] [Google Scholar]
- 43.Sanchorawala V. Light-chain (AL) amyloidosis: diagnosis and treatment. Clin. J. Am. Soc. Nephrol. 2006;1:1331–1341. doi: 10.2215/CJN.02740806. [DOI] [PubMed] [Google Scholar]
- 44.Hou X, Aguilar MI, Small DH. Transthyretin and familial amyloidotic polyneuropathy. Recent progress in understanding the molecular mechanism of neurodegeneration. FEBS. J. 2007;274:1637–1650. doi: 10.1111/j.1742-4658.2007.05712.x. [DOI] [PubMed] [Google Scholar]
- 45.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 46.Wickner RB, Edskes HK, Ross ED, Pierce MM, Baxa U, Brachmann A, Shewmaker F. Prion genetics: new rules for a new kind of gene. Annu Rev Genet. 2004;38:681–707. doi: 10.1146/annurev.genet.38.072902.092200. [DOI] [PubMed] [Google Scholar]
- 47.Becker RE, Greig NH. Alzheimer's disease drug development in 2008 and beyond: problems and opportunities. Curr. Alzheimer Res. 2008;5:346–357. doi: 10.2174/156720508785132299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrianantoandro E, Blanchoin L, Sept D, McCammon JA, Pollard TD. Kinetic mechanism of end-to-end annealing of actin filaments. J. Mol. Biol. 2001;312:721–730. doi: 10.1006/jmbi.2001.5005. [DOI] [PubMed] [Google Scholar]
- 49.Binger KJ, Pham CL, Wilson LM, Bailey MF, Lawrence LJ, Schuck P, Howlett GJ. Apolipoprotein C-II amyloid fibrils assemble via a reversible pathway that includes fibril breaking and rejoining. J. Mol. Biol. 2008;376:1116–1129. doi: 10.1016/j.jmb.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 50.Pallito MM, Murphy RM. A mathematical model of the kinetics of beta-amyloid fibril growth from the denatured state. Biophys J. 2001;81:1805–1822. doi: 10.1016/S0006-3495(01)75831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
Appendix References
- (A1).Smith JF, Knowles TP, Dobson CM, Macphee CE, Welland ME. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A2).Nguyen HD, Hall CK. Kinetics of Fibril Formation by Polyalanine Peptides. J. Biol. Chem. 2005;280:9074–9082. doi: 10.1074/jbc.M407338200. [DOI] [PubMed] [Google Scholar]
- (A3).Hall D, Hirota N. Multi-scale modelling of amyloid formation from unfolded proteins using a set of theory derived rate constants. Biophys Chem. 2009;140:122–128. doi: 10.1016/j.bpc.2008.11.013. (2009) [DOI] [PubMed] [Google Scholar]
- (A4).Reddy G, Straub JE, Thirumalai D. Influence of Preformed Asp23-Lys28 Salt Bridge on the Conformational Fluctuations of Monomers and Dimers of A-β Peptides with Implications for Rates of Fibril Formation. J. Phys. Chem. B. 2009;113:1162–1172. doi: 10.1021/jp808914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A5).Wang J, Tan C, Chen HF, Luo R. All-Atom Computer Simulations of Amyloid Fibril Disaggregation. Biophys. J. 2008;95:5037–5047. doi: 10.1529/biophysj.108.131672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A6).Murray MM, Krone MG, Bernstein SL, Baumetner A, Condron MM, Lazo ND, Teplow DB, Wyttenbach T, Shea JE, Bowers MT. Amyloid-β-Protein: Experiment and Theory on the 21–30 Fragment. J. Phys. Chem. B. 2009;113:6041–6046. doi: 10.1021/jp808384x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A7).Standley DM, Yonezawa Y, Goto Y, Nakamura H. Flexible docking of an amyloid-forming peptide from β2-microglobulin. FEBS Letters. 2006;580:6199–6205. doi: 10.1016/j.febslet.2006.10.022. [DOI] [PubMed] [Google Scholar]
- (A8).Zheng J, Jang H, Ma B, Nussinov R. Annular Structures as Intermediates in Fibril Formation of Alzheimer A-β. J. Phys. Chem. B. 2008;112:6856–6865. doi: 10.1021/jp711335b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A9).Song W, Wei G, Mousseau N, Derreumaux P. Self-Assembly of the β2-Microglobulin NHVTLSQ Peptide Using a Coarse-Grained Protein Model Reveals a β-Barrel Species. J. Phys. Chem. B. 2008;112:4410–4418. doi: 10.1021/jp710592v. [DOI] [PubMed] [Google Scholar]
- (A10).Friedman R, Pellarin R, Caflisch A. Amyloid Aggregation on Lipid Bilayers and its Impact on Membrane Permability. J. Mol. Biol. 2009;387:407–415. doi: 10.1016/j.jmb.2008.12.036. [DOI] [PubMed] [Google Scholar]
- (A11).Jang H, Zheng J, Lal R, Nussinov R. New Structures help the modelling of toxic amyloid-β ion channels. 2008;33:91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- (A12).Baskakov IV. FEBS Journal. ‘Branched chain mechanism of polymerization and ultrastructure of prion protein amyloid fibrils’. 2007;274:2756–3765. doi: 10.1111/j.1742-4658.2007.05916.x. [DOI] [PubMed] [Google Scholar]
- (A13).Guerin G, Wang H, Manners I, Winnik MA. Fragmentation of Fiberlike Structures: Sonication Studies of Cylindrical Block Copolymer Micelles and Behavioral Comparisons to Biological Fibrils. J. Am. Chem. Soc. 2008;130:14763–14771. doi: 10.1021/ja805262v. [DOI] [PubMed] [Google Scholar]
- (A14).Binger KJ, Pham CL, Wilson LM, Bailey MF, Lawrence LJ, Schuck P, Howlett GJ. Apolipoprotein C-II amyloid fibrils assemble via a reversible pathway that includes fibril breaking and rejoining. J. Mol. Biol. 2008;376:1116–1129. doi: 10.1016/j.jmb.2007.12.055. [DOI] [PubMed] [Google Scholar]
- (A15).Ohhashi Y, Kihara M, Naiki H, Goto Y. Ultrasonication-induced amyloid fibril formation of beta2-microglobulin. J. Biol. Chem. 2005;280:32843–32848. doi: 10.1074/jbc.M506501200. [DOI] [PubMed] [Google Scholar]
- (A16).Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- (A17).Kushnirov VV, Vishnevskaya AB, Alexandrov IM, Ter-Avanesyan MD. Prion and nonprion amyloids: a comparison inspired by the yeast Sup35 protein. Prion. 2007;1:179–184. doi: 10.4161/pri.1.3.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A18).Cox B, Ness F, Tuite M. Analysis of the gerneration and separtation of propagons: entities that propagate the [PSI+]prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (A19).Chernoff YO, Lindquist SL, Ono B, Ingevecthtomov SG, Liebmann SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- (A20).Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- (A21).Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–999. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (A22).Chiang PK, Lam MA, Luo Y. The many faces of amyloid-β in Alzheimer’s disease. Curr. Mol. Med. 2008;8:580–584. doi: 10.2174/156652408785747951. [DOI] [PubMed] [Google Scholar]