Abstract
Alphaviruses are small, spherical, enveloped, positive-sense ssRNA viruses responsible for a considerable number of human and animal diseases. Alphavirus members include Chikungunya virus, Sindbis virus, Semliki Forest virus, the western, eastern and Venezuelan equine encephalitis viruses, and the Ross River virus. Alphaviruses can cause arthritic diseases and encephalitis in humans and animals and continue to be a worldwide threat. The viruses are transmitted by blood-sucking arthropods, and replicate in both arthropod and vertebrate hosts. Alphaviruses form spherical particles (65–70 nm in diameter) with icosahedral symmetry and a triangulation number of four. The icosahedral structures of alphaviruses have been defined to very high resolutions by cryo-electron microscopy and crystallographic studies. In this review, we summarize the major events in alphavirus infection: entry, replication, assembly and budding. We focus on data acquired from structural and functional studies of the alphaviruses. These structural and functional data provide a broader perspective of the virus lifecycle and structure, and allow additional insight into these important viruses.
Keywords: arbovirus, replicon, RNA synthesis, RNA virus, togavirus, virion assembly
The alphaviruses are small, enveloped, plus-strand RNA viruses. The genus has more than 40 recognized members, and is responsible for human and animal diseases that cause symptoms such as fever, rash and arthritis [1]. Well-studied members include Sindbis virus (SINV), Semliki Forest virus (SFV), Venezuelan equine encephalitis virus (VEEV) and Ross River virus (RRV). Recently, the Chikungunya species, characterized by symptoms such as rashes and joint pain, has received attention owing to increasing global spread, including cases in Europe [2]. Alphaviruses are transmitted by blood-sucking arthropods, typically the mosquito, and replicate in both arthropod and vertebrate hosts worldwide. Members of the genus are categorized into either the New World or Old World group based upon the area in which they are found and the disease they cause. Alphaviruses are considered a model system for structural studies of enveloped viruses. Alphaviruses form spherical particles of 65–70 nm in diameter, and the icosahedral structures of many alphaviruses have been defined to very high resolutions by cryo-electron microscopy (cryo-EM) and crystallographic studies, revealing details of the interactions between the structural proteins [3,4]. The genome is composed of a single strand of positive-sense RNA approximately 11.5 kb in length. The RNA encodes four nonstructural proteins involved in virus replication and pathogenesis, and five structural proteins that compose the virion [5]. Recent in vitro and in vivo biochemical and functional studies have allowed additional elucidation of the virus life-cycle and pathogenesis. Furthermore, the efficiency with which many of these viruses replicate, coupled with the broad range of susceptible and permissive hosts, has allowed these viruses to be used as tools in heterologous gene expression and gene therapy delivery vectors [6]. Taken together, recent discoveries within the alphavirus field have provided a broader perspective of the virus lifecycle and may allow for additional control as well as further exploitation of these important viruses.
Classification
The family Togaviridae is comprised of two genera: Alphavirus and Rubivirus. The Rubivirus genus is composed of a single member, Rubella virus, and is not discussed here. Members of the genus Alphavirus can be classified antigenically into six complexes [1,7]. The alphaviruses have classically been described as either Old World or New World viruses, depending on their geographic distribution, and it is probable that several transoceanic exchanges, likely mediated by birds, have occurred [1]. Old World viruses can often cause fever, rash, and arthritic symptoms and diseases, while the host infected with New World viruses may succumb to encephalitis. In humans and other mammals, alphavirus infection is acute and in many cases characterized by high-titer viremia, rash, fever and encephalitis until the death of the infected host or clearance of the virus by the immune system. The encephalitogenic alphaviruses, including VEEV, eastern and western equine encephalitis viruses, represent a continuous public health threat in the USA [7]. Many Old World viruses, including the Ross River, Barmah Forest, Mayaro, O'nyong-nyong, Chikungunya and Sindbis viruses, cause an arthralgia syndrome, while Highlands J virus causes dramatic decreases in egg production and mortality in domestic birds [1].
Alphaviruses are maintained in natural cycles by transmission between susceptible vectors and vertebrate hosts [5]. The vector for most alphaviruses are arthropods (typically the mosquito) in which they cause a persistent, lifelong infection with minimal effect on biological functions. The salmonid viruses, salmon pancreas disease virus and sleeping disease virus (which infects rainbow trout), present examples of alphaviruses for which arthropod transmission is unlikely [8,9]. Southern elephant seal virus, isolated from the coast of Australia [10] and grouped genetically with the SFV complex, has been isolated from the louse Lepidophthirus macrorhini [9,11]. This isolation demonstrates not only that alphaviruses can be transmitted by lice, but also that they can infect marine mammals. Although the alphaviruses and Rubella virus have been classified within the same family, the evolutionary relationship between them is obscure [12].
Structure of the virion
The 3D structure of alphaviruses has been studied by a variety of biophysical methods over many years. Structural studies of alphaviruses have consisted primarily of cryo-EM analyses of the whole virus and x-ray crystallographic examination of the component structural proteins [13]. Since the crystallization of SINV and SFV have yielded crystals that diffract to only approximately 3 nm [14], recent work that has advanced the field has come from cryo-EM and image reconstruction techniques [15]. Cryo-EM has been used to study the structure of several alphaviruses, including SINV, SFV, RRV, VEEV and Aura virus. The studies with SFV and SINV are the most advanced, with a resolution of 9 Å reported for both viruses [3,16–18]. Alphavirus particles are icosahedral structures (Figure 1A) with a diameter of approximately 700 Å, a molecular mass of 5.2 × 106 Da and a density of 1.22 g/cm3 [16,19]. The virions are composed of multiple organized shells of molecules (Figure 1B) that effectively protect and deliver the viral RNA to susceptible host cells. Alphaviruses contain one copy of an approximately 11.5-kb, positive-strand, genomic RNA that is encapsidated by capsid proteins forming an icosahedral nucleocapsid (NC). The NC is composed of the capsid protein together with the genomic RNA. The NC is enveloped in a host-derived lipid bilayer on which the viral glycoproteins are arranged in an icosahedral lattice.
Figure 1. Virion structure and structural proteins.
(A) Surface-shaded view of Sindbis virus at 9 Å resolution and viewed down at the icosahedral twofold axis. The flower-like trimeric spikes are seen in blue, and small portions of the lipid bilayer are seen in green. (B) A central cross-section of the virus particle showing the organization of the particle with the glycoproteins (blue), skirt region of the envelope (turquoise), the lipid bilayer (green) penetrated by the transmembrane helices of glycoproteins, ordered protease domain of the capsid protein (yellow), disordered protein–RNA region (orange) and RNA region (red). (C) Radial section at 208 Å of the 9 Å structure showing the nucleocapsid core viewed down at the icosahedral twofold axis. (D) Structure of the Semliki Forest virus envelope protein E1 in the monomeric conformation (protein data bank identification [PDB ID]: 2ala). The domains (DI, DII and DIII) are colored according to the scheme DI: red; DII: orange; and DIII: blue. (E) Structure of the ordered C-terminal domain of the Sindbis virus capsid protein (PDB ID: 1svp) 106–264.
Virion envelope
The virion envelope consists of a host-derived lipid bilayer in which 240 copies of E1 and E2 are embedded [5]. Smaller amounts of the membrane-associated protein, 6K (55 amino acid residues), are also found in the virus particle [20,21]. E1 and E2 interact to form a rigid structure across the membrane with a one-to-one relationship. The lipid bilayer is enriched in cholesterol and sphingolipid molecules, which are required for entry and budding [22]. E1 and E2 each have one transmembrane helix that traverses the lipid bilayer. Both El and E2 are glycosylated, but the number and position of the attached chains are not absolutely conserved among alphaviruses. The alphavirus envelope proteins are modified by palmitoylation. El and E2 of SINV and SFV have been shown to contain covalently attached palmitic acids in or near the membrane-spanning anchors [23,24]. The glycoproteins of the virus form an icosahedral lattice with T = 4 symmetry [15,25–27], and the E1 and E2 heterodimers on the surface of the virus assemble into 80 spikes (Figure 1A). The x-ray crystal structures of the ectodomain of the E1 protein (residues 1–383) of SFV have been determined [4]. The E1 ectodomain consists of three β-barrel domains (Figure 1D). Domain I contains the amino terminus and is spatially located between domains II and III. The carboxy terminus lies within domain III, and the fusion peptide is at the distal end of domain II. The E1 monomers were found to lie at the base of each of the surface spikes and form a lattice on the virus surface. Residues from Pro383 to Trp409 comprise the E1 stem region, and the transmembrane helix of E1 enters the bilayer at residue Trp409 and exits at residue Met433. The six carboxy-terminal residues of E1 extend past the inner lipid leaflet into the interior cavity of the virus [3].
E2 is a long, thin molecule, with a highly exposed leaf-like structure at the top of the spike followed by the narrower stem, which twists around the more tangentially disposed E1 molecule [13]. The first 260 amino acids of E2 constitute the ectodomain, followed by approximately 100 amino acids that form the stem region and a 30-amino-acid transmembrane helix. The carboxy-terminal domain of E2 (cdE2) consists of 33 amino acids that interact with the NC core [3,28]. There are contacts between the leaf-like structure of E2 and the distal end of the E1 glycoprotein domain II, and between the stalk portion of E2 and domains I and III of E1 [3]. The receptor attachment site near residue 218 and the carbohydrate associated with residue 216 of E2 are situated in the large, protruding, external, leaf-like surface [29]. This surface-accessible site is also the binding site for heparan sulfate (HS) in a RRV mutant [30] as well as the Fab binding site for SINV- and RRV-neutralizing antibodies [29,31–33].
pE2/E2 glycoprotein
The E1 and pE2 (precursor to the E3 and E2 proteins prior to furin cleavage) glycoproteins are assembled as heterodimers in the endoplasmic reticulum (ER). E3 is cleaved from pE2 by furin in the Golgi, and the resultant E1–E2 heterodimers are then transported to the plasma membrane. These heterodimers self-assemble into 80 trimeric spikes on the virus surface [25,34]. E1 is responsible for fusion of the viral membrane with the endosomal membrane, and E2 is involved in receptor binding and the subsequent receptor-mediated endocytosis. The 64-amino-acid E3 mediates proper folding of pE2 and controls the spike functions by interacting with the fusion protein E1. Thus, E3 is required for efficient particle assembly, mediating both spike folding and spike activation for viral entry.
The structure of the immature virus containing an uncleaved pE2 has been solved using cryo-EM [35–37]. Mutant versions of both SINV and SFV were used for independent structure determinations and yielded similar structures, essentially confirming the location of the E3 domain. The extra density corresponding to the E3 protein was found predominantly between the petals of the spike resulting in a dual-lobed petal [35]. Except for this spike morphology, the icosahedral structure of the wild-type and mutant particles is essentially similar, suggesting that following cleavage of pE2 and release of E3, no significant conformational changes occur in the general organization of virus structure. E3 has been hypothesized to have an enzymatic or functional role in virus assembly but as yet there is no evidence for this [38]. Furthermore, E3 has a central role in pE2/E1 complex formation and transport of the viral structural components to the site of budding. Replacement of SFV E3 with an artificial signal peptide abolished the spike heterodimerization and surface expression of E1 [39]. E3 has been proposed to stabilize the fusion protein as the pE2/E1 complex transits the mildly acidic environment of the Golgi [39–41]. Cleavage of E3 from the assembled spike is required to make the virus particles fusion competent. SINV virions incorporating uncleaved pE2 bind efficiently to cell-surface HS, but they are nonviable [42]. The lethality associated with the incorporation of pE2 into SINV due to mutagenic introduction of an N-linked glycosylation consensus sequence immediately adjacent to the pE2 cleavage site is known to be suppressed by second-site mutations in E3 and E2 [43].
Each SINV glycoprotein has two sites for N-linked glycosylation (E1–139, E1–245, E2–196 and E2–318) and their locations were identified by cryo-EM [28]. Elimination of either E2 glycosylation site increased replication owing to increased efficiency of binding to HS on mammalian cells [44]. HS binding can increase virulence, presumably through enhancing the replication of SINV within specific host tissues such as the brain [45]. Amino acid substitutions affecting binding to proteoglycans may differ in importance for CNS infection and viremia [46]. Depending on the strain of virus, SINV causes encephalitis, paralysis or no observable disease in mice, and the amino acid sequence of the E2 glycoprotein is an important determinant of virulence [47]. In addition, E2 has a role in determining mosquito infectivity. For example, a single Ser to Asn substitution in the cell-receptor-binding domain of E2 from VEEV caused an increased vector infectivity phenotype [48]. Functional characterization of the SINV E2 glycoprotein using transposon linker-insertion mutagenesis has described domains of E2 critical for the structural integrity of the protein, transport to the plasma membrane and virus–host interactions [49]. The organization of the E2 glycoprotein has also been studied by surface biotinylation of intact virions [50]. Seven sites of modification were identified in the E2 and one in the E1, confirming that the E1 protein is almost completely buried in the virus structure. Given that the biotinylation maintained wild-type levels of infectivity, the labeled lysines are predicted not to be directly involved in the virus–host interface or in conformational changes involved in the infection process.
E1 glycoprotein
The ectodomain of SFV E1 protein was purified as a soluble hemagglutinin and crystallized [51–54]. The prefusion structure of the SFV E1 polypeptide chain derived from a 3.5-Å electron density map [4] was fitted into the 9-Å cryo-EM reconstruction and defined the location of E1 and E2 in the viral surface. E1 makes a continuous icosahedral protein shell on the virion, covering most of the lipid membrane. The overall fold is related to the flavivirus E protein, with three structural domains disposed in the same primary sequence arrangement [4,55,56].
The E1 ectodomain postfusion homotrimer from SFV was produced by acidic treatment of E1 ectodomains in the presence of cholesterol and sphingolipid-containing target membranes [57]. Subsequently, the crystal structure of the ectodomain of the SFV E1 was determined in its low-pH-induced trimeric form [58].
The alphavirus E1 protein converts the viral surface proteins into ion-permeable pores at the plasma membrane [59] and is responsible for fusion of the viral envelope with the host endosomal membrane during virus entry. These pores are believed to have a permeability for Na+, K+ and Ca2+ ions, and allow endosomal protons to flow into the cytoplasm in exchange for K+ ions. This physiological mechanism can lead to a low-pH region in the vicinity of the endosomes, resulting in localized core disassembly and translation of the viral genome. E1 is made fusion competent during acidification of the endosome. The acidic environment causes a rearrangement in the viral glycoproteins, exposing a previously hidden fusion peptide in E1. Insertion of the E1 fusion peptide into the host endosome membrane is thought to be a cooperative process, resulting in rings composed of five–six homotrimers [60]. Crosslinking studies suggest that the postfusion trimers are maintained by E1–E1 interactions [61], a result consistent with the finding that, upon fusion of the viral envelope with a cell membrane, the E1–E2 heterodimer disassembles to give rise to E2 monomers and E1 homotrimers [62]. Other E1 regions that are normally hidden at neutral pH (aside from the fusion peptide) are also exposed in the fusion-active conformation [63]. A histidine residue at position three acts to regulate the low-pH-dependent refolding of E1 during membrane fusion [64]. Mutations in the fusion loop block cell–cell fusion (G91D), owing to a block in a late step in membrane fusion involving the lack of efficient formation of the E1 homotrimer [65,66]. The exposure to low pH during entry may not be an obligatory step in the process of infection of mosquito cells [67].
6K protein
6K is a small, 6000-Da polypeptide that is incorporated into virions in small amounts (7–30 copies) despite being translated in equimolar amounts relative to the other structural proteins [20,21]. The presence and location of 6K have yet to be identified in any cryo-EM virion structures. The presence of the 6K protein in the virion is considered as a necessary structural component of alphavirus particles [68]. Several alterations introduced into the 6K region, including mutations that reduced palmitoylation of 6K [20,69] or deletion of the sequence encoding this peptide [70], resulted in a greatly reduced yield of infectious virus. However, isolated virus particles formed that were totally devoid of 6K and appeared to be structurally indistinguishable from wild-type particles. The growth of an SFV mutant that lacks the 6K protein has a strong dependency on the host cell that is infected, with mammalian cells being much more affected in assembly than insect cells [71].
The 6K protein associates with the pE2/E1 heterodimer soon after synthesis and is thereafter transported to the site of viral assembly at the plasma membrane [21]. Owing to the presence of a signal sequence at the C-terminus of the pE2 protein, the 6K protein is cotranslationally translocated across the ER membrane such that it contains a 16-amino-acid lumenal domain and two transmembrane domains linked by a short (eight amino acid) cytoplasmic loop [72]. The cytoplasmic loop contains three palmitoylated cysteines [20], and the removal of this palmitoylation resulted in aberrant particle formation [69]. E2 and 6K appear to interact because mutations in 6K can be suppressed by mutations in E2 [73], and chimeric viruses containing a SINV glycoprotein and a RRV 6K are highly defective for virus formation [74]. Although 6K has been implicated in the budding process and in the formation of virions, the removal of 6K from the genome of SFV did not influence the formation of the E1–E2 heterodimer or its transport to the cell surface [20,69,72]. Other studies have shown that mutations in 6K can influence glycoprotein trafficking and virion assembly [75]. The mechanism by which the 6K protein affects transport and assembly is thus obscure.
The alphavirus 6K protein is involved in membrane modification both in Escherichia coli and mammalian cells [76,77]. 6K can form cation-selective ion channels in planar lipid bilayers, and has hence been characterized as a viroporin [78–80]. The permeability sequence for RRV 6K channels was found to be Na+ > K+ > Ca2+ [78]. Viroporins are not essential for the replication of viruses, but they do participate in release of viral particles from cells, glycoprotein trafficking and membrane permeability [81], and induce caspase-dependent programmed cell death [80]. SINV release has long been known to be sensitive to the ionic strength of the medium, and final maturation of the particles is very inefficient at low ionic strength [82,83], suggesting that the 6K protein exerts its ion channel function during virus budding [70]. HIV type 1 Vpu, an integral membrane protein that forms oligomeric structures in membranes, shares some analogous functions with 6K, and its presence in trans in the 6K-deleted virus facilitates infectious virus particle production [84].
Nucleocapsid core
The NC core density is divided into a protein region and an internal region probably consisting of protein and RNA (Figure 1B). The NC core has a T = 4 arrangement of capsid proteins, and this leads to an ordered array of projections that are seen as capsomeres on the core (Figure 1C). The carboxy-terminal domain of the capsid contains a hydrophobic pocket that binds the short cdE2. cdE2 links the surface spikes to the internal NC core [85,86]. The cdE2 density extends from the end of the E2 transmembrane helix into the NC shell, ending close to the hydrophobic pocket in the capsid protein (CP), and places E2 residues 400–402 in the pocket [85–87]. The first 100 amino acids of the CP are highly basic and are presumed to bind to the genomic RNA [88]. The genomic RNA does not appear to assume regular symmetry within the NC core and is not ordered in the reconstructions.
The crystal structure of the carboxy-terminal region of the CP (residues 114–264) of SINV was determined [89]. A similar observation was made for the SFV CP [90]. The polypeptide fold from residue 114 to residue 264 of CP is homologous to that of chymotrypsin-like serine proteinases, with catalytic residues of the core protein positioned as in other serine proteinases [89]. The chymotrypsin-like structure of the C-terminal domain of CP consists of two subdomains, with each subdomain having a six- or seven-stranded, antiparallel, β-barrel (‘Greek key’) structure (Figure 1e) [91]. The capsid functions as a serine protease [89] with His141, Asp163 and Ser215 forming the catalytic triad [91,92]. During translation, CP cleaves itself from its own polyprotein, leaving the C-terminal Trp residue in the substrate-binding pocket, thus inhibiting further proteolytic activity.
Alphavirus lifecycle
Entry
The process of entering a susceptible cell begins with engagement of a host receptor by the virus (Figure 2). Specific host receptors vary amongst different alphavirus species, and are believed to be proteinaceous, although nonprotein attachment factors (e.g., HS) may be utilized by the virus to aid in initial binding to the cell [93]. The viral E2 glycoprotein is primarily responsible for receptor interaction, although the E1 protein may also play a role in receptor engagement.
Figure 2. Alphavirus lifecycle.
The lifecycle starts with the attachment of a virion to the cellular receptor (top left), after which receptor-mediated endocytosis, fusion of the viral envelope, disassembly of the core and release of the genomic RNA occur. The replication proteins are then translated and processed (bottom left). These replication proteins enable the replication of the input genomic RNA and translation of the subgenomic mRNA into structural proteins (bottom center). Glycoproteins are translocated across the ER, processed and transported through the Golgi to the plasma membrane (right). Cytoplasmic assembly of genomic RNA and capsid produces the nucleocapsid core, which associates with processed glycoproteins at the plasma membrane, resulting in budding (top right). Scale varies. ER: Endoplasmic reticulum; nsP: Nonstructural protein; PM: Plasma membrane.
Because alphavirus virions are capable of infecting many distinct vertebrate and invertebrate hosts, a mechanism must exist to facilitate entry into different host cells. One hypothesis suggests that the virus utilizes a conserved receptor displayed on cells of multiple host types. For example, the eukaryotic laminin receptor is found on both insect and mosquito cells and may be used by some alphaviruses for entry [94,95]. A second hypothesis proposes that the virus is capable of interacting with multiple discrete cellular receptors [96]. It has been shown that small, even single, changes in the amino acid sequence of either the E2 or E1 glycoproteins can alter the cellular receptor utilized by the virus [97,98]. Both hypotheses may account for the diversity in host range among different alphavirus species.
Engagement of the virus with the host receptor induces conformational changes in the E2 and E1 glycoproteins, as monoclonal antibodies can be raised that recognize so-called transitional epitopes displayed only when the virus is bound to a receptor [32,99]. Some evidence exists that disulfide bond reduction and exchange may play a role in this rearrangement [100,101]; however, utilization of thiol-blocking reagents did not cause a significant inhibition of infection [102]. Virions bound to a receptor molecule are endocytosed in a clathrin-dependent manner [103,104]. Cells in which the ability to form clathrin-coated pits has been ablated are not susceptible to infection [105]. As the virus-containing endosome matures, the pH in the vesicle becomes mildly acidic. This drop in pH triggers destabilization of the E1–E2 heterodimer and subsequent display of a previously hidden fusion loop at the distal tip of an E1 glycoprotein [4,60,63,106]. The fusion peptide inserts into the late endosomal membrane and subsequently trimerizes [62,107]. In mammalian cell culture at least, cholesterol is required in the target membrane, although the reason is unknown [108–110]. As a consequence of fusion-peptide insertion, viral and endosomal membranes mix, yielding a fusion pore and allowing the NC to be deposited in the host cell cytoplasm.
Once in the cytoplasm, the NC must somehow disassemble to expose the encapsidated genome for translation. Thin-section electron micrographs have shown that incoming NCs enter the cytoplasm intact, but disassemble within 5 min [111]. A model for NC disassembly must encompass the fact that newly synthesized NCs are stable within the cell, yet NCs from entering virus are apparently not. It has been shown that CP can bind to ribosomes, and one model has been developed in which interaction of incoming NCs with ribosomes facilitates disassembly. The same model proposes that ribosomes are saturated with newly synthesized capsid during infection, and are thus unavailable to disassemble newly formed NC [112]. Others have suggested that the exposure of the virion to low pH during entry could prime the NC for disassembly once it enters the cytoplasm. Putative ion channel properties in the E1 and 6K viral membrane proteins have been proposed to allow a flow of protons into the interior of the virion particle during residence within the endosome [59,78,113].
Translation of & the role of replication proteins
The nonstructural proteins are initially translated from the incoming full-length genomic viral RNA. For most alphaviruses, translation of the genomic RNA yields two different polyproteins – P123 or the larger P1234. In SINV, the latter occurs as a result of translational readthrough of an opal termination codon after codon 1897 of the nonstructural open reading frame. Readthrough is estimated to occur with 10–20% efficiency [114]. Thus, the typical and predominant polyprotein product is P123. However, some alphaviruses, encompassing both Old World and New World species, lack the opal codon, and only the polyprotein P1234 is produced [115]. The resultant polyproteins are processed exclusively by the virus-encoded protease, located within the nonstructural protein (nsP)2 protein. Both fully cleaved, individual proteins as well as cleavage intermediates have roles in virus replication. Characterization of the activities of the four nonstructural proteins (and polyprotein precursors) has been analyzed via biochemical assays, identification of enzymatic sequence motifs and by examining the effects of nonstructural protein mutations on virus replication. While these robust studies have elucidated many functions of the nonstructural proteins, it is anticipated that novel roles for these proteins have yet to be found.
Nonstructural protein 1 possesses both guanine-7-methyltransferase and guanyltransferase enzymatic activities [116–118]. Both activities are required for the capping and cap methylation of newly synthesized viral genomic and subgenomic RNAs. nsP1 is unique among the nonstructural proteins in that it has been reported to be a membrane-associated protein. This association has been shown to be mediated by cysteine palmitoylation as well as a distinct patch of basic and hydrophobic residues within the protein [119,120]. Thus, nsP1 may anchor replication complexes to cellular membranes. In addition to known enzymatic activities, phenotype analyses of nsP1 mutants have demonstrated that the protein may be required for specific initiation or maintenance of minus-strand replicative intermediates [121].
The nsP2 protein has multiple known enzymatic activities and roles. The N-terminal domain of nsP2 contains helicase activity required for RNA duplex unwinding during RNA replication and transcription [122]. The N-terminal domain also contains RNA triphosphatase and nucleoside triphosphatase activity [123,124]. The C-terminal domain contains a papain-like cysteine protease as well as an enzymatically nonfunctional methyltransferase (MTase) domain [125,126]. The inactive MTase domain has been proposed to regulate minus-strand synthesis and is likely involved in the development of cellular cytopathic effects [127]. The nsP2 protease domain is responsible for cleaving the viral nonstructural polyprotein into intermediate and final component proteins [125]. The crystal structure of the MTase and protease domains has recently been solved to 2.5 Å and revealed that the nsP2 protease fold is novel relative to the structure of other known proteases [128]. The protease cleaves at distinct sites within the viral nonstructural polyprotein, correlating to the junctions between individual proteins, and the amino acid sequences of these cleavage sites are similar [125]. Both the individual nsP2 protein, as well as nsP2 resident within the polyprotein, are proteolytically active, although substrate preferences are different. nsP2, when present in a polyprotein, can cleave in cis only at the nsP3/nsP4 junction [129]. Cleavage at other sites within the polyprotein can only occur via cleavage in trans. As such, processing order, and thus nonstructural protein makeup within the infected cell, is temporally regulated during an infection [129]. Specifically, early in infection, P123 and P1234 are produced via translation of the viral genomic RNA. The nsP3/nsP4 junction of the P1234 polyprotein is cleaved in cis, yielding P123 plus nsP4. Cleavage sites within P123 can only be cleaved in trans and hence, early in infection, the P123 and nsP4 proteins predominate. As the infection proceeds, however, the concentration of P123 increases and one P123 molecule can cleave in trans at the nsP1/nsP2 bond of a second P123 molecule, yielding nsP1 plus P23 or P234. The resultant P23 or P234 polyproteins are only then capable of the final polyprotein cleavage at the nsP2/nsP3 bond in trans. Interestingly, the nsP2 protein contains multiple nuclear localization signals, and approximately 50% of nsP2 resides in the nucleus [130]. Because RNA replication and polyprotein processing occur in the cytoplasm, it is not known what role nsP2 may play within the nucleus, but abolishment of nsP2 translocation to the nucleus yields an attenuated phenotype [131,132].
To date, relatively little is known about the nsP3 protein. Mutational analyses have shown that nsP3 is required for minus-strand and subgenomic RNA synthesis, both as the individual protein and in the context of the P123 polyprotein [133–135]. Mutational and chimeric analyses have also revealed a role for nsP3 in modulation of pathogenicity in mice [136,137]. Alignment of the amino acid sequence of nsP3 from a variety of alphaviruses has shown the primary structure of the protein is composed of two discrete domains. The N-terminal domain is well conserved among the genus, whereas the C-terminal domain varies in both sequence and length [5]. The N-terminal domain shares sequence similarity with the so-called ‘macro’ domain displayed by proteins from all kingdoms of life [138,139]. The Chikungunya virus and VEEV macro domains have been crystallized and were shown to possess both ADP-ribose 1″-phosphate phosphatase and RNA-binding activity [140]. The former activity may be related to induction of apoptosis in infected cells. nsP3 is heavily phosphorylated, particularly on serine and threonine residues within the nonconserved C-terminus, and it is the hyperphosphorylated form of the protein that may play a role in RNA synthesis [141,142]. A fraction of nsP3 within infected mammalian cells has also been shown to localize to the nuclear envelope, distinct from nsP3 found in cytoplasmic replication complexes [143].
The requisite RNA-dependent RNA polymerase (RdRP) is represented by nsP4. The common GDD motif observed in many viral RdRPs is found in nsP4, and the motif is required for activity [144,145]. While the C-terminus of nsP4 maintains the homology of other viral RdRPs, the N-terminus of nsP4 contains a small region whose primary structure is not conserved among other viral RdRPs [145]. This N-terminal region may serve as a scaffold for interaction with other nsPs, particularly nsP1, in the formation of a replication complex, or may serve to interact with unidentified host proteins [146,147]. The cellular concentration of nsP4 is lower than that of the other nsPs for two reasons. First, for those alphavirus species that employ opal codon readthrough, P1234, and thus nsP4, is only manufactured at a fraction of P123, and thus the other nonstructural proteins. Second, the N-terminal residue of nsP4 is a conserved tyrosine. This tyrosine is a primary destabilizing residue, which results in rapid degradation of nsP4 within the cell by the N-end rule pathway [148]. Interestingly, although the N-terminal tyrosine leads to rapid degradation of nsP4 within the cell, replacement of the conserved tyrosine residue with a nonaromatic residue leads to poor RNA replication [149]. nsP4 has also been shown, in vitro, to possess terminal adenylyltransferase activity via addition of nontemplated adenine to the 3′ end of an acceptor RNA [150]. This enzymatic activity may aid in maintenance of the viral genomic poly(A) tail.
Replication & transcription of viral RNAs
Replication of viral RNA takes place in cytoplasmic vacuoles derived from lysosomal and endosomal membranes [151]. The nonstructural protein products form replicase complexes on these membranes [152]. In addition, both host proteins and viral structural proteins have been implicated in replicase complex formation and activity [153,154]. It has long been believed that the differential and temporal processing of the nonstructural polyprotein by nsP2 yields different replication complexes of distinct composition, and is the basis for the switch from minus- to plus-strand synthesis. However, recent evidence has been presented that demonstrates that SINV mutants incapable of P23 or P123 cleavage in IFN-α/β-competent mammalian cells are capable of the switch from minus- to plus-strand synthesis [155,156]. Further investigation of this phenomenon in other alphaviruses and in animal models will prove interesting. Minus-strand synthesis utilizes genomic RNA as a template, and requires a replication complex composed of P123 and nsP4, the former of which is only present in high concentrations early in infection [125,135,157]. This complex is effective at synthesizing minus-strand RNAs from genomic templates, but does not efficiently synthesize plus-strand RNAs. Thus, minus-strand synthesis predominates early in infection. As the infection proceeds, minus-strand synthesis wanes as P123 polyproteins are efficiently cleaved in trans to yield the individual nonstructural proteins. The plus-strand replicase is composed of the individual nonstructural proteins (plus host factors), presumably in a conformation different from that of the P123/nsP4 replication complex [5]. An intermediate replication complex, composed of nsP1, P23 and nsP4, may exist transiently, and may be capable of both plus- and minus-strand synthesis [155,158,159]. Plus-strand and subgenomic RNAs are synthesized exclusively later in infection. Plus strands are synthesized from either the genomic or subgenomic promoter. In SINV, transcription from the subgenomic promoter yields an approximately threefold excess of subgenomic RNAs relative to full-length genomic RNA [160]. It is not known if two distinct plus-strand replication/transcription complexes exist to manufacture these two different RNA species.
Structural elements within the viral RNA
Important RNA structural elements, including four separate conserved sequence elements (CSEs), have been characterized in the alphavirus genome. These structural elements and CSEs have been shown to interact with both viral and host proteins to promote replication, transcription and packaging of viral RNAs. The 5′ nontranslated region of the genome has been implicated as a component of the promoter for both minus- and plus-strand RNA synthesis [161]. A CSE found at the 5′ end of the genomic RNA (CSE 1) forms a stem-loop structure containing approximately the first 44 nt of the genome, and is believed to function, in the context of the minus-strand, as a promoter for plus-strand synthesis from minus-strand templates [162]. A second 51 nt CSE (CSE 2) also resident near the 5′ end of the genome within the coding sequence for nsP1 (beginning around nt 155) may act as a promoter for initiation of minus-strand synthesis from a genomic RNA template, and has been implicated in enhancing both minus- and plus-strand RNA synthesis [161]. The absence of CSE 2 in the subgenomic RNA may explain why only full-length, genomic RNAs can act as templates for minus-strand synthesis. A 19-nt CSE found just upstream of the polyA tract in the 3′ untranslated region of the genome (CSE 4) is also believed to act as a copromoter for initiation of minus-strand synthesis, perhaps via interaction of the 5′ and 3′ ends of the full-length genomic RNA [161,163,164]. Transcription of the subgenomic RNA occurs efficiently only when a separate 24-nt CSE (CSE 3), located in the junction region between the coding sequence for the nonstructural and structural proteins, is present [165]. This subgenomic promoter element is exceptionally efficient, and has seen widespread use in heterologous gene expression [166].
Most alphaviruses contain 40–60-nt repeated sequence elements upstream of the polyA sequence in the 3′ untranslated region of the genome [167]. These repeated sequences have been implicated in binding host proteins important for translation of viral RNAs [153]. Alphavirus RNAs also have a specific packaging signal utilized such that only viral RNA is encapsidated during NC formation [168]. The packaging signal is recognized by the CP, and this interaction is likely an early step in NC formation [169–171]. The position of the packaging signal in the viral RNA varies by species, but is typically found in the 5′ half of the genome such that only full-length RNA is packaged.
Nucleocapsid assembly
The alphavirus NC contains a single copy of the RNA genome complexed with 240 copies of CP. NCs isolated from alphavirus-infected cells are stable and have a sedimentation coefficient of 140 S. Purified capsid proteins oligomerize into core-like particles in vitro in the presence of single-stranded nucleic acid, and resemble the native NCs [172]. Core-like particles can also be generated from truncated crosslinked CP dimers [173]. NCs devoid of RNA have not been found either by analyzing infected cells or by using in vitro assembly assays [169,172]. One of the difficulties in dissecting the assembly pathway of SINV NC from CP and RNA is the inability to isolate assembly intermediates in vivo. The assembly of the alphavirus NC is a multistep event involving a nucleic acid-bound dimer of CP as an early step in the assembly pathway [169]. A model of SINV NC assembly based on in vitro results suggests that the NC is formed from a nucleation event in which the amino acid region from residues 81 to 112 recognizes an encapsidation signal on the genomic RNA and leads to CP interactions and the encapsidation of this RNA [168,170,174]. A deletion of residues 97–106 in SINV CP results in a virus that has lost its ability to recognize the genomic RNA, and the replacement of leucine residues at positions 108 and 110 results in a failure to accumulate NCs in the cytoplasm [85,175]. Residues 99, 103 and 105 promote the formation of a CP dimer between two capsid molecules. This dimerization presumably also requires the interaction of helix I from each individual CP to stabilize the dimer [176] through coiled–coil interactions [177], and may play a regulatory role during the early steps of the assembly process [178]. In the crystal structure of truncated CP, the amino acids 108–111 of the ‘N-terminal arm’ of CP bind to a specific hydrophobic pocket in neighboring CP molecules and mimic the binding of cdE2 in the pocket [85]. The mutation of residues corresponding to SINV CP positions 108 and 110 to alanines in SFV CP also results in the inhibition of NC accumulation [179]. Mutations in the CP can result in the assembly of the virus structural proteins into icosahedra of different triangulation numbers [180].
Structural protein processing
The structural proteins of alphaviruses are translated as a polyprotein from the subgenomic RNA in the order of CP–pE2–6K–E1 [160]. The CP is translated first and is released from the polyprotein by autoproteolysis (Figure 3). Once the CP is released from the nascent polypeptide chain, the new N-terminus of the polyprotein now contains a signal sequence for translocation of the pE2 sequence across the ER membrane [181]. The signal also has a carbohydrate attachment site, which may be responsible for the retention of the signal sequence in pE2. E1 and pE2 undergo a complex series of folding intermediates that require chaperones and disulfide bond formation and exchange [101]. Oligomerization of pE2 and E1 is also a requirement for the transport of the glycoprotein to the cell surface, but the presence of the CP is not required. SINV glycoproteins can first be detected at the cell surface at 1.5 h and virus budding is evident by 3 h post-infection [182].
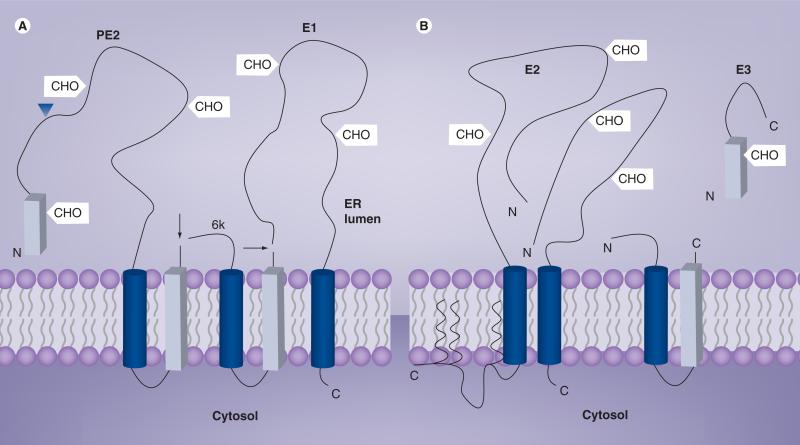
Figure 3. Model of glycoprotein configuration on the endoplasmic reticulum and plasma membranes.
(A) Configuration of pE2, 6K and E1 after signalase cleavage on the ER membrane. The arrows represent the site of signalase cleavage in the ER. The blue triangle represents the location of pE2 furin cleavage. (B) Configuration of glycoproteins on the plasma membrane after the furin cleavage of pE2 into E3 and E2. Only the palmitoylation sites on the cytoplasmic side of E2 are shown as thicker, wavy lines on the membrane bilayer. Signal sequences in the polypeptide are indicated as rectangular blocks and stop-transfer sequences are indicated as cylinders. The schematic does not represent the native glycoprotein configuration.
ER: Endoplasmic reticulum.
pE2 and E1 molecules that were translocated into the ER are processed and undergo post-translational modifications. High-mannose chains are added to all potential N-linked glycosylation sites, and the oligosaccharide chains are trimmed depending on the availability of the site [183]. The addition of carbohydrate chains is usually required for proper folding and solubility of the protein and to prevent aggregation. High-mannose carbohydrate or complex oligosaccharide is added at a glycosylation site of envelope proteins depending on the type and growth state of the infected cell. When SINV was grown in mosquito cells, high-mannose sugars were added to all glycosylation sites that were previously shown to have complex glycans when the virus was grown in vertebrate cells [184,185].
The C terminus of pE2 is thought to lie in the lumen of the ER when first synthesized, with subsequent reorientation of the C-terminal region into the cytoplasm being required for virus budding. The reorientation of the pE2 tail (cdE2) takes place during transport of the protein complex to the cell surface and is associated with phosphorylation, which is believed to occur on Thr398 and Tyr400 of the conserved tripeptide TPY [186]. Site-directed mutagenesis of these two potential phosphorylation sites in cdE2 resulted in the lack of virus production, although NCs are formed and the envelope proteins are exported from the ER [187].
The translocation of the glycoproteins across the ER membrane is regulated by various signal sequences and further post-translational modifications, including palmitoylation of some or all of the conserved cysteine residues of cdE2 (C396, C416 and C417) [72,188,189]. Palmitoylation of pE2 in vertebrate cells occurs after exit from the ER, but probably before arrival in the cis Golgi, and is presumably associated with the reorientation process of cdE2. This might serve to orientate the cdE2 along the bilayer since the palmitic acid residues will be present within the lipid bilayer, rather than in the cytosol. Mutagenesis to reduce the amount of palmitoylation did not interfere with the transport of the glycoproteins to the cell surface, but virus budding was affected.
During transport of the pE2–E1 complex, after the heterodimer reaches the trans-Golgi network but prior to arrival at the plasma membrane, pE2 is cleaved by furin to form E3 and E2 [20]. The cleavage of pE2 is required for entry and fusion activation in new cells [43,190]. Uncleaved pE2 can also be efficiently transported and incorporated into virus particles; however, the pE2 cleavage activation event is formally similar to the furin cleavage that occurs in flaviviruses where the glycoprotein precursor prM must be cleaved to activate virus infectivity [191]. Thus, the cleavage of pE2 is required to generate infectious particles.
Virus budding
The final stage of the alphavirus lifecycle is the budding of virions from the plasma membrane. This process requires effective interaction between the NC and the glycoproteins [192,193]. Specifically, the interaction between the NC and cdE2 [186,194], but not E1, residues is essential for viral budding [195–197]. NCs assembled in the cell cytoplasm are thought to diffuse or otherwise transit to the plasma membrane, where they are bound by cdE2. This binding provides the free energy to propel the capsid across the plasma membrane, during which it acquires a complete complement of glycoprotein spikes [198]. Lateral interactions between the glycoproteins are also important for virus assembly [34], and there appears to be a requirement for cholesterol in the membrane during the budding process [199].
The interaction between the CP and the envelope glycoproteins has been extensively investigated, although an authentic in vitro assay has not yet been established. A synthetic peptide of 31 residues corresponding to the SFV cdE2 binds to SFV capsids in vitro [195]. Synthetic peptides corresponding to the entire 31 residues and eight carboxy-terminal residues of the cytoplasmic domain of SFV E2 inhibited viral budding in microinjection experiments and when conjugated to colloidal gold are bound specifically to nucleocapsids in infected cells [197]. These peptides, which could inhibit virion formation, may have interfered with the attachment of E2 to the NC [5]. Interaction between the C-terminal 16 amino acids of cdE2 and CP was determined by using a phospholipid membrane system that incorporated a variety of SINV cdE2 domains [87]. The peptides included sites subsequently identified by mutagenesis as important for budding. Site-specific mutagenesis of the CP itself identified Tyr180 and/or Glu183 as probable participants in the binding of the NC to E2 [200,201]. Deletions that disrupt the accumulation of NCs in the cytoplasm do not prevent virus budding [202], so NC assembly at the plasma membrane may be possible. Deletion of amino acids 408–415 of cdE2 resulted in the failure to assemble virions in mammalian cells [203]. Binding to the NC by the glycoproteins has been directly demonstrated in studies in which SFV virions were treated with octylglucoside. When isolated SFV virions were treated with the nonionic detergent β-d-octylglucoside at neutral pH and low ionic strength, the lipid bilayer membrane could be removed, leaving most of the spike glycoproteins attached to the NCs. This interaction between capsid and membrane protein was sensitive to elevated pH and ionic strength, suggesting that there is a noncovalent binding of the glycoprotein spikes by the CP [204]. Site-specific mutagenesis of Tyr400 in the cdE2 identified the approximate site of interaction with the NC [205]. In addition, cryo-EM studies have shown that cdE2 clearly extends down into the core to the site of the hydrophobic pocket. Chimeric viruses of SINV and RRV demonstrated that Ala403 and Asn405 in the cdE2 are also important for budding [206]. The interaction of the transmembrane domains of E2 and E1 is required for efficient budding and assembly of infectious virions [207]. Lateral interactions between the glycoproteins are also important in virus assembly [34,208].
Host cell effects
One hallmark of alphavirus infection in the vertebrate cell is the ability to shutdown host transcription and translation processes without affecting viral protein and nucleic acid synthesis. Importantly, these shutdowns result in a decrease in IFN-α/β production in the host cell, and the ability of the innate immune system to attenuate the infection is diminished. These functions appear to map, in part, to nsP2 in Old World viruses, and to the CP in New World viruses [209–211]. The shutoff of host-cell protein synthesis in both groups may be mediated by phosphorylation of the host-cell translation initiation factor eIF2α, which is likely the result of detection of viral dsRNA by the host protein kinase R. Translation of the viral subgenomic RNA does not require host eIF2α [212]. In concert with shutdown of host macromolecular synthesis, the development of cellular cytopathic effect typically arises. Alphavirus infection of the vertebrate cell usually leads to the induction of the apoptotic pathway [213]. This pathway has proven important for the development of neuronal pathogenesis in infected organisms [214]. In some alphaviruses, apoptosis has been shown to occur via the process of membrane fusion and entry, via expression of the viral envelope proteins or via expression of nsP2 and RNA replication [215–217]. Interestingly, mutants that replicate, but do not induce apoptosis, have been identified [127], and transfection of cells with replicating but budding-deficient viral RNA, thus removing the process of virion entry, also induces apoptosis. Thus, multiple mechanisms have been proposed to be involved in the induction of apoptosis by alphavirus infection, and probably involve both host- and virus-directed initiation of apoptosis. The topic of alphaviruses and apoptosis has been recently reviewed [218,219]. Alphaviruses utilize host membranes during the course of infection. Endosomal and lysosomal membranes are rearranged into vacuolar structures. These have been termed ‘cytopathic vacuoles’, are 1–2 μm in diameter, and are presumed to be the site of viral replication complexes [151].
Future perspective
Alphaviruses have been studied extensively for the past 40 years and have become a model animal virus system. Yet, while the genus may appear simple from a superficial point of view, we have much to learn about alphaviruses. The role of host proteins in the virus lifecycle has not been studied to the same extent as the role of the viral proteins. Detailed analyses of the function of host factors in entry, replication, assembly and modulation of the host antiviral activities continue to be undertaken and should provide a better understanding of host–virus interplay. The solution of a detailed structure for the E2 glycoprotein will further our understanding of virion structure, and elucidation of structures for the remaining nonstructural proteins may lead to novel insights about their activity and interactions. The use of the alphaviruses as reagents in molecular biology and therapeutics is also evolving. Virus constructs containing fusion of fluorescent proteins to individual nonstructural proteins have been produced, and may aid to further probe the function and localization of nonstructural proteins and replication complexes [220]. Similarly, a viable SINV clone has been produced in which E2 has been fused to the red fluorescent protein (RFP) [Jose J & Kuhn RJ, Unpublished Data]. Reconstruction revealed the incorporation of RFP into the virus particle in stoichiometric amounts, and in vivo data have shown that both E2 and RFP retain activity. Alphaviruses have already proven useful in the expression of heterologous proteins in mammalian systems, and alphavirus replicons will continue to provide a vehicle for vaccine and gene-therapy delivery [6,221]. Recent epidemics of alphaviruses, specifically Chikungunya, have raised global health concerns, and the further development of both human and animal vaccines are underway.
Executive summary.
Alphavirus classification
■ Alphaviruses belong to the family Togaviridae.
■ Two virus member categories have been characterized: Old World and New World alphaviruses.
■ Several important human and animal pathogens are represented by the alphaviruses, including Chikungunya and western and eastern equine enchephalitis viruses.
■ The viruses are transmitted by an insect vector, typically the mosquito.
Alphavirus virion structure
■ The icosahedral T = 4 virion is 700 Å in diameter and contains one copy of an approximately 11.5-kb, positive strand, genomic RNA that is encapsidated by capsid proteins forming an icosahedral nucleocapsid (NC). The NC is enveloped in a host-derived lipid bilayer that contains viral glycoproteins. The envelope contains 240 E1/E2 heterodimers on the surface of the virus assembled into 80 spikes, and substoichiometric amounts of the 6K protein.
Alphavirus virally encoded proteins
- ■ The virus encodes five structural proteins that are processed from a single structural polypeptide.
- The capsid protein is the sole protein component of the NC. Capsid cotranslationally cleaves itself out of a nascent structural polypeptide.
- The E2 glycoprotein is present in the envelope and is responsible for receptor attachment. Mature E2 is formed by furin cleavage of the pE2 precursor protein late in virus maturation, yielding E3 and E2. Most alphaviruses do not retain E3 in the virion.
- 6K may contain proton-specific ion-channel activity necessary for virus budding and/or entry.
- The E1 glycoprotein is present in the envelope and is responsible for membrane fusion during virus entry.
- ■ The virus encodes four nonstructural proteins that are processed from a single nonstructural polypeptide.
- Nonstructural protein (nsP)1 contains both guanine-7-methyltransferase and guanyltransferase enzymatic activities. Helicase activity may be required for unwinding dsRNA replicative intermediates. Protease activity is required for cleavage of the nonstructural polypeptide. 50% of nsP2 is found in the host cell nucleus.
- nsP3 is required for minus-strand and subgenomic RNA synthesis and possesses both ADP-ribose 1″-phosphate phosphatase and RNA-binding activity. nsP3 has multiple phosphoacceptor sites and may be heavily phosphorylated.
- nsP4 contains the RNA-dependent RNA polymerase activity required for viral RNA replication. In many alphavirues, nsP4 is produced at approximately 10% of the other nonstructural proteins owing to translational readthrough of an opal codon at the end of nsP3.
Alphavirus lifecycle
■ The virus binds to a host receptor via the E1 and E2 glycoproteins.
■ The virus is endocytosed in a clathrin-dependent manner. The low pH of the endosome causes a structural rearrangement of the virion, and exposes the fusion peptide of E1.
■ The fusion peptide inserts into the endosomal membrane, forming a fusion pore and allowing the NC into the cytoplasm. The NC disassembles, exposing the viral RNA for translation.
■ Nonstructural proteins are translated first from the full-length viral RNA, allowing assemblage of viral replicase complexes on endosomal and lysosomal membranes.
■ Minus-strand synthesis predominates early in infection, and plus-strand and subgenomic RNA synthesis occurs exclusively later in infection.
■ Conserved sequence elements within the viral RNA aid in transcription initiation, act as translational enhancers and function as packaging signals.
■ The structural proteins are translated from the subgenomic RNA. The capsid protein assembles into NCs with the full-length viral RNA. pE2, 6K and E1 insert into the endoplasmic reticulum membrane and transit to the host plasma membrane via the secretory pathway. Host signalase and furin are responsible for cleavage and maturation of the structural proteins. NCs interact with the cytoplasmic domain of E2, become enveloped by the E1/E2 icosohedral lattice and bud from the plasma membrane.
Alphavirus effects on the host cell
■ Viral infection leads to host transcription and translation shutdown. nsP2 in the Old World viruses and capsid in the New World viruses appear to be responsible.
■ Alphavirus infection leads to apoptosis of the mammalian host cell via multiple mechanisms.
■ Alphavirus infection leads to rearrangement of host lysosomal and endosomal membranes into vacuolar structures. These vacuolar structures are presumed to be the site of viral replication complexes.
Alphaviruses as biological tools
■ The alphaviruses are a model enveloped virus system, and structural and functional studies have led to many discoveries in virology and cell biology. Alphavirus replicons have been employed extensively in heterologous gene expression systems. Alphaviruses have also proven valuable as a vehicle for vaccine and gene-therapy delivery.
Acknowledgements
We thank Thomas J Edwards for help with the figures and Rushika Perera for useful discussions. We also acknowledge the assistance of Anita Robinson with the preparation of this review.
Financial & competing interests disclosure
The authors acknowledge support from the NIH through a NIGMS award GM56279. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Joyce Jose, Department of Biological Sciences, Bindley Bioscience Center, Lilly Hall of Life Sciences, 915 West State St., Purdue University, West Lafayette, IN 47907, USA Tel.: +1 765 494 4407 Fax: +1 765 494 0876 jjose@purdue.edu.
Jonathan E Snyder, Department of Biological Sciences, Bindley Bioscience Center, Lilly Hall of Life Sciences, 915 West State St., Purdue University, West Lafayette, IN 47907, USA Tel.: +1 765 494 4407 Fax: +1 765 494 0876 jonsnyder@purdue.edu.
Richard J Kuhn, Department of Biological Sciences, Bindley Bioscience Center, Lilly Hall of Life Sciences, 915 West State St., Purdue University, West Lafayette, IN 47907, USA Tel.: +1 765 494 4407 Fax: +1 765 494 0876 kuhnr@purdue.edu.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Powers AM, Brault AC, Shirako Y, et al. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 2001;75(21):10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enserink M. Infectious diseases. Chikungunya: no longer a third world disease. Science. 2007;318(5858):1860–1861. doi: 10.1126/science.318.5858.1860. [DOI] [PubMed] [Google Scholar]
- 3■■.Mukhopadhyay S, Zhang W, Gabler S, et al. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure. 2006;14(1):63–73. doi: 10.1016/j.str.2005.07.025. [■■ of considerable interestStructure of an alphavirus virion is reconstructed.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lescar J, Roussel A, Wien MW, et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105(1):137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 5■.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [■ of interestExceptionally detailed, global overview of the alphaviruses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6■.Atkins GJ, Fleeton MN, Sheahan BJ. Therapeutic and prophylactic applications of alphavirus vectors. Expert Rev. Mol. Med. 2008;10:e33. doi: 10.1017/S1462399408000859. [■ of interestReview of the applications and potential use of alphaviruses in gene therapy and therapeutics.] [DOI] [PubMed] [Google Scholar]
- 7.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 2007;81(5):2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weston J, Villoing S, Bremont M, et al. Comparison of two aquatic alphaviruses, salmon pancreas disease virus and sleeping disease virus, by using genome sequence analysis, monoclonal reactivity, and cross-infection. J. Virol. 2002;76(12):6155–6163. doi: 10.1128/JVI.76.12.6155-6163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weston JH, Welsh MD, McLoughlin MF, Todd D. Salmon pancreas disease virus, an alphavirus infecting farmed Atlantic salmon, Salmo salar L. Virology. 1999;256(2):188–195. doi: 10.1006/viro.1999.9654. [DOI] [PubMed] [Google Scholar]
- 10.La Linn M, Gardner J, Warrilow D, et al. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J. Virol. 2001;75(9):4103–4109. doi: 10.1128/JVI.75.9.4103-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villoing S, Bearzotti M, Chilmonczyk S, Castric J, Bremont M. Rainbow trout sleeping disease virus is an atypical alphavirus. J. Virol. 2000;74(1):173–183. doi: 10.1128/jvi.74.1.173-183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frey TK. Molecular biology of rubella virus. Adv. Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in Sindbis virus. J. Virol. 2002;76(22):11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison SC, Strong RK, Schlesinger S, Schlesinger MJ. Crystallization of Sindbis virus and its nucleocapsid. J. Mol. Biol. 1992;226(1):277–280. doi: 10.1016/0022-2836(92)90141-6. [DOI] [PubMed] [Google Scholar]
- 15.Fuller SD. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell. 1987;48(6):923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 16■■.Cheng RH, Kuhn RJ, Olson NH, et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80(4):621–630. doi: 10.1016/0092-8674(95)90516-2. [■■ of considerable interestStructure of the capsid protein is solved.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell. 2000;5(2):255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Fisher BR, Olson NH, Strauss JH, Kuhn RJ, Baker TS. Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins. J. Virol. 2002;76(14):7239–7246. doi: 10.1128/JVI.76.14.7239-7246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paredes AM, Simon MN, Brown DT. The mass of the Sindbis virus nucleocapsid suggests it has T = 4 icosahedral symmetry. Virology. 1992;187(1):329–332. doi: 10.1016/0042-6822(92)90322-g. [DOI] [PubMed] [Google Scholar]
- 20.Gaedigk-Nitschko K, Schlesinger MJ. The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology. 1990;175(1):274–281. doi: 10.1016/0042-6822(90)90209-a. [DOI] [PubMed] [Google Scholar]
- 21.Lusa S, Garoff H, Liljestrom P. Fate of the 6K membrane protein of Semliki Forest virus during virus assembly. Virology. 1991;185(2):843–846. doi: 10.1016/0042-6822(91)90556-q. [DOI] [PubMed] [Google Scholar]
- 22.Kielian M, Chatterjee PK, Gibbons DL, Lu YE. Specific roles for lipids in virus fusion and exit. Examples from the alphaviruses. Subcell. Biochem. 2000;34:409–455. doi: 10.1007/0-306-46824-7_11. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt MF. Acylation of viral spike glycoproteins: a feature of enveloped RNA viruses. Virology. 1982;116(1):327–338. doi: 10.1016/0042-6822(82)90424-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt MF, Bracha M, Schlesinger MJ. Evidence for covalent attachment of fatty acids to Sindbis virus glycoproteins. Proc. Natl Acad. Sci. USA. 1979;76(4):1687–1691. doi: 10.1073/pnas.76.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Bonsdorff CH, Harrison SC. Sindbis virus glycoproteins form a regular icosahedral surface lattice. J. Virol. 1975;16(1):141–145. doi: 10.1128/jvi.16.1.141-145.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel RH, Provencher SW, von Bonsdorff CH, Adrian M, Dubochet J. Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature. 1986;320(6062):533–535. doi: 10.1038/320533a0. [DOI] [PubMed] [Google Scholar]
- 27.Paredes AM, Brown DT, Rothnagel R, et al. Three-dimensional structure of a membrane-containing virus. Proc. Natl Acad. Sci. USA. 1993;90(19):9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pletnev SV, Zhang W, Mukhopadhyay S, et al. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell. 2001;105(1):127–136. doi: 10.1016/s0092-8674(01)00302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith TJ, Cheng RH, Olson NH, et al. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl Acad. Sci. USA. 1995;92(23):10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, Heil M, Kuhn RJ, Baker TS. Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology. 2005;332(2):511–518. doi: 10.1016/j.virol.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis NL, Pence DF, Meyer WJ, Schmaljohn AL, Johnston RE. Alternative forms of a strain-specific neutralizing antigenic site on the Sindbis virus E2 glycoprotein. Virology. 1987;161(1):101–108. doi: 10.1016/0042-6822(87)90175-9. [DOI] [PubMed] [Google Scholar]
- 32.Meyer WJ, Johnston RE. Structural rearrangement of infecting Sindbis virions at the cell surface: mapping of newly accessible epitopes. J. Virol. 1993;67(9):5117–5125. doi: 10.1128/jvi.67.9.5117-5125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss EG, Stec DS, Schmaljohn AL, Strauss JH. Identification of antigenically important domains in the glycoproteins of Sindbis virus by ana lysis of antibody escape variants. J. Virol. 1991;65(9):4654–4664. doi: 10.1128/jvi.65.9.4654-4664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Bonsdorff CH, Harrison SC. Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol. 1978;28(2):578–583. doi: 10.1128/jvi.28.2.578-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu SR, Haag L, Sjoberg M, Garoff H, Hammar L. The dynamic envelope of a fusion class II virus. E3 domain of glycoprotein E2 precursor in Semliki Forest virus provides a unique contact with the fusion protein E1. J. Biol. Chem. 2008;283(39):26452–26460. doi: 10.1074/jbc.M801470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferlenghi I, Gowen B, de Haas F, et al. The first step: activation of the Semliki Forest virus spike protein precursor causes a localized conformational change in the trimeric spike. J. Mol. Biol. 1998;283(1):71–81. doi: 10.1006/jmbi.1998.2066. [DOI] [PubMed] [Google Scholar]
- 37.Paredes AM, Heidner H, Thuman-Commike P, Prasad BV, Johnston RE, Chiu W. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J. Virol. 1998;72(2):1534–1541. doi: 10.1128/jvi.72.2.1534-1541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrott MM, Sitarski SA, Arnold RJ, Picton LK, Hill RB, Mukhopadhyay S. Role of conserved cysteines in the alphavirus E3 protein. J. Virol. 2009;83(6):2584–2591. doi: 10.1128/JVI.02158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lobigs M, Zhao HX, Garoff H. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J. Virol. 1990;64(9):4346–4355. doi: 10.1128/jvi.64.9.4346-4355.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J. Virol. 1990;64(3):1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahlberg JM, Boere WA, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 1989;63(12):4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryman KD, Klimstra WB, Johnston RE. Attenuation of Sindbis virus variants incorporating uncleaved PE2 glycoprotein is correlated with attachment to cell-surface heparan sulfate. Virology. 2004;322(1):1–12. doi: 10.1016/j.virol.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Heidner HW, McKnight KL, Davis NL, Johnston RE. Lethality of PE2 incorporation into Sindbis virus can be suppressed by second-site mutations in E3 and E2. J. Virol. 1994;68(4):2683–2692. doi: 10.1128/jvi.68.4.2683-2692.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knight RL, Schultz KL, Kent RJ, Venkatesan M, Griffin DE. Role of N-linked glycosylation for Sindbis virus infection and replication in vertebrate and invertebrate systems. J. Virol. 2009;83(11):5640–5647. doi: 10.1128/JVI.02427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryman KD, Gardner CL, Burke CW, Meier KC, Thompson JM, Klimstra WB. Heparan sulfate binding can contribute to the neurovirulence of neuroadapted and nonneuroadapted Sindbis viruses. J. Virol. 2007;81(7):3563–3573. doi: 10.1128/JVI.02494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bear JS, Byrnes AP, Griffin DE. Heparin-binding and patterns of virulence for two recombinant strains of Sindbis virus. Virology. 2006;347(1):183–190. doi: 10.1016/j.virol.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 47.Tucker PC, Lee SH, Bui N, Martinie D, Griffin DE. Amino acid changes in the Sindbis virus E2 glycoprotein that increase neurovirulence improve entry into neuroblastoma cells. J. Virol. 1997;71(8):6106–6112. doi: 10.1128/jvi.71.8.6106-6112.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myles KM, Pierro DJ, Olson KE. Deletions in the putative cell receptor-binding domain of Sindbis virus strain MRE16 E2 glycoprotein reduce midgut infectivity in Aedes aegypti. J. Virol. 2003;77(16):8872–8881. doi: 10.1128/JVI.77.16.8872-8881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navaratnarajah CK, Kuhn RJ. Functional characterization of the Sindbis virus E2 glycoprotein by transposon linker-insertion mutagenesis. Virology. 2007;363(1):134–147. doi: 10.1016/j.virol.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharp JS, Nelson S, Brown D, Tomer KB. Structural characterization of the E2 glycoprotein from Sindbis by lysine biotinylation and LC-MS/MS. Virology. 2006;348(1):216–223. doi: 10.1016/j.virol.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 51.Helenius A, von Bonsdorff CH. Semliki Forest virus membrane proteins. Preparation and characterization of spike complexes soluble in detergent-free medium. Biochim. Biophys. Acta. 1976;436(4):895–899. doi: 10.1016/0005-2736(76)90421-1. [DOI] [PubMed] [Google Scholar]
- 52.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell. Biol. 1985;101(6):2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wengler G, Rey FA. The isolation of the ectodomain of the alphavirus E1 protein as a soluble hemagglutinin and its crystallization. Virology. 1999;257(2):472–482. doi: 10.1006/viro.1999.9661. [DOI] [PubMed] [Google Scholar]
- 54■■.Roussel A, Lescar J, Vaney MC, Wengler G, Rey FA. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure. 2006;14(1):75–86. doi: 10.1016/j.str.2005.09.014. [■■ of considerable interestCrystal structure of E1 is described.] [DOI] [PubMed] [Google Scholar]
- 55.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375(6529):291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 56.Strauss JH, Strauss EG. Virus evolution: how does an enveloped virus make a regular structure? Cell. 2001;105(1):5–8. doi: 10.1016/S0092-8674(01)00291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibbons DL, Reilly B, Ahn A, et al. Purification and crystallization reveal two types of interactions of the fusion protein homotrimer of Semliki Forest virus. J. Virol. 2004;78(7):3514–3523. doi: 10.1128/JVI.78.7.3514-3523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibbons DL, Vaney MC, Roussel A, et al. Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427(6972):320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 59.Wengler G, Koschinski A, Dreyer F. Entry of alphaviruses at the plasma membrane converts the viral surface proteins into an ion-permeable pore that can be detected by electrophysiological analyses of whole-cell membrane currents. J. Gen. Virol. 2003;84(Pt 1):173–181. doi: 10.1099/vir.0.18696-0. [DOI] [PubMed] [Google Scholar]
- 60.Gibbons DL, Erk I, Reilly B, et al. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell. 2003;114(5):573–583. doi: 10.1016/s0092-8674(03)00683-4. [DOI] [PubMed] [Google Scholar]
- 61.Anthony RP, Brown DT. Protein–protein interactions in an alphavirus membrane. J. Virol. 1991;65(3):1187–1194. doi: 10.1128/jvi.65.3.1187-1194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wahlberg JM, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 1992;66(12):7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn A, Klimjack MR, Chatterjee PK, Kielian M. An epitope of the Semliki Forest virus fusion protein exposed during virus–membrane fusion. J. Virol. 1999;73(12):10029–10039. doi: 10.1128/jvi.73.12.10029-10039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qin ZL, Zheng Y, Kielian M. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J. Virol. 2009;83(9):4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kielian M, Klimjack MR, Ghosh S, Duffus WA. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell. Biol. 1996;134(4):863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duffus WA, Levy-Mintz P, Klimjack MR, Kielian M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 1995;69(4):2471–2479. doi: 10.1128/jvi.69.4.2471-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hernandez R, Luo T, Brown DT. Exposure to low pH is not required for penetration of mosquito cells by Sindbis virus. J. Virol. 2001;75(4):2010–2013. doi: 10.1128/JVI.75.4.2010-2013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McInerney GM, Smit JM, Liljestrom P, Wilschut J. Semliki Forest virus produced in the absence of the 6K protein has an altered spike structure as revealed by decreased membrane fusion capacity. Virology. 2004;325(2):200–206. doi: 10.1016/j.virol.2004.04.043. [DOI] [PubMed] [Google Scholar]
- 69.Gaedigk-Nitschko K, Ding MX, Levy MA, Schlesinger MJ. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology. 1990;175(1):282–291. doi: 10.1016/0042-6822(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 70.Liljestrom P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 1991;65(8):4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loewy A, Smyth J, von Bonsdorff CH, Liljestrom P, Schlesinger MJ. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J. Virol. 1995;69(1):469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liljestrom P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 1991;65(1):147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ivanova L, Lustig S, Schlesinger MJ. A pseudo-revertant of a Sindbis virus 6K protein mutant, which corrects for aberrant particle formation, contains two new mutations that map to the ectodomain of the E2 glycoprotein. Virology. 1995;206(2):1027–1034. doi: 10.1006/viro.1995.1025. [DOI] [PubMed] [Google Scholar]
- 74.Yao JS, Strauss EG, Strauss JH. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J. Virol. 1996;70(11):7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanz MA, Carrasco L. Sindbis virus variant with a deletion in the 6K gene shows defects in glycoprotein processing and trafficking: lack of complementation by a wild-type 6K gene in trans. J. Virol. 2001;75(16):7778–7784. doi: 10.1128/JVI.75.16.7778-7784.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanz MA, Madan V, Carrasco L, Nieva JL. Interfacial domains in Sindbis virus 6K protein. Detection and functional characterization. J. Biol. Chem. 2003;278(3):2051–2057. doi: 10.1074/jbc.M206611200. [DOI] [PubMed] [Google Scholar]
- 77.Sanz MA, Perez L, Carrasco L. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J. Biol. Chem. 1994;269(16):12106–12110. [PubMed] [Google Scholar]
- 78.Melton JV, Ewart GD, Weir RC, Board PG, Lee E, Gage PW. Alphavirus 6K proteins form ion channels. J. Biol. Chem. 2002;277(49):46923–46931. doi: 10.1074/jbc.M207847200. [DOI] [PubMed] [Google Scholar]
- 79.Madan V, Sanz MA, Carrasco L. Requirement of the vesicular system for membrane permeabilization by Sindbis virus. Virology. 2005;332(1):307–315. doi: 10.1016/j.virol.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Madan V, Castello A, Carrasco L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 2008;10(2):437–451. doi: 10.1111/j.1462-5822.2007.01057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gonzalez ME, Carrasco L. Viroporins. FEBS Lett. 2003;552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 82.Strauss EG, Lenches EM, Stamreich-Martin MA. Growth and release of several alphaviruses in chick and BHK cells. J. Gen. Virol. 1980;49(2):297–307. doi: 10.1099/0022-1317-49-2-297. [DOI] [PubMed] [Google Scholar]
- 83.Waite MR, Brown DT, Pfefferkorn ER. Inhibition of Sindbis virus release by media of low ionic strength: an electron microscope study. J. Virol. 1972;10(3):537–544. doi: 10.1128/jvi.10.3.537-544.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez ME, Carrasco L. Human immunodeficiency virus type 1 VPU protein affects Sindbis virus glycoprotein processing and enhances membrane permeabilization. Virology. 2001;279(1):201–209. doi: 10.1006/viro.2000.0708. [DOI] [PubMed] [Google Scholar]
- 85.Lee S, Owen KE, Choi HK, et al. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4(5):531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 86.Skoging U, Vihinen M, Nilsson L, Liljestrom P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4(5):519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 87.Wilkinson TA, Tellinghuisen TL, Kuhn RJ, Post CB. Association of sindbis virus capsid protein with phospholipid membranes and the E2 glycoprotein: implications for alphavirus assembly. Biochemistry. 2005;44(8):2800–2810. doi: 10.1021/bi0479961. [DOI] [PubMed] [Google Scholar]
- 88.Coombs KM, Brown DT. Form-determining functions in Sindbis virus nucleocapsids: nucleosomelike organization of the nucleocapsid. J. Virol. 1989;63(2):883–891. doi: 10.1128/jvi.63.2.883-891.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi HK, Tong L, Minor W, et al. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354(6348):37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 90.Choi HK, Lu G, Lee S, Wengler G, Rossmann MG. Structure of Semliki Forest virus core protein. Proteins. 1997;27(3):345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 91.Hahn CS, Strauss JH. Site-directed mutagenesis of the proposed catalytic amino acids of the Sindbis virus capsid protein autoprotease. J. Virol. 1990;64(6):3069–3073. doi: 10.1128/jvi.64.6.3069-3073.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Melancon P, Garoff H. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol. 1987;61(5):1301–1309. doi: 10.1128/jvi.61.5.1301-1309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith AL, Tignor GH. Host cell receptors for two strains of Sindbis virus. Arch. Virol. 1980;66(1):11–26. doi: 10.1007/BF01315041. [DOI] [PubMed] [Google Scholar]
- 94.Ludwig GV, Kondig JP, Smith JF. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J. Virol. 1996;70(8):5592–5599. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66(8):4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Strauss JH, Rümenapf T, Weir RC, Kuhn RJ, Wang K, Strauss EG. Cellular receptors for alphaviruses. In: Wimmer E, editor. Cellular Receptors for Animal Viruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1994. pp. 141–163. [Google Scholar]
- 97.Lustig S, Jackson AC, Hahn CS, Griffin DE, Strauss EG, Strauss JH. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 1988;62(7):2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tucker PC, Griffin DE. Mechanism of altered Sindbis virus neurovirulence associated with a single-amino-acid change in the E2 glycoprotein. J. Virol. 1991;65(3):1551–1557. doi: 10.1128/jvi.65.3.1551-1557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flynn DC, Meyer WJ, Mackenzie JM, Jr, Johnston RE. A conformational change in Sindbis virus glycoproteins E1 and E2 is detected at the plasma membrane as a consequence of early virus–cell interaction. J. Virol. 1990;64(8):3643–3653. doi: 10.1128/jvi.64.8.3643-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abell BA, Brown DT. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J. Virol. 1993;67(9):5496–5501. doi: 10.1128/jvi.67.9.5496-5501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anthony RP, Paredes AM, Brown DT. Disulfide bonds are essential for the stability of the Sindbis virus envelope. Virology. 1992;190(1):330–336. doi: 10.1016/0042-6822(92)91219-k. [DOI] [PubMed] [Google Scholar]
- 102.Glomb-Reinmund S, Kielian M. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology. 1998;248(2):372–381. doi: 10.1006/viro.1998.9275. [DOI] [PubMed] [Google Scholar]
- 103.Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of Semliki forest virus into BHK-21 cells. J. Cell. Biol. 1980;84(2):404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marsh M, Bolzau E, Helenius A. Penetration of Semliki Forest virus from acidic prelysosomal vacuoles. Cell. 1983;32(3):931–940. doi: 10.1016/0092-8674(83)90078-8. [DOI] [PubMed] [Google Scholar]
- 105.DeTulleo L, Kirchhausen T. The clathrin endocytic pathway in viral infection. EMBO J. 1998;17(16):4585–4593. doi: 10.1093/emboj/17.16.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hammar L, Markarian S, Haag L, Lankinen H, Salmi A, Cheng RH. Prefusion rearrangements resulting in fusion peptide exposure in Semliki Forest virus. J. Biol. Chem. 2003;278(9):7189–7198. doi: 10.1074/jbc.M206015200. [DOI] [PubMed] [Google Scholar]
- 107.Gibbons DL, Ahn A, Chatterjee PK, Kielian M. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 2000;74(17):7772–7780. doi: 10.1128/jvi.74.17.7772-7780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kielian MC, Helenius A. Role of cholesterol in fusion of Semliki Forest virus with membranes. J. Virol. 1984;52(1):281–283. doi: 10.1128/jvi.52.1.281-283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ahn A, Gibbons DL, Kielian M. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 2002;76(7):3267–3275. doi: 10.1128/JVI.76.7.3267-3275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J. Cell. Biol. 1991;112(4):615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Helenius A. Semliki Forest virus penetration from endosomes: a morphological study. Biol. Cell. 1984;51(2):181–185. doi: 10.1111/j.1768-322x.1984.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 112.Wengler G, Wurkner D. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992;191(2):880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 113.Wengler G, Gros C. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology. 1996;222(1):123–132. doi: 10.1006/viro.1996.0403. [DOI] [PubMed] [Google Scholar]
- 114.Li G, Rice CM. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol. 1993;67(8):5062–5067. doi: 10.1128/jvi.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Takkinen K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986;14(14):5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cross RK. Identification of a unique guanine-7-methyltransferase in Semliki Forest virus (SFV) infected cell extracts. Virology. 1983;130(2):452–463. doi: 10.1016/0042-6822(83)90099-5. [DOI] [PubMed] [Google Scholar]
- 117.Laakkonen P, Hyvonen M, Peranen J, Kaariainen L. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J. Virol. 1994;68(11):7418–7425. doi: 10.1128/jvi.68.11.7418-7425.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mi S, Stollar V. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology. 1991;184(1):423–427. doi: 10.1016/0042-6822(91)90862-6. [DOI] [PubMed] [Google Scholar]
- 119.Laakkonen P, Ahola T, Kaariainen L. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 1996;271(45):28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 120.Ahola T, Lampio A, Auvinen P, Kaariainen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18(11):3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang YF, Sawicki SG, Sawicki DL. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 1991;65(2):985–988. doi: 10.1128/jvi.65.2.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gomez de Cedron M, Ehsani N, Mikkola ML, Garcia JA, Kaariainen L. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 1999;448(1):19–22. doi: 10.1016/s0014-5793(99)00321-x. [DOI] [PubMed] [Google Scholar]
- 123.Rikkonen M, Peranen J, Kaariainen L. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 1994;68(9):5804–5810. doi: 10.1128/jvi.68.9.5804-5810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vasiljeva L, Merits A, Auvinen P, Kaariainen L. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 2000;275(23):17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- 125.Strauss EG, De Groot RJ, Levinson R, Strauss JH. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology. 1992;191(2):932–940. doi: 10.1016/0042-6822(92)90268-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vasiljeva L, Valmu L, Kaariainen L, Merits A. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem. 2001;276(33):30786–30793. doi: 10.1074/jbc.M104786200. [DOI] [PubMed] [Google Scholar]
- 127.Mayuri, Geders TW, Smith JL, Kuhn RJ. Role for conserved residues of Sindbis virus nonstructural protein 2 methyltransferase-like domain in regulation of minus-strand synthesis and development of cytopathic infection. J. Virol. 2008;82(15):7284–7297. doi: 10.1128/JVI.00224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Russo AT, White MA, Watowich SJ. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure. 2006;14(9):1449–1458. doi: 10.1016/j.str.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 129.de Groot RJ, Hardy WR, Shirako Y, Strauss JH. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9(8):2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peranen J, Rikkonen M, Liljestrom P, Kaariainen L. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 1990;64(5):1888–1896. doi: 10.1128/jvi.64.5.1888-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaariainen L, Ahola T. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:187–222. doi: 10.1016/S0079-6603(02)71044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tamm K, Merits A, Sarand I. Mutations in the nuclear localization signal of nsP2 influencing RNA synthesis, protein expression and cytotoxicity of Semliki Forest virus. J. Gen. Virol. 2008;89(Pt 3):676–686. doi: 10.1099/vir.0.83320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hahn YS, Strauss EG, Strauss JH. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 1989;63(7):3142–3150. doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lemm JA, Rumenapf T, Strauss EG, Strauss JH, Rice CM. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13(12):2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 1994;68(3):1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Park E, Griffin DE. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology. 2009;388(2):305–314. doi: 10.1016/j.virol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tuittila M, Hinkkanen AE. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J. Gen. Virol. 2003;84(Pt 6):1525–1533. doi: 10.1099/vir.0.18936-0. [DOI] [PubMed] [Google Scholar]
- 138.Allen MD, Buckle AM, Cordell SC, Lowe J, Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330(3):503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- 139.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34(Database issue):D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140■.Malet H, Coutard B, Jamal S, et al. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83(13):6534–6545. doi: 10.1128/JVI.00189-09. [■ of interestFirst presentation of an enzymatic activity for nonstructural protein 3.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vihinen H, Saarinen J. Phosphorylation site ana lysis of Semliki Forest virus nonstructural protein 3. J. Biol. Chem. 2000;275(36):27775–27783. doi: 10.1074/jbc.M002195200. [DOI] [PubMed] [Google Scholar]
- 142.Vihinen H, Ahola T, Tuittila M, Merits A, Kaariainen L. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 2001;276(8):5745–5752. doi: 10.1074/jbc.M006077200. [DOI] [PubMed] [Google Scholar]
- 143.Gorchakov R, Garmashova N, Frolova E, Frolov I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 2008;82(20):10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hahn YS, Grakoui A, Rice CM, Strauss EG, Strauss JH. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 1989;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shirako Y, Strauss EG, Strauss JH. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology. 2000;276(1):148–160. doi: 10.1006/viro.2000.0544. [DOI] [PubMed] [Google Scholar]
- 147.Fata CL, Sawicki SG, Sawicki DL. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J. Virol. 2002;76(17):8641–8649. doi: 10.1128/JVI.76.17.8641-8649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.de Groot RJ, Rumenapf T, Kuhn RJ, Strauss EG, Strauss JH. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl Acad. Sci. USA. 1991;88(20):8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shirako Y, Strauss JH. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J. Virol. 1998;72(3):2310–2315. doi: 10.1128/jvi.72.3.2310-2315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tomar S, Hardy RW, Smith JL, Kuhn RJ. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 2006;80(20):9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell. Biol. 1988;107(6 Pt 1):2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kujala P, Ikaheimonen A, Ehsani N, Vihinen H, Auvinen P, Kaariainen L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75(8):3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kuhn RJ, Niesters HG, Hong Z, Strauss JH. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182(2):430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 154.Barton DJ, Sawicki SG, Sawicki DL. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 1991;65(3):1496–1506. doi: 10.1128/jvi.65.3.1496-1506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. A new role for ns polyprotein cleavage in Sindbis virus replication. J. Virol. 2008;82(13):6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mai J, Sawicki S, Sawicki D. Fate of minus strand templates and replication complexes produced by a P23-cleavage defective mutant of Sindbis virus. J. Virol. 2009;83(17):8553–8564. doi: 10.1128/JVI.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shirako Y, Strauss JH. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990;177(1):54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- 158.Lemm JA, Bergqvist A, Read CM, Rice CM. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 1998;72(8):6546–6553. doi: 10.1128/jvi.72.8.6546-6553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang YF, Sawicki SG, Sawicki DL. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 1994;68(10):6466–6475. doi: 10.1128/jvi.68.10.6466-6475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Raju R, Huang HV. Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters. J. Virol. 1991;65(5):2501–2510. doi: 10.1128/jvi.65.5.2501-2510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Frolov I, Hardy R, Rice CM. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA. 2001;7(11):1638–1651. doi: 10.1017/s135583820101010x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ou JH, Strauss EG, Strauss JH. The 5′-terminal sequences of the genomic RNAs of several alphaviruses. J. Mol. Biol. 1983;168(1):1–15. doi: 10.1016/s0022-2836(83)80319-2. [DOI] [PubMed] [Google Scholar]
- 163.Kuhn RJ, Hong Z, Strauss JH. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J. Virol. 1990;64(4):1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hardy RW. The role of the 3′ terminus of the Sindbis virus genome in minus-strand initiation site selection. Virology. 2006;345(2):520–531. doi: 10.1016/j.virol.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 165.Pushko P, Parker M, Ludwig GV, Davis NL, Johnston RE, Smith JF. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239(2):389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 166.Frolov I, Hoffman TA, Pragai BM, et al. Alphavirus-based expression vectors: strategies and applications. Proc. Natl Acad. Sci. USA. 1996;93(21):11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ou JH, Trent DW, Strauss JH. The 3′-non-coding regions of alphavirus RNAs contain repeating sequences. J. Mol. Biol. 1982;156(4):719–730. doi: 10.1016/0022-2836(82)90138-3. [DOI] [PubMed] [Google Scholar]
- 168.Weiss B, Nitschko H, Ghattas I, Wright R, Schlesinger S. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J. Virol. 1989;63(12):5310–5318. doi: 10.1128/jvi.63.12.5310-5318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tellinghuisen TL, Kuhn RJ. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 2000;74(9):4302–4309. doi: 10.1128/jvi.74.9.4302-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Linger BR, Kunovska L, Kuhn RJ, Golden BL. Sindbis virus nucleocapsid assembly: RNA folding promotes capsid protein dimerization. RNA. 2004;10(1):128–138. doi: 10.1261/rna.5127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wengler G. Identification of a transfer of viral core protein to cellular ribosomes during the early stages of alphavirus infection. Virology. 1984;134(2):435–442. doi: 10.1016/0042-6822(84)90310-6. [DOI] [PubMed] [Google Scholar]
- 172.Tellinghuisen TL, Hamburger AE, Fisher BR, Ostendorp R, Kuhn RJ. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 1999;73(7):5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tellinghuisen TL, Perera R, Kuhn RJ. In vitro assembly of Sindbis virus core-like particles from crosslinked dimers of truncated and mutant capsid proteins. J. Virol. 2001;75(6):2810–2817. doi: 10.1128/JVI.75.6.2810-2817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Weiss B, Geigenmuller-Gnirke U, Schlesinger S. Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 1994;22(5):780–786. doi: 10.1093/nar/22.5.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Owen KE, Kuhn RJ. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 1996;70(5):2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Warrier R, Linger BR, Golden BL, Kuhn RJ. Role of Sindbis virus capsid protein region II in nucleocapsid core assembly and encapsidation of genomic RNA. J. Virol. 2008;82(9):4461–4470. doi: 10.1128/JVI.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Perera R, Navaratnarajah C, Kuhn RJ. A heterologous coiled coil can substitute for helix I of the Sindbis virus capsid protein. J. Virol. 2003;77(15):8345–8353. doi: 10.1128/JVI.77.15.8345-8353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hong EM, Perera R, Kuhn RJ. Alphavirus capsid protein helix I controls a checkpoint in nucleocapsid core assembly. J. Virol. 2006;80(18):8848–8855. doi: 10.1128/JVI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Skoging-Nyberg U, Liljestrom P. M-X-I motif of Semliki Forest virus capsid protein affects nucleocapsid assembly. J. Virol. 2001;75(10):4625–4632. doi: 10.1128/JVI.75.10.4625-4632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ferreira D, Hernandez R, Horton M, Brown DT. Morphological variants of Sindbis virus produced by a mutation in the capsid protein. Virology. 2003;307(1):54–66. doi: 10.1016/s0042-6822(02)00034-x. [DOI] [PubMed] [Google Scholar]
- 181.Garoff H, Huylebroeck D, Robinson A, Tillman U, Liljestrom P. The signal sequence of the p62 protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J. Cell. Biol. 1990;111(3):867–876. doi: 10.1083/jcb.111.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Johnson DC, Schlesinger MJ, Elson EL. Fluorescence photobleaching recovery measurements reveal differences in envelopment of Sindbis and vesicular stomatitis viruses. Cell. 1981;23(2):423–431. doi: 10.1016/0092-8674(81)90137-9. [DOI] [PubMed] [Google Scholar]
- 183.Sefton BM. Immediate glycosylation of Sindbis virus membrane proteins. Cell. 1977;10(4):659–668. doi: 10.1016/0092-8674(77)90099-x. [DOI] [PubMed] [Google Scholar]
- 184.Hsieh P, Robbins PW. Regulation of asparagine-linked oligosaccharide processing. Oligosaccharide processing in Aedes albopictus mosquito cells. J. Biol. Chem. 1984;259(4):2375–2382. [PubMed] [Google Scholar]
- 185.Knight RL, Schultz KL, Kent RJ, Venkatesan M, Griffin DE. Role of N-linked glycosylation for Sindbis virus infection and replication in vertebrate and invertebrate systems. J. Virol. 2009;83(11):5640–5647. doi: 10.1128/JVI.02427-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu N, Brown DT. Phosphorylation and dephosphorylation events play critical roles in Sindbis virus maturation. Virology. 1993;196(2):703–711. doi: 10.1006/viro.1993.1527. [DOI] [PubMed] [Google Scholar]
- 187.Liu LN, Lee H, Hernandez R, Brown DT. Mutations in the endo domain of Sindbis virus glycoprotein E2 block phosphorylation, reorientation of the endo domain, and nucleocapsid binding. Virology. 1996;222(1):236–246. doi: 10.1006/viro.1996.0414. [DOI] [PubMed] [Google Scholar]
- 188.Gaedigk-Nitschko K, Schlesinger MJ. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183(1):206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 189.Ivanova L, Schlesinger MJ. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 1993;67(5):2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Salminen A, Wahlberg JM, Lobigs M, Liljestrom P, Garoff H. Membrane fusion process of Semliki Forest virus. II: Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell. Biol. 1992;116(2):349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yu IM, Zhang W, Holdaway HA, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 192.Zhao H, Garoff H. Role of cell surface spikes in alphavirus budding. J. Virol. 1992;66(12):7089–7095. doi: 10.1128/jvi.66.12.7089-7095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Strauss JH, Strauss EG, Kuhn RJ. Budding of alphaviruses. Trends Microbiol. 1995;3(9):346–350. doi: 10.1016/s0966-842x(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 194.Liu N, Brown DT. Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J. Cell. Biol. 1993;120(4):877–883. doi: 10.1083/jcb.120.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Metsikko K, Garoff H. Oligomers of the cytoplasmic domain of the p62/E2 membrane protein of Semliki Forest virus bind to the nucleocapsid in vitro. J. Virol. 1990;64(10):4678–4683. doi: 10.1128/jvi.64.10.4678-4683.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Suomalainen M, Liljestrom P, Garoff H. Spike protein–nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 1992;66(8):4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kail M, Hollinshead M, Ansorge W, et al. The cytoplasmic domain of alphavirus E2 glycoprotein contains a short linear recognition signal required for viral budding. EMBO J. 1991;10(9):2343–2351. doi: 10.1002/j.1460-2075.1991.tb07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Garoff H, Simons K. Location of the spike glycoproteins in the Semliki Forest virus membrane. Proc. Natl Acad. Sci. USA. 1974;71(10):3988–3992. doi: 10.1073/pnas.71.10.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lu YE, Cassese T, Kielian M. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 1999;73(5):4272–4278. doi: 10.1128/jvi.73.5.4272-4278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lee H, Ricker PD, Brown DT. The configuration of Sindbis virus envelope proteins is stabilized by the nucleocapsid protein. Virology. 1994;204(1):471–474. doi: 10.1006/viro.1994.1557. [DOI] [PubMed] [Google Scholar]
- 201.Lee H, Brown DT. Mutations in an exposed domain of Sindbis virus capsid protein result in the production of noninfectious virions and morphological variants. Virology. 1994;202(1):390–400. doi: 10.1006/viro.1994.1355. [DOI] [PubMed] [Google Scholar]
- 202.Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. Membrane proteins organize a symmetrical virus. EMBO J. 2000;19(19):5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.West J, Hernandez R, Ferreira D, Brown DT. Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly. J. Virol. 2006;80(9):4458–4468. doi: 10.1128/JVI.80.9.4458-4468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Helenius A, Kartenbeck J. The effects of octylglucoside on the Semliki Forest virus membrane. Evidence for a spike-protein–nucleocapsid interaction. Eur. J. Biochem. 1980;106(2):613–618. doi: 10.1111/j.1432-1033.1980.tb04609.x. [DOI] [PubMed] [Google Scholar]
- 205.Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13(18):4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lopez S, Yao JS, Kuhn RJ, Strauss EG, Strauss JH. Nucleocapsid–glycoprotein interactions required for assembly of alphaviruses. J. Virol. 1994;68(3):1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Strauss EG, Lenches EM, Strauss JH. Molecular genetic evidence that the hydrophobic anchors of glycoproteins E2 and E1 interact during assembly of alphaviruses. J. Virol. 2002;76(20):10188–10194. doi: 10.1128/JVI.76.20.10188-10194.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hernandez R, Lee H, Nelson C, Brown DT. A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly. J. Virol. 2000;74(9):4220–4228. doi: 10.1128/jvi.74.9.4220-4228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Garmashova N, Gorchakov R, Frolova E, Frolov I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 2006;80(12):5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 2005;79(15):9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Aguilar PV, Weaver SC, Basler CF. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 2007;81(8):3866–3876. doi: 10.1128/JVI.02075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20(1):87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Levine B, Huang Q, Isaacs JT, Reed JC, Griffin DE, Hardwick JM. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature. 1993;361(6414):739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 214.Lewis J, Wesselingh SL, Griffin DE, Hardwick JM. Alphavirus-induced apoptosis in mouse brains correlates with neurovirulence. J. Virol. 1996;70(3):1828–1835. doi: 10.1128/jvi.70.3.1828-1835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Glasgow GM, McGee MM, Tarbatt CJ, Mooney DA, Sheahan BJ, Atkins GJ. The Semliki Forest virus vector induces p53-independent apoptosis. J. Gen. Virol. 1998;79(Pt 10):2405–2410. doi: 10.1099/0022-1317-79-10-2405. [DOI] [PubMed] [Google Scholar]
- 216.Joe AK, Foo HH, Kleeman L, Levine B. The transmembrane domains of Sindbis virus envelope glycoproteins induce cell death. J. Virol. 1998;72(5):3935–3943. doi: 10.1128/jvi.72.5.3935-3943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jan JT, Griffin DE. Induction of apoptosis by Sindbis virus occurs at cell entry and does not require virus replication. J. Virol. 1999;73(12):10296–10302. doi: 10.1128/jvi.73.12.10296-10302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Griffin DE, Hardwick JM. Regulators of apoptosis on the road to persistent alphavirus infection. Annu. Rev. Microbiol. 1997;51:565–592. doi: 10.1146/annurev.micro.51.1.565. [DOI] [PubMed] [Google Scholar]
- 219.Li ML, Stollar V. Alphaviruses and apoptosis. Int. Rev. Immunol. 2004;23(1–2):7–24. doi: 10.1080/08830180490265529. [DOI] [PubMed] [Google Scholar]
- 220.Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 2007;81(10):5046–5057. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lundstrom K. Alphavirus vectors for gene therapy applications. In: Hunt KK, Vorburger SA, Swisher SG, editors. Cancer Drug Discovery and Development: Gene Therapy for Cancer. Human Press; Totowa, NJ, USA: 2007. pp. 109–119. [Google Scholar]