Abstract
The dipeptidyl peptidase IV (DPIV) enzyme family contains both potential and proven therapeutic targets. Recent reports indicate the presence of DP8 and DP9 in peripheral blood lymphocytes, testis, lung, and brain. For a more comprehensive understanding of DP8 and DP9 tissue and cellular expression, mRNA and enzyme activity were examined. Many organs from C57BL/6 wild-type and DPIV gene-knockout mice were examined; DP8/9 enzyme activity was detected in the immune system, brain, testis, muscle, and epithelia. In situ hybridization localized DP8 and DP9 mRNA to lymphocytes and epithelial cells in liver, gastrointestinal tract, lymph node, spleen, and lung. DP8 and DP9 mRNA was detected in baboon and mouse testis, and DP9 expression was elevated in human testicular cancers. DP8 and DP9 mRNA were ubiquitous in day 17 mouse embryo, with greatest expression in epithelium (skin and gastrointestinal tract) and brain. Thus, DP8 and DP9 are widely expressed enzymes. Their expression in lymphocytes and epithelia indicates potential for roles in the digestive and immune systems. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials. (J Histochem Cytochem 57:1025–1040, 2009)
Keywords: dipeptidyl peptidase, testis, liver cirrhosis, epithelium, lymphocyte
Dipeptidyl peptidase IV (DPIV) family enzymes have emerged as clinically important in recent years, with DPIV a therapeutic target for type 2 diabetes and fibroblast activation protein (FAP) a potential cancer therapeutic target (Wang et al. 2008; LeBeau et al. 2009; Kirby et al. in press). DPIV (CD26) is a ubiquitous aminopeptidase and has a variety of roles in the fields of metabolism, immunology, endocrinology, and cancer biology (Gorrell 2005). These advances and the discovery of the related enzymes DP8 and DP9 (Abbott et al. 2000; Qi et al. 2003; Ajami et al. 2004; Pitman et al. 2009) highlight a need to re-examine the tissue expression of this peptidase family, particularly the newer peptidases DP8 and DP9.
Little is known about the functions of the cytoplasmic enzymes DP8 and DP9. Both are ubiquitously expressed at the mRNA level in humans and rodents (Abbott et al. 2000; Ajami et al. 2004; Gorrell and Yu 2005). DP8 mRNA levels are elevated in activated and transformed human lymphocytes (Abbott et al. 2000), and a DP8 splice variant is upregulated in human adult testis compared with fetal testis (Zhu et al. 2005). DP8/9 enzymatic activity has been reported in human leukocytes (Maes et al. 2007), rat brain (Frerker et al. 2007), bovine testis (Dubois et al. 2009), and mouse lung (Schade et al. 2008). DP8 and DP9 are able to cleave some known DPIV substrates, including neuropeptide Y (NPY) and CXCL12 (Bjelke et al. 2006; Ajami et al. 2008). DP8 and DP9 overexpression can downregulate cell adhesion and migration and increase apoptosis, but these effects are independent of their enzyme activity (Yu et al. 2006). Studies that used isoleucyl thiazolidine as a DPIV inhibitor, which we now know primarily inhibits DP8 and DP9 (Lankas et al. 2005), suggest that inhibition of DP8 and DP9 suppresses inflammation in the DPIV-deficient rat (Tanaka et al. 1997; von Bonin et al. 1998). This is supported by a recent report that a DP8/9-selective inhibitor suppresses DNA synthesis in mitogen-stimulated splenocytes from wild-type and DPIV gene-knockout (gko) mice (Reinhold et al. in press). Thus, DP8 and DP9 are likely to have functions in leukocytes.
DPIV gko mice generally have a normal, healthy phenotype. Furthermore, in metabolism studies, DPIV gko mice have increased glucose clearance after a glucose challenge, in comparison to wild-type mice (Marguet et al. 2000) and are resistant to high-fat diet–induced obesity and associated insulin resistance (Conarello et al. 2003). DPIV gko mice have altered inflammatory responses, with more knee joint inflammation in experimental arthritis (Busso et al. 2005) and increased experimental autoimmune encephalomyelitis severity (Preller et al. 2007), but whether these roles relate to DPIV enzyme activity or extra-enzymatic activities (Wang et al. 2005) is unknown. Moreover, these phenomena in the DPIV gko mouse might relate to compensatory DP8 and DP9 overexpression.
Before the discovery of DP8 and DP9, DPIV-like enzyme activity in cells and organs was attributed solely to DPIV (Gossrau 1979; Hartel et al. 1988; Gorrell et al. 1991,2001). However, in the DPIV-deficient Fischer rat substrain, a non-DPIV–derived residual DPIV-like enzyme activity remains in certain organs (Smith et al. 1992), which might include activity derived from DP8 or DP9. FAP mRNA distribution has been reported as ubiquitous at very low levels (Dolznig et al. 2005), but FAP protein is not detected in normal tissue (Wang et al. 2008). Here we provide further insight into the distribution of the newer DPIV gene family members, DP8 and DP9. An ex vivo enzymatic approach was undertaken using the DP8/DP9 inhibitor N-ethylmaleimide (NEM) (Park et al. 2008) on C57BL/6 wild-type and DPIV gko animals. Because DPIV is widely expressed and often masks the presence of other DPs, the DPIV gko mouse was particularly useful in detecting DP8 and DP9 enzyme activity. DP8 and DP9 mRNA was localized in cell types by in situ hybridization (ISH).
Materials and Methods
Tissue and Cell Samples
Human liver transplant recipient paraffin sections (Table 1) were obtained in accordance with National Health and Medical Research Council (NHMRC) guidelines under Royal Prince Alfred Hospital Human Ethics Committee approvals. Human liver, lymph node, and colon samples were obtained under a single protocol. Human testicular tumor samples were obtained from juvenile male patients (aged 11–18) undergoing orchidectomy for testicular cancer at Westmead Children's Hospital under institutional approvals. Tissue samples for RNA preparation were snap frozen immediately after collection and stored at −80C.
Table 1.
Human tissues
| Tissue | Condition/diagnosis | Histopathology |
|---|---|---|
| Liver (Patient 1) | Alcoholic cirrhosis and HCC | Established cirrhosis with nodules of regenerating hepatocytes, surrounded by bands of fibrosis. Prominent bile ductular proliferation within fibrous bands. Lymphoplasmacytic infiltrate within fibrous bands. |
| Liver (Patient 2) | Cirrhosis; alcoholic, HCV, and HCC | Established cirrhosis consistent with HCV. Well-differentiated HCC. Lymphoid aggregates within portal tracts. Mild lobular inflammation with occasional scattered apoptotic bodies. |
| Colon | PSC and mild ulcerative colitis | Mild mucosal crypt abnormalities: crypt shortening and focal branching. Mild active mucosal inflammation with crypt abscesses. |
| Lymph node | HCV cirrhosis and enlarged hilar lymph nodes | Prominent sinus histiocytosis: massive lymphadenopathy with marked proliferation of histiocytes within distended lymph node sinuses. |
| Gallbladder | HBV | Chronic cholecystitis |
HCC, hepatocellular carcinoma; PSC, primary sclerosing cholangitis; HCV, hepatitis C virus; HBV, hepatitis B virus.
Baboon (aged 8–13 years) and mouse organs were collected after euthanasia. Baboon tissues for ISH (Table 2) were fixed in 10% neutral-buffered formalin and paraffin embedded. C57BL/6 wild-type and DPIV gko mice have been described (Marguet et al. 2000; Wang et al. 2002). Adult (8–16 weeks old) mouse organs were collected onto ice and homogenized on ice in 1:10 wet weight/volume in 1 × Tris-EDTA (TE) (pH 8.0)/1% Triton X-114/2 mM dithiothreitol (DTT)/10% glycerol with the addition of protease inhibitors [20 μg/ml phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml aprotinin, 1 μM leupeptin, 2 μM pepstatin, 2 mM 4-(2-aminoethyl)benzenesulphonylfluoride] using a PT2100 Polytron homogenizer (Kinematica; Lucerne, Switzerland) and spun at 4C at 10,000 × g for 5 min. Supernatants, discarding lipid where possible, were stored at −20C. Glycerol was necessary to preserve DP8/9 enzyme activity (data not shown). Mouse blood (from tail or by heart puncture after CO2 euthanasia) was collected into EDTA tubes for plasma, and peripheral blood mononuclear cells (PBMCs) were isolated by red blood cell lysis then homogenized as above. Bone marrow was collected from mouse femurs by flushing with PBS and washed in PBS before homogenization. Skin samples were collagenase treated to obtain cell suspensions (Lynch et al. 2003). Swiss mice were obtained from the University of Newcastle Central Animal Facility and killed by decapitation (juveniles) or CO2 asphyxiation (adults) before tissue removal. All investigations conformed to the NHMRC regulations and were approved by the University of Sydney or University of Newcastle Animal Care and Ethics Committees.
Table 2.
Baboon tissues
| Baboon | Tissue condition and histopathology |
|---|---|
| G | Chronic respiratory wheeze, enlarged spleen and lymph nodes. |
| Histology: liver, mild hepatitis and steatosis, mild lobular inflammation; lung, lymphoid infiltrate. | |
| S | Chronic respiratory infection, enlarged liver and spleen. |
| Histology: lung, focal congestion and edema; spleen, reactive follicular hyperplasia in white pulp. | |
| M | Chronic elevated white blood cell count. |
| Histology: lung, chronic inflammation in bronchial mucosa. | |
| J | Chronic respiratory wheeze, type 1 diabetic animal (Birrell et al. 2002). |
| Histology: lung, chronic inflammatory infiltrate in the small bronchi walls. |
Cell lines were grown in RPMI (Jurkat and Raji) or DMEM [293T, human hepatoma lines Huh-7 and HepG2, human skin squamous carcinoma line Colo-16, human skin fibroblast line, mouse fibroblast 3T3, and Chinese hamster ovary (CHO)] with 10% fetal calf serum. The human skin fibroblast line was a gift from Ian Frazer, and was derived from a human punch biopsy (with consent) immortalized with an E6/E7 retrovirus (Xiong et al. 1996). Cells were harvested with trypsin because DPIV is trypsin resistant.
Labeling of Riboprobes
Anti-sense and sense digoxigenin (DIG)-labeled RNA probes were synthesized by transcribing 0.5 μg of a PCR product that contained artificially introduced T7 phage promoter sequences. Human 332 bp DP8 (578–910 bp, accession number AF221634) and 288 bp DP9 (2152–2439 bp, accession number AY374518) templates were produced under standard PCR conditions using AmpliTaq-Gold (Applied Biosystems; Foster City, CA). To produce templates for anti-sense probes, the T7_DP8_Reverse (5′-TATGCATAATACGACTCACTATAGGGAGAGGTTGTTGCGTAAATCCTTGTGG-3′) and the T7_DP9_Reverse (5′-TATGCATAATACGACTCACTATAGGGAGAAGGACCAGCCATGGATGGCAACTC-3′) primers were paired with standard DP8_Forward (5′-CCAGATGGACCTCATTCAGACAG-3′) and DP9_Forward (5′-AGAAGCACCCCACCGTCCTCTTTG-3′) primers. To produce templates for sense probes, the T7_DP8_Forward (5′-gccttctaatacgactcactatagggagaCCAGATGGACCTCATTCAGACAG-3′) and the T7_DP9_Forward (5′-gccttctaatacgactcactatagggagaAGAAGCACCCCACCGTCCTCTTTG-3′) primers were paired with DP8_Reverse (5′-GGTTGTTGCGTAAATCCTTGTGG-3′) and DP9_Reverse (5′-AGGACCAGCCATGGATGGCAACTC-3′) primers. In vitro transcription of the amplified DNA template used the DIG system RNA labeling kit (Roche Applied Science; Mannheim, Germany).
ISH
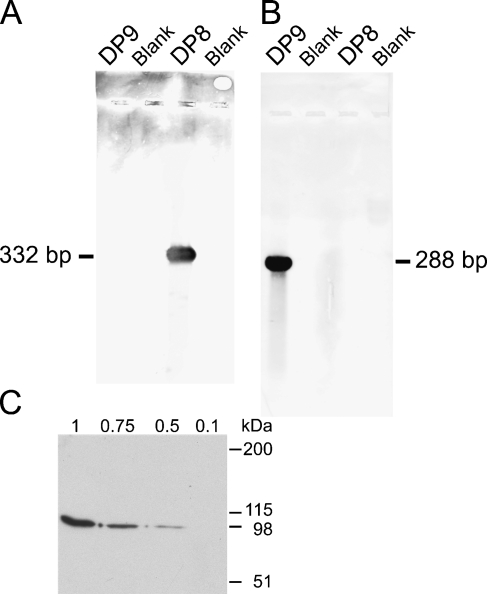
Serial 5-μm paraffin sections were dewaxed in histolene and re-hydrated. Sections were postfixed in 4% paraformaldehyde, then serially treated with 0.3% Triton X-100, 18 ng/ml proteinase K (37C for 30 min), and 0.1 M triethanolamine/0.25% acetic anhydride. Prehybridization and hybridization buffers included 50% deionized formamide. The 2-hr prehybridization and overnight hybridization were at 37C and 58C for DP8 and DP9, respectively. Anti-sense and sense probes were at 1 and 2 ng/μl for DP8 and DP9, respectively. Slides were then washed twice in 2 × saline-sodium citrate (SSC) for 15 min, 1 × SSC for 15 min, and 0.1 × SSC for 30 min. DIG-labeled RNA was detected using anti-DIG antibody (Roche Applied Science) and 4-nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate (Roche Applied Science) chromogenic substrate. The probes were sequenced, and their specificity was verified using Southern blotting with DIG detection (Figures 1A and 1B).
Figure 1.
In situ hybridization (ISH) probe specificity Southern blots (A,B). Digoxigenin-labeled dipeptidyl peptidase 8 (DP8) and DP9 riboprobes were hybridized onto full-length human DP8 and DP9 cDNAs on Southern blots. The DP8 probe bound only to DP8 (A), and the DP9 probe bound only to DP9 (B), thus showing their specificity. Western blot using the rabbit anti-DP8 antibody on 0.1 to 1 μg of recombinant human DP8 (C).
Enzyme Assays
To differentiate DPs, assays were designed considering inhibitor and substrate specificity, and optimum temperature and pH. DP8 and DP9, but not DPIV and FAP, are non-competitively inhibited by the thiol reagent NEM, due to alkylation of cysteine residues (Park et al. 2008). The non-selective dipeptidyl peptidase (DP) inhibitor ValboroPro (PT100) and the DPIV selective inhibitor sitagliptin (MK-0431) have been described (Flentke et al. 1991; Kim et al. 2005). The substrates H-GlyPro-p-nitroanilide (H-GlyPro-pNA) and H-AlaPro-p-nitroanilide (H-AlaPro-pNA) are efficiently cleaved by DPIV, DP8, and DP9, but FAP poorly hydrolyses H-GlyPro substrates (Rettig et al. 1994; Aertgeerts et al. 2005). To exclude DPII activity, all assays were performed at pH 8.0 (Maes et al. 2005). Whereas DPIV-derived enzyme activity is detectable immediately at room temperature, DP8- and DP9-mediated hydrolysis is more efficient at 37C (Abbott et al. 2000; Ajami et al. 2004).
Each assay used 10 μl of homogenized sample in triplicate in the presence/absence of inhibitor (5 mM NEM or 10 μM sitagliptin or ValboroPro), in a total of 50 μl TE buffer, pH 8.0, and was preincubated for 10 min at room temperature. Following the time zero 405-nm absorbance measurement, 50 μl of 1 mM substrate, 1 mM DTT in TE was added. Enzyme activity calculations were made following subtraction of lysis buffer control (or TE buffer for plasma) and converted to enzyme units, where one enzyme unit catalyses the release of 1 μmol pNA from the substrate per minute.
To discriminate between DPIV and DP8/9, three types of assays were used. Assay types 1 and 2 employed NEM and sitagliptin as inhibitors and were applied to two substrates, H-GlyPro-pNA and H-AlaPro-pNA. Assay types 1 and 2 were performed with time zero at room temperature, then the assay was gradually warmed to 37C in the plate reader during the assay. Two time intervals were analyzed: the first at 20 min (assay type 1) for detecting predominantly DPIV, and the second at 15–35 min (assay type 2), when the assay is warm, for detecting predominantly DP8/9. Assay type 1 recorded high backgrounds while the lysis buffer warmed from room temperature to 37C. This was overcome by using assay type 3, described below. Assay type 2 measurements were above the instrument limit on wild-type organs that exhibited very high DPIV activity, but in these organs, DPIV gko mouse organs generated readable data.
Assay type 3 was designed to mainly detect DPIV, and used NEM, sitagliptin, and ValboroPro as inhibitors. Assays were performed at room temperature (where DPIV is more active than DP8/9) with the substrate H-GlyPro-pNA (this substrate excludes FAP). The absorbance change at 405 nm from 5–15 min was measured, and background absorbance (lysis buffer–derived absorbance) at 572 nm was subtracted.
Immunohistochemistry
Immunohistochemistry was performed as previously described (McCaughan et al. 1990). Briefly, 5-μm human or baboon paraffin sections underwent deparaffinization and rehydration. Antigen retrieval was performed using a pressure cooker and Universal Decloaker solution (Biocare Medical; Concord, CA). Sections were treated with 3% H2O2 for 10 min, rinsed in PBS, then treated with 1% human or baboon serum/1% goat serum/1% BSA for 10 min to minimize nonspecific binding. Sections were incubated for 1 hr each with a rabbit polyclonal antibody against DP9 human/mouse peptide (Abcam; Cambridge, UK, #ab42080, 1:50) with 0.2% saponin and 1% BSA in PBS, followed by goat anti-rabbit conjugated to horseradish peroxidase (HRPO) (Dako; Glostrup, Denmark, #P0448, 1:100). The chromogenic reaction utilized either 3,3-diaminobenzidine (DAB)/H2O2 or NovaRed substrate kit (Vector Laboratories; Burlingame, CA).
Immunoblot
Tissue homogenates were prepared from samples stored at −70C, 1:10 wet weight/volume in 1 × TE, pH 7.6/0.15 M NaCl/1% Triton X-114/2 mM DTT, with protease inhibitors added. Homogenates were DNase I treated at 10 μg/ml for 10 min at room temperature. A further incubation of 10 min on ice allowed clumps to settle and be removed before centrifugation at 4C, 10,000 × g for 15 min. SDS-PAGE and Western blot were performed using standard techniques in reducing conditions (McCaughan et al. 1990), with samples preheated at 70C for 8 min. Blots were stained with a rabbit anti-DP8 anti-serum diluted 1:500, generated to the human/mouse DP8 peptides TGYTERYMGHPDQNEQG and GKPYDLQIYPQERHS or control normal rabbit serum. These antigenic peptides were chosen using the MacVector software (Accelrys Software Inc.; San Diego, CA) and were conjugated to diphtheria toxin using added N- and C-terminal cysteines (Auspep; Melbourne, Australia). The secondary antibody was goat anti-rabbit immunoglobulin-HRPO (Dako) diluted 1:500 to 1:800. Using dot immunoblot and Western blot, the DP8 antiserum was shown to contain detectable antibodies to both of the immunizing DP8 peptides and not bind to DPIV (data not shown). The anti-DP8 polyclonal antibody bound to recombinant human DP8 protein, made as described previously (Ajami et al. 2008; Park et al. 2008), in Western blot (Figure 1C) and binding to DP8, was completely ablated by blocking with purified recombinant DP8 (not shown).
Germ Cell Isolation
For isolation of spermatogonia, testes from 20 8-day-old Swiss mice were pooled, whereas for pachytene spermatocytes and round spermatids, mice 8 or more weeks old were used. Testes were isolated, decapsulated, and incubated for 15 min each in 0.5 mg/ml collagenase/DMEM with agitation and then in 0.25% trypsin/EDTA in DMEM. Tubules were dissociated manually by pipetting and washed in 0.5% BSA in DMEM by centrifugation. Cell pellets were resuspended in DMEM and filtered twice through a 70-μm membrane, then separated over a BSA gradient. Purified germ cells were identified by GCNA1 staining for spermatogonia and Arachis hypogaea lectin staining of the developing acrosome for spermatocytes and spermatids (Baleato et al. 2005).
CodeLink Microarrays
Gene expression analysis was performed utilizing CodeLink mouse whole-genome array slides (GE Healthcare; Chalfont St Giles, UK) according to the manufacturer's instructions. Briefly, cDNA was generated from ∼2 μg of total RNA from neonatal isolated mouse germ cells and testes. In vitro transcription was performed, incorporating biotinylated uridine 5′-triphosphate in the resulting amplified RNA (aRNA). Ten micrograms of aRNA were hybridized with the mouse whole-genome slide, and detection of hybridization was carried out by probing with Cy5-streptavidin. Slides were scanned in an Axon scanner, and data were analyzed with proprietary CodeLink Expression Analysis Software (GE Healthcare) (Holt et al. 2006). Relative signals of the following mRNAs were compared: DPIV (NM_010074), DP8 (NM_028906), DP9 (NM_172624), and the unrelated enzymes, carboxypeptidase DPII (NM_031843) and metalloproteinase DP3 (NM_133803). In addition, a DPIV-like expressed sequence tag (EST; RIKEN BB005242) was analyzed; this EST had greatest identity (84%) with the 3′ non-coding region of mouse DPIV mRNA (NM_010074).
Total RNA was extracted from human testicular tumor samples and subjected to DNase treatment, (6 units RQ DNase I; Promega, Madison, WI) at 37C for 60 min. The RNA was precipitated, resuspended, and then reverse transcribed using M-MLV reverse transcriptase (200 units, Promega) for 60 min at 42C. Quantitative PCR was conducted twice in triplicate using the resulting cDNA and the RT control with DP9 PCR primer pair 5′-AAGTACTCGGGCCTCATT-3′, 3′-TCTTGGGAATCTCAGAGTAG-5′ (product of 155 bp). The quantitative PCR (qPCR) parameters were: 1 cycle at 95C (15 min), and 35 cycles at 95C (30 sec), 55C (30 sec), and 72C (40 sec) on an Opticon 2 (Baleato et al. 2005).
Statistical Methods
Results are expressed as mean ± standard error. Differences among groups were analyzed using Student's t-test. P values <0.05 were considered significant.
Results
DP Distribution in Mouse Organs
Immune System (Thymus, Lymph Node, Spleen, PBMCs)
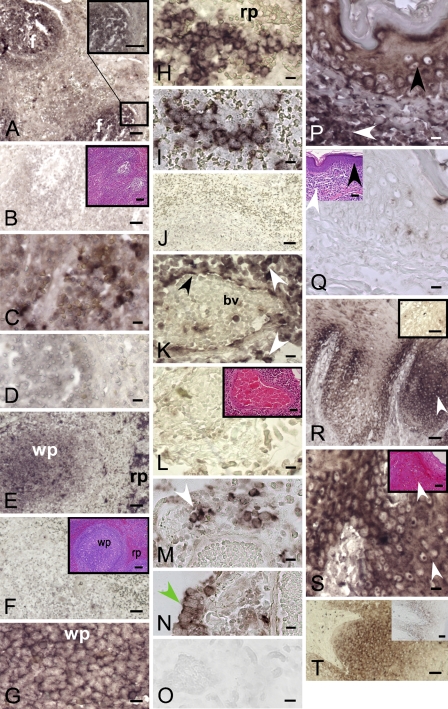
ISH for DP8 and DP9 revealed positive staining for lymphocytes in mantle and paracortical zones of human lymph node and baboon spleen (Figures 2A–2D). In baboon spleen, marginal zone small lymphocytes were also positive (Figure 2E). Large lymphoid cells in red pulp sinusoids were strongly positive, whereas sinusoidal endothelium was negative (Figures 2E–2J).
Figure 2.
DP8 and DP9 mRNA expression in leukocytes and epithelium detected by ISH. Human lymph node follicular lymphocytes (f) were DP8-ISH (A) and DP9-ISH (C) positive compared with the sense controls [(B) DP8 with hematoxylin and eosin (H and E) stain inset; (D) DP9]. Baboon spleen: intense DP8-specific (E,G,H) and DP9-specific (I) staining in white pulp (wp) follicular lymphocytes and red pulp (rp) inter-follicular lymphocytes (Table 2, Animal S). DP8 (F) and DP9 (J) baboon spleen sense control and H and E staining in inset. DP8-positive (K) and DP9-positive (M,N) staining in lymphoid aggregates (white arrow), blood vessel (bv) endothelium (black arrow), and epithelial cells (green arrow) in pneumonic baboon lung (Animal S). Lung DP8 (L) and DP9 (O) sense control and H and E staining inset showing interstitial pneumonia. DP8 in squamous epithelial cells (keratinocytes; black arrow) and dermis (white arrow) of baboon skin (Animal S) (P). Negative, sense control, and H and E staining, inset (Q). DP8 (R,S) and DP9 (T) positivity in stratified squamous epithelium of baboon tongue (white arrow) (Animal G). DP8 (R) and DP9 (T) insets are negative controls. Magnified area of R with H and E stain, inset (S). Bars: A,B,E,F,J,R,T, and all insets = 50 μm; C,D,G–I,K–Q,S = 10 μm.
DP8/9 activity was detected in all immune system tissues examined using assay type 2 and the DP8/9 inhibitor NEM, in lymph node, PBMCs, thymus, and spleen. In H-GlyPro assays, NEM inhibition was significant in wild-type PBMCs and in DPIV gko lymph node, thymus, and spleen (Table 3). Using H-AlaPro, NEM inhibition was significant in wild-type lymph node and PBMC, and in DPIV gko lymph node and thymus (Table 4). Assay type 1 did not detect NEM inhibition (see supplementary Tables ST1 and ST2).
Table 3.
Assay type 2, H-GlyPro hydrolysis expressed as enzyme activity (mU/g, or mU/ml for plasma) at OD405 (mean ± SE) on five to eight animals
| Wild type
|
DPIV gko
|
|||
|---|---|---|---|---|
| N-ethylmaleimide | − | + | − | + |
| Immune system | ||||
| Lymph node | 581 ± 25 | 502 ± 29 | 209 ± 11 | 116 ± 4.3a |
| PBMCs | 97.2 ± 14 | 52.8 ± 6.0a | 44.7 ± 14 | 17.0 ± 5.4 |
| Thymus | 502 ± 56 | 444 ± 47 | 130 ± 17 | 61.5 ± 8.2a |
| Spleen | X | X | 132 ± 24 | 71.0 ± 8.2a |
| Epithelial organs | ||||
| Colon | 245 ± 15 | 178 ± 11a | 132 ± 4.1 | 66.3 ± 6.8a |
| Small intestine | 444 ± 52 | 391 ± 43 | 96.7 ± 12 | 98.6 ± 13 |
| Skin | 357 ± 29 | 336 ± 22 | 45.1 ± 8.7 | 33.3 ± 5.6 |
| Lung | X | X | 171 ± 20.6 | 96.0 ± 12a |
| Liver | X | X | 442 ± 20 | 315 ± 15a |
| Kidney | X | X | 190 ± 11 | 163 ± 8.3 |
| Central nervous system | ||||
| Brain | 162 ± 7.7 | 79.9 ± 4.4a | 121 ± 7.0 | 55.0 ± 3.1a |
| Midbrain and hindbrain | 141 ± 6.0 | 82.4 ± 6.1a | 119 ± 15 | 61.0 ± 4.3a |
| Muscle | ||||
| Skeletal | 125 ± 7.5 | 105 ± 8.2 | 77.8 ± 4.9 | 57.1 ± 3.7a |
| Heart | 153 ± 9.4 | 127 ± 9.4a | 87.6 ± 8.5 | 64.1 ± 10 |
| Uterus | 305 ± 16 | 216 ± 21a | 201 ± 24 | 119 ± 17a |
| Reproductive/endocrine/other | ||||
| Testis | 148 ± 11 | 82.8 ± 16a | 139 ± 7.3 | 77.8 ± 6.8a |
| Bone marrow | 87.2 ± 7.7 | 86.7 ± 9.7 | 32.9 ± 8.7 | 34.5 ± 7.8 |
| Plasma | 11.0 ± 1.5 | 9.82 ± 1.4 | 1.7 ± 0.6 | 2.73 ± 0.74 |
| Pancreas | 440 ± 27 | 381 ± 23.7 | 387 ± 28 | 295 ± 19a |
| Adipose | 131 ± 11 | 101 ± 12 | 22.8 ± 3.8 | 3.60 ± 1.5a |
| Adrenal | 166 ± 12 | 147 ± 12 | 137 ± 7.9 | 104 ± 10a |
| Ovary | 257 ± 51 | 242 ± 50 | 177 ± 9.8 | 160 ± 20 |
| Prostate | 327 ± 39 | 283 ± 33 | 89.5 ± 9.8 | 71.3 ± 14 |
| Submaxillary gland | X | X | 97.9 ± 8.6 | 92.9 ± 7.2 |
N-ethylmaleimide (NEM) inhibition is significant compared to no inhibitor, indicating the detection of DP8/9 activity.
+, presence of NEM inhibitor; −, absence of NEM inhibitor; gko, gene knockout; X, exceeded the maximum measurable optical density (OD). PBMCs, peripheral blood mononuclear cells.
Table 4.
Assay type 2, H-AlaPro hydrolysis expressed as enzyme activity (mU/g, or mU/ml for plasma) at OD405
| Wild type
|
DPIV gko
|
|||
|---|---|---|---|---|
| NEM | − | + | − | + |
| Immune system | ||||
| Lymph node | 605 ± 20 | 581 ± 24a | 377 ± 16 | 286 ± 16a |
| PBMCs | 109 ± 7.7 | 68.7 ± 5.0a | 55.4 ± 10 | 36.2 ± 6.9 |
| Thymus | 523 ± 28 | 471 ± 32 | 235 ± 21 | 135 ± 12a |
| Spleen | X | X | 252 ± 32 | 225 ± 23 |
| Epithelial organs | ||||
| Colon | 362 ± 18 | 312 ± 14 | 278 ± 20 | 213 ± 20a |
| Small intestine | 471 ± 25 | 476 ± 16 | 233 ± 27 | 257 ± 33 |
| Skin | 329 ± 35 | 324 ± 33 | 86.0 ± 18 | 71.0 ± 19 |
| Lung | X | X | 290 ± 21 | 216 ± 20a |
| Liver | X | X | 670 ± 34 | 632 ± 29 |
| Kidney | X | X | 391 ± 7.7 | 406 ± 7.5 |
| Central nervous system | ||||
| Brain | 240 ± 12 | 159 ± 8.2a | 201 ± 13 | 128 ± 10a |
| Midbrain and hindbrain | 201 ± 4.8 | 161 ± 6.9a | 183 ± 10 | 157 ± 6.2a |
| Muscle | ||||
| Skeletal | 156 ± 13 | 141 ± 11 | 129 ± 7.9 | 105 ± 6.3a |
| Heart | 161 ± 9.4 | 129 ± 7.4a | 129 ± 11 | 82.4 ± 6.5a |
| Uterus | 411 ± 26 | 374 ± 29 | 341 ± 39 | 305 ± 39 |
| Reproductive/endocrine/other | ||||
| Testis | 189 ± 14 | 139 ± 14a | 202 ± 9.9 | 155 ± 9.1a |
| Bone marrow | 104 ± 5.8 | 108 ± 13 | 70.3 ± 11 | 83.5 ± 12 |
| Plasma | 8.59 ± 0.8 | 13.1 ± 0.9 | 2.16 ± 0.5 | 5.48 ± 0.6 |
| Pancreas | 578 ± 16 | 559 ± 16 | 595 ± 8.2 | 557 ± 16 |
| Adipose | 94.1 ± 19 | 75.2 ± 20 | 29.3 ± 10 | 24.2 ± 8.9 |
| Submaxillary gland | X | X | 194 ± 8.7 | 214 ± 11 |
NEM inhibition is significant compared to no inhibitor, indicating the detection of DP8/9 activity.
Mean ± SE on five to eight animals. +, presence of NEM inhibitor; −, absence of NEM inhibitor. X, exceeded the maximum measurable OD.
DPIV activity was also detected in all immune system tissues examined using the DPIV-specific inhibitor sitagliptin, using assay type 3 in lymph node, PBMCs, thymus, and spleen, with strong sitagliptin inhibition of H-GlyPro hydrolysis in wild type (see supplementary Table ST3). The DP non-selective inhibitor ValboroPro almost completely inhibited all activity in all immune samples tested. DPIV activity levels were generally far less in DPIV gko mouse samples, compared with wild type, except in pancreas (Tables 3 and 4; see supplementary Tables ST1–ST3).
Epithelial Organs (Colon, Small Intestine, Stomach, Liver, Lung, Skin, Tongue, Kidney)
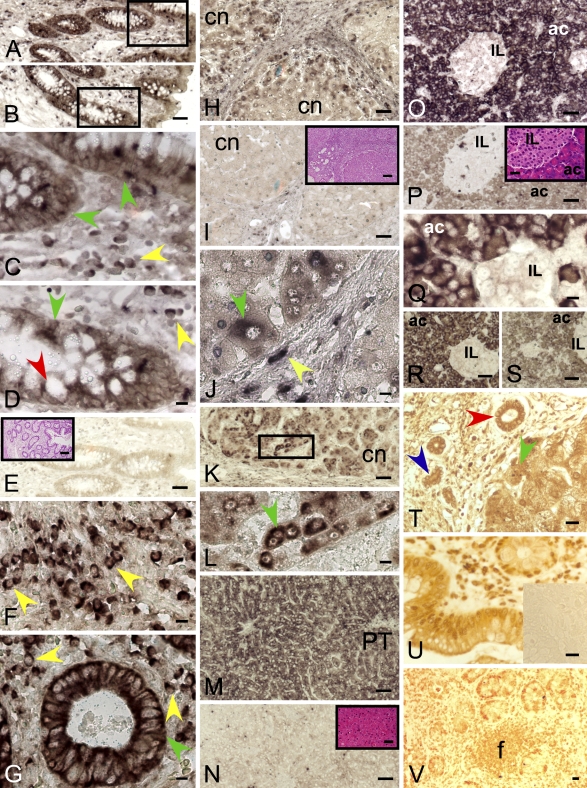
DP8 ISH stained goblet cells as well as enterocytes of both the apical and crypt epithelium of human and baboon colon (Figures 3A–3D) and small intestine. Lymphocytes in lamina propria were also positive for DP8, as were capillary endothelial cells in the intervening vessels. Muscularis propria (smooth muscle) cells were DP8 negative. The baboon liver showed signs of mild lobular and interface hepatitis, and DP8 staining was detected in all hepatocytes, with the most-intense signals in periportal hepatocytes (Figure 3M). Bile ducts and portal lymphocytes were negative. In human cirrhotic liver, hepatocytes were DP8 positive, especially in the periseptal area of regenerative nodules (Figures 3H–3J). Bile duct epithelium was negative, and lymphocytes were positive for DP8 in the portal tracts. In inflamed baboon lung, bronchiolar lymphocytes and endothelium in small vessels were positive, and it is likely that alveolar pneumocytes were DP8 positive (Figure 2K). In baboon skin, epidermal basal keratinocytes and dermal lymphoid cells were DP8 positive (Figure 2P). In baboon tongue, the basal cells in the squamous epithelium of the tongue were strongly DP8 positive, whereas stromal cells and lymphocytes in lamina propria were negative (Figures 2R and 2S). DP9 ISH patterns were similar to those of DP8 on human colon (Figures 3F and 3G), baboon and human liver (Figures 3K and 3L), baboon lung (Figures 3M and 3N), and baboon tongue (Figure 2T). The anti-DP9 antibody showed staining patterns similar to those of ISH in human colon (goblet cells, enterocytes, lymphocytes) (Figures 3U and 3V). In human cirrhotic liver, hepatocytes and bile ducts, including ductular reactions (Figure 3T), were DP9 immunopositive. In human gallbladder, epithelial cell cytoplasm was DP9 immunopositive (Figure 4W).
Figure 3.
DP8 and DP9 expression in leukocytes, enterocytes, hepatocytes, and pancreatic acinar cells. Epithelium, goblet cell, and leukocyte DP8 ISH positivity in human colonic crypt; low- (A,B) and high-power (C,D) views. DP8 sense control with H and E staining in inset (E). DP9 ISH of human colon (F,G). The most-intense DP8 and DP9 staining was in plasma cells (yellow arrow), goblet cells (red arrow), and enterocytes (green arrow). DP8 (H,J) (Table 1, Patient 1) and DP9 (K,L) (Patient 2) ISH positivity in human hepatocytes (green arrow) at the periphery of cirrhotic nodules (cn) and in some septal cells. L is high-power view of boxed area in K. Negative control sense probe with H and E stain, inset (I). The small septal DP8+ cells (yellow arrow) are probably lymphocytes. DP8 ISH in baboon liver (Table 2, Animal G) showing positivity in hepatocytes (M) compared with negative sense control (N; inset is H and E stain). PT, portal tract. Baboon pancreas showing acinar cells (ac), DP8- (O,Q) and DP9-positive (R) and islets of Langerhans (IL), negative (Table 2, Animal M). DP8 sense control with H and E stain inset (P). DP9 sense control (S). DP9-immunostained human cirrhotic liver showing staining in hepatocytes (green arrow), bile ducts (red arrow) and ductular reactions (blue arrow) (T) (Table 1, Patient 1). DP9-immunostained human colon showed staining similar to that of DP9 ISH in epithelia and lymphocytes, including lymphoid follicles (f) (U,V); negative control, inset (U). Bars: A,B,E,H,I,K,M–P,R,S, and all H and E insets = 50 μm; C,D,F,G,J,L,Q,T–V = 10 μm.
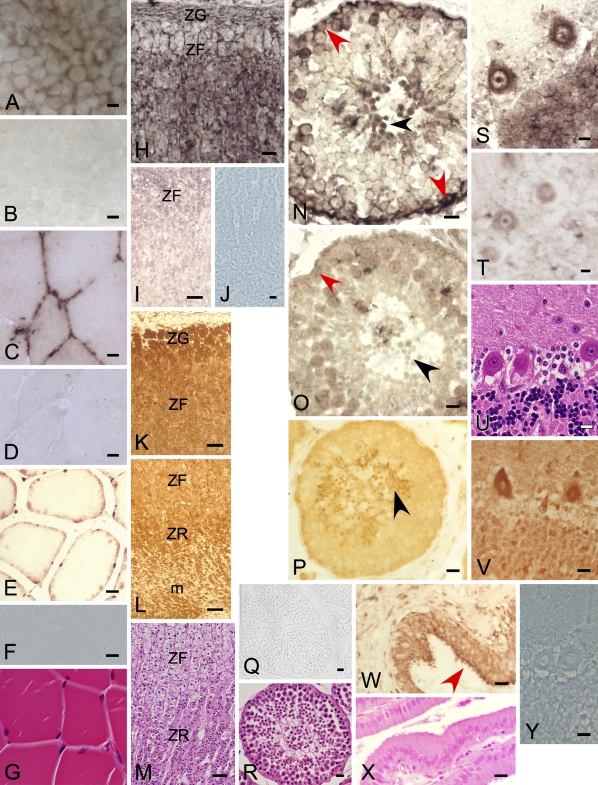
Figure 4.
DP8 and DP9 expression in the 293T cell line, human gallbladder, and baboon muscle, adrenal, testis, and brain detected by ISH and immunohistochemistry. ISH of DP8 mRNA on the cell line 293T showed intense perinuclear staining from the antisense probe (A). Sense control (B). DP8 ISH staining in baboon muscle (Table 2, Animal M) (C), sense control (D), H and E stain (G). DP9 immunopositivity in baboon muscle (E). Immunostain control (F). DP8 ISH (H) and DP9 immunostain (K,L) in adrenal gland showing staining in zona glomerulosa (ZG), zona fasciculata (ZF), zona reticularis (ZR), and medulla (m). DP8 ISH negative (I), DP9 immunostain control (J), H and E stain (M). DP8 ISH staining in baboon spermatogonia (red arrow) and spermatids (black arrow) (N) compared with negative control (O) (Table 2, Animal J). DP9 immunostaining in baboon spermatids (P). DP9 immunostain control (Q); cross-section of a seminiferous tubule (R, H and E stain). DP8 ISH (S) and DP9 immunostain (V) in baboon cerebellum (Table 2, Animal G) showing immunopositivity in Purkinje cells and neuronal cells in the granular layer, DP8 sense control (T), DP9 immunostain control (Y), H and E stain (U). DP9 immunopositivity in the cytoplasm of epithelial cells of human gallbladder (red arrow); lamina propria negative (W); H and E stain (X). Bars: H,I,K–O = 50 μm; A–G,J,P–R,S–Y = 10 μm.
DP8/9 enzyme activity was detected in colon, lung, and liver using assay type 2. NEM inhibition was significant in wild-type and DPIV gko colon, in DPIV gko lung and liver with H-GlyPro (Table 3), and in DPIV gko colon and lung with H-AlaPro (Table 4).
DPIV enzyme activity was detected in all epithelial organs using assay type 3 (see supplementary Table ST3). All wild-type organs demonstrated strong sitagliptin inhibition, with some H-GlyPro hydrolysis remaining, particularly in colon, lung, liver, and kidney that could be attributed, at least in part, to DP8/9. ValboroPro almost completely inhibited H-GlyPro activity.
Central Nervous System (Brain, Midbrain/hindbrain)
In baboon brain, DP8 ISH staining was in Purkinje cells and neuronal cells in the granular layer, as well as neurites in the molecular layer, but not in cortex, forebrain, or midbrain (Figure 4S). DP9 immunostaining revealed a similar pattern (Figure 4V). Both DP8 and DP9 were detected in fetal mouse brain; cerebellar neural cells and neural tube epithelium were intensely ISH positive, whereas weaker staining was observed in glial and neuronal cells of cerebral cortex (Figure 5).
Figure 5.
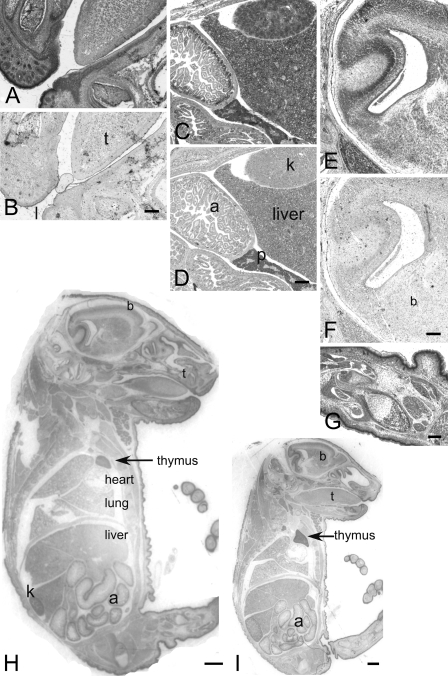
DP8 and DP9 ISH of C57BL/6 mouse at embryonic day 17. DP8 ISH in mouth (A), abdomen (C), brain (E), leg (G), whole embryo (H), and sense controls (B,D,F). DP9 ISH in whole embryo (I). Positive staining in many organs, including intestine (a), brain (b), liver and kidney (k), thymus and lips (l), tongue (t), and skin. Heart and pancreas (p) are negative. Bars: A–G = 50 μm; H–I =1 mm.
DP8/9 enzyme activity was detected in cortical and pooled midbrain/hindbrain homogenates using assay type 2 (Tables 3 and 4). DPIV gko and wild-type samples had similar levels of activity with both H-GlyPro and H-AlaPro. DPIV enzyme activity was detected in wild-type cortical samples using assay type 3, with a small but significant sitagliptin inhibition in H-GlyPro activity (see supplementary Table ST3). ValboroPro inhibition completely ablated activity. Taken together, this suggests that a large proportion of enzyme activity in brain was derived from DP8 and/or DP9, as has been reported by others (Frerker et al. 2007; Stremenova et al. 2007).
Muscle (Skeletal, Heart, Uterus)
Baboon skeletal muscle was DP8 ISH positive between individual muscle cells (endothelium) (Figure 4C). DP9 muscle immunohistochemistry showed strong positivity in nuclei, whereas cytoplasm was negative (Figure 4E). No DP8 or DP9 ISH staining was detected in baboon heart muscle (data not shown).
DP8/9 enzyme activity was detected in mouse skeletal, heart, and uterine muscle using assay type 2 (Tables 3 and 4). With H-GlyPro, significant NEM inhibition was found in DPIV gko samples for skeletal and uterine muscle, and in wild-type heart and uterine muscle (Table 3). Similar data emerged from DPIV gko skeletal muscle and heart and wild-type heart using H-AlaPro and NEM (Table 4). Thus, DP8/9 activity was more readily detected using H-GlyPro than H-AlaPro.
DPIV enzyme activity was detected in all muscle types using assay type 3, with significant sitagliptin inhibition in wild type (see supplementary Table ST3). ValboroPro ablated enzyme activity.
Reproductive/endocrine/other (Testis, Pancreas, Adrenal, Ovary, Adipose Tissue, Submaxillary Gland, Bone Marrow, Plasma)
Baboon testis was DP8 ISH positive in spermatogonia and spermatids, whereas Sertoli cells were negative (Figure 4N). The nonspecific stain on spermatocytes appeared to be an artifact of underfixation. DP9 immunopositivity was intense in spermatids, weak in spermatocytes, and negative in spermatogonia and stroma (Figure 4P). Testis has been reported to express DP8 and splice variant mRNA (Abbott et al. 2000; Zhu et al. 2005). In pancreas, acini were intensely DP8 and DP9 mRNA positive, whereas islets of Langerhans were negative (Figures 3O–3S). Adrenal medulla and cortex were DP8 ISH positive and DP9 immunopositive (Figures 4H–4M). Ovary was ISH negative (data not shown).
DP8/9 enzyme activity was detected in mouse testis, pancreas, visceral non-omental adipose tissue, and adrenal gland using assay type 2 (Tables 3 and 4). In testis, significant NEM inhibition was detected consistently, and DPIV gko testis had levels of activity similar to those of wild type. This suggests that DPIV enzyme activity is predominantly derived from DP8 and/or DP9 in testis. In assays using H-GlyPro (Table 3), significant NEM inhibition was detected in DPIV gko pancreas, adipose tissue, and adrenal gland. No DP8/9 activity was detected in plasma, ovary, bone marrow, or prostate.
DPIV enzyme activity was generally detected in all organs in this category using assay types 1 and 3 (see supplementary Tables ST1–ST3), with significant sitagliptin inhibition in all wild-type organs. ValboroPro ablated activity except in pancreas, where some residual activity remained (see supplementary Table ST3).
Mouse Embryo ISH
DP8 and DP9 mRNA was detected in 17-day-old wild-type mouse embryo (Figure 5). The most-intense staining was observed in skin, gastrointestinal tract, liver, and some brain regions (Figure 5H). Intense staining occurred in the basal region of gut where enterocytes proliferate (Figure 5C), in proliferating keratinocytes around hair follicles (Figure 5A), and in maturing/ossifying chondrocytes in the developing limbs (Figure 5G). The renal tubules in the kidney, hepatocytes and lung, tongue and skin epithelium were ISH positive, whereas heart and pancreas were negative (Figure 5).
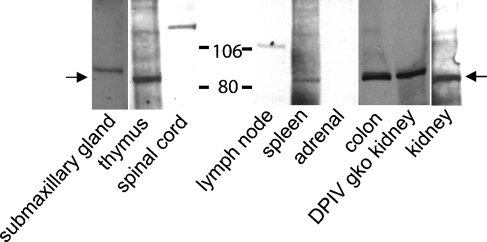
Immunoblot
A selection of mouse organs was screened by immunoblot for DP8 protein expression. A 90-kDa band was detected in C57BL/6 submaxillary gland, thymus, colon, spleen, and kidney, and DPIV gko kidney (Figure 6). Bands of 125 kDa and 110 kDa were seen in spinal cord and lymph node, respectively. None of these bands appeared on blots probed with control rabbit serum.
Figure 6.
Western blot of anti-DP8 antiserum on mouse tissue homogenates (1.2 mg tissue wet weight per lane). C57BL/6 or DPIV gene-knockout (gko) mouse. DP8 bands at 85–90 kDa are arrowed. Representative data from four animals.
DP Distribution in Testis
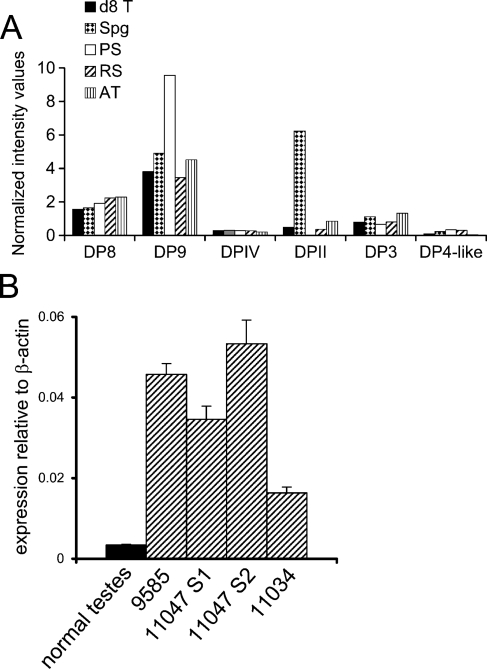
To gain a greater understanding of the distribution of DPs in various stages of testis development, microarray data from both intact testes and isolated germ cells pooled from multiple animals were analyzed. DP8 and DP9 mRNA predominated over DPIV in all samples (Figure 7A). Examination of isolated mouse germ cells revealed that DPII was abundant in premeiotic spermatogonia (Figure 7A). Interestingly, qPCR showed that DP9 was upregulated in human testicular tumors (Figure 7B).
Figure 7.
Dipeptidyl peptidase (DP) mRNA expression in testis. (A) DNA microarray intensities of mouse testis samples: d8T, postnatal day 8 neonatal testis; Spg, spermatogonia; PS, pachytene spermatocytes; RS, round spermatids; AT, adult testis. Representative data from four animals. (B) DP9 in normal human testes (pooled from 23 individuals) and four human testicular tumors; real-time PCR data (mean ± SE) as multiples of the β-actin signal.
Cell Lines
DP8 ISH on the epithelial cell line 293T showed intense specific staining in the perinuclear region (Figures 4A and 4B).
DP8/9 activity was detected in cell lines using assay type 2 in the cell lines Jurkat, Raji, 293T, Huh7, and HepG2 (see supplementary Table ST4). Jurkat, Raji, and 293T cells had the greatest proportions of DP8/9 activity. Jurkat cells have been reported to contain DP8/9 activity (Maes et al. 2007), and to be DPIV negative (Tanaka et al. 1992; Hegen et al. 1993; Tanaka et al. 1993; Aytac et al. 2001). The human skin fibroblast line had the greatest amount of activity of all cell lines in assay 1 and assay 2 (see supplementary Table ST4), and this activity was not significantly inhibited by NEM. This suggested that DPIV predominates in the skin fibroblast line. DP8/9 activity was also detected in cell lines Colo-16, 3T3, and CHO (see supplementary Table ST4).
To further examine the proportion of DPIV-derived activity, assay type 2 was applied to some cell lines in the presence of sitagliptin using H-GlyPro (see supplementary Table ST5). In Jurkat and 293T cells, there was significant NEM but not sitagliptin inhibition, and the sitagliptin-resistant activity was further inhibited by NEM, suggesting that DP8/9 but not DPIV activity was present. In contrast, the human skin fibroblast line had significant inhibition by sitagliptin but not NEM, and NEM did not significantly decrease the residual sitagliptin-resistant activity, suggesting that DPIV but not DP8/9 activity was detected. In Huh-7 cells, both NEM and sitagliptin significantly inhibited activity, suggesting that both DPIV and DP8/9 activity were present.
Discussion
DP8 and DP9 were shown to be predominantly expressed in lymphoid and epithelial cells. Some other lymphoid molecules, CD75, CD138, and CD167a, have a similar distribution. Several approaches were applied to the question of DP8 and DP9 in vivo expression, and a high level of concordance was obtained. Enzyme assay and ISH were the two central complementary approaches. Most organs were found to have detectable DP8/9 with both approaches, and included immune organs (lymph node, thymus, spleen, and PBMC), lymphocytes within lymphocyte-rich organs such as gut and liver, epithelial organs (lung, liver, and intestine), muscle (skeletal muscle and uterus), testis, brain, and pancreas. The ISH data showed that DP8 and DP9 mRNA expression generally includes immune cells, proliferating cells (colonic mucosa and goblet cells, testis, embryo), epithelial cells (skin, lung, tongue, stomach, small intestine, colon, bile duct cells, hepatocytes), some endothelial cells, and some brain and testis cells. Large amounts of DP8/9 enzyme activity were detected in testis, brain, uterus, and muscle. No differences between DP8 and DP9 distribution were observed. These data are consistent with previous Northern blots (Abbott et al. 2000; Ajami et al. 2004; Zhu et al. 2005).
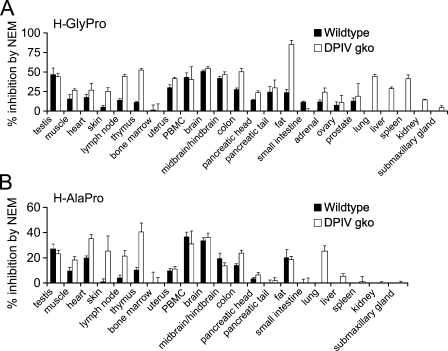
ISH localized DP8 and DP9 mRNA expression to specific cell types within organs. Detecting mRNA would include enzyme inactive forms and splice variants that may be present (Abbott et al. 2000). ISH staining is less readily detected when mRNA density is low within a cell or organ, such as in muscle, which could explain the absence of ISH signals in heart muscle. In contrast, enzyme assays in the presence of the DP8/9 inhibitor NEM indicated the average relative abundance of DP8/9 activity in homogenized tissue (Figure 8). A limitation to this method of DP8/9 detection was the large abundance of DPIV in some organs. Assaying samples from DPIV gko mice overcame this limitation.
Figure 8.
Summary of DP8/9 enzymatic activity in wild-type and DPIV gko mouse organs using assay type 2 presented as percentage inhibition by N-ethylmaleimide (NEM) (mean ± SE). H-GlyPro (A) and H-AlaPro (B). For lung, liver, spleen, kidney, and submaxillary gland, inhibition by NEM was measurable only in the DPIV gko. Representative data from five to eight animals.
Two organs, small intestine and skin, were detectably positive for DP8 ISH but not DP8/9 enzyme activity. As well as the possibility of enzyme inactive forms, the heterogeneity of cell types in these two organs may make it more difficult to detect enzyme activity in tissue homogenates, compared with cell-specific detection in ISH. However, ISH is more technically difficult, dependent on tissue quality and postmortem degradation, and is prone to false positives and negatives. The baboon genome is 96% identical to human, and mouse DP8 and DP9 cDNA are 89.2% and 84.1%, respectively, identical to human. Consequently, the two methods complemented well across species, suggesting that for DP8 and DP9, mRNA expression generally, but not always, correlates with enzyme activity detection.
DP8/9 activity in human PBMCs has been previously reported (Maes et al. 2007). In our studies, DP8/9 activity was present in all major immunological organs, including thymus, PBMC, spleen, and lymph node, and ISH indicated that lymphocyte subpopulations within these organs express DP8 and DP9. In baboon spleen, DP8 was localized to both B cell–rich follicles and the red pulp, which contains mature T cells. These data concur with an enzymology study of DPIV-deficient Fischer rats (Smith et al. 1992), which found that DPIV-deficient immunological organs retain some H-AlaPro hydrolytic activity (thymus 17%, spleen 13–14%, and peripheral mononuclear cells 45–50% of wild-type levels) that may be DP8 and/or DP9 derived. In our ISH, DP8 and DP9 mRNA was localized to lymphoid cells within colon, lung, small intestine, stomach, and cirrhotic liver. This range of locations of lymphoid cells expressing DP8/9 suggests a general role for DP8/9 in many immune cells.
DPs are abundant in epithelia, including gut and liver. Our previous report suggested non-DPIV DP expression in DPIV gko mouse liver (Wang et al. 2002). We now show that DP8 and DP9 are expressed in hepatocytes and hepatoma cell lines. In kidney, DP8/9 activity was not detected, but DP8 protein was detected by Western blot. DPIV-deficient rat kidney contains only 1% the H-AlaPro hydrolytic activity of control rats (Smith et al. 1992), so little DP8/9 activity may be present in kidney. Our studies indicate that colon is a rich source of DP8 and DP9. Pancreatic acinar cells appear to express DP8 and DP9; however, the residual activity remaining in DPIV gko mouse pancreas in the presence of ValboroPro, which inhibits DPIV, DP8, DP9, and DPII, suggests the presence of additional unknown enzymes that have DP activity.
In agreement with our findings, other studies using DP8/9 inhibitors have indicated that DP8/9 activity is greater than that of DPIV in normal brain (Frerker et al. 2007; Stremenova et al. 2007) and in testis (Dubois et al. 2009). NPY is a potential DP8 and/or DP9 substrate within brain (Frerker et al. 2007). Our ISH data localized DP8 to Purkinje cells and granular layer in cerebellum, whereas DPIV is found in vascular endothelial structures (Hartel et al. 1988; McCaughan et al. 1990). DP8 and DP9 mRNA and protein in testis have previously been reported by our group and others (Abbott et al. 2000; Zhu et al. 2005; Dubois et al. 2009). We showed that in testis at various stages of spermatogenesis, DP8 and DP9 mRNA and enzyme activity predominate among the DP molecules. Thus, both DP8 and DP9 might have functions in spermatogenesis. DP8 and DP9 have a redox-sensing ability (Park et al. 2008) and may respond to the presence of reactive oxygen species in the testis, to which spermatozoa are highly sensitive. Peptidase expression by germ cells may be linked to the activity of target substrates in both embryonic and postnatal mouse testis (Holt et al. 2006) and human testis and testicular tumors (Gilbert et al. 2009).
We found that the distribution of DP8 and DP9 is similar overall to DPIV expression. This agrees with previous studies comparing DPIV immunostaining with H-AlaPro enzyme histochemistry that have found generally corresponding patterns (Heike et al. 1988; Hegen et al. 1990; McCaughan et al. 1990; Gorrell et al. 1991). Our finding that freezing depletes DP8/9 activity suggests a reason for our observation that DP8 and DP9 are difficult to detect by enzyme cytochemistry on frozen sections (D. Yu, M. Gall, S. Chowdhury, unpublished data). A recent report on testis similarly detected DP8 and DP9 by immunohistochemistry and by enzyme assay on tissue homogenates but not by enzyme cytochemistry on frozen sections (Dubois et al. 2009). Therefore, enzyme cytochemistry probably detects DPIV rather than related enzymes.
The phenomenon of compensation (upregulation of a compensatory enzyme in a gko mouse) can occur when enzymes have overlapping roles. No evidence of DP8/9 upregulation in the absence of DPIV was seen in these unchallenged mice. This could suggest that DP8 and DP9 roles have little overlap with DPIV, consistent with DPIV roles being extracellular whereas DP8 and DP9 roles are cytoplasmic (Kirby et al. in press), but this question should be examined in disease models.
DP8 enzyme inactive forms as well as active forms may be detected on immunoblots. Human DP8 has a range of mRNA splice variant forms, some of which lack catalytically vital components, predicted to produce enzymatically inactive protein (Abbott et al. 2000; Zhu et al. 2005). In the immunoblots, it is not known how the observed sizes relate to the potential DP8 mRNA splice variants (Abbott et al. 2000). In addition, the larger protein sizes detected in some tissues could be oligomers or DP8 associated with other proteins, as occurs with DPIV (Ajami et al. 2003).
In conclusion, DP8 and DP9 are widely expressed in lymphocytes and epithelial cells of many organs, and so may have functions in the immune system and in epithelia, possibly in proliferation. Because the multi-functional DPs are known to have both enzymatic and extra-enzymatic properties (Wang et al. 2005; Yu et al. 2006), DP8 and DP9 are likely to have diverse roles depending on tissue and cell type and biological and pathological processes.
Acknowledgments
We thank Dr. Didier Marguet of Centre d'Immunologie de Marseille Luminy for DPIV gko mice, Professor Ian Frazer of the University of Queensland Diamantina Institute for the human skin fibroblast line, Dr. Sally Thompson for samples from the National Baboon Colony, and Jené Roestorf, Francine Portelli, Brenna Osborne, and Barbara Fraser for technical assistance. E.A.M. thanks The Children's Hospital Westmead Paediatric Tumour Bank for kind provision of samples.
This work was supported by the National Health and Medical Research Council of Australia (NHMRC) project grant 512282 (to MDG), NHMRC program grant 358398 (to GWM), a Dora Lush postgraduate scholarship (to KA), and a Flinders University research scholarship (to MRP). E.A.M. acknowledges funding from the Australian Research Council Centre of Excellence in Biotechnology and Development.
References
- Abbott CA, Yu DMT, Woollatt E, Sutherland GR, McCaughan GW, Gorrell MD (2000) Cloning, expression and chromosomal localization of a novel human dipeptidyl peptidase (DPP) IV homolog, DPP8. Eur J Biochem 267:6140–6150 [DOI] [PubMed] [Google Scholar]
- Aertgeerts K, Levin I, Shi L, Snell GP, Jennings A, Prasad GS, Zhang Y, et al. (2005) Structural and kinetic analysis of the substrate specificity of human fibroblast activation protein alpha. J Biol Chem 280:19441–19444 [DOI] [PubMed] [Google Scholar]
- Ajami K, Abbott CA, McCaughan GW, Gorrell MD (2004) Dipeptidyl peptidase 9 has two forms, a broad tissue distribution, cytoplasmic localization and DPIV-like peptidase activity. Biochim Biophys Acta 1679:18–28 [DOI] [PubMed] [Google Scholar]
- Ajami K, Abbott CA, Obradovic M, Gysbers V, Kähne T, McCaughan GW, Gorrell MD (2003) Structural requirements for catalysis, expression and dimerisation in the CD26/DPIV gene family. Biochemistry 42:694–701 [DOI] [PubMed] [Google Scholar]
- Ajami K, Pitman MR, Wilson CH, Park J, Menz RI, Starr AE, Cox JH, et al. (2008) Inflammatory protein-10, interferon-inducible T cell chemo-attractant and stromal cell-derived factors 1α and 1β are novel substrates of dipeptidyl peptidase 8. FEBS Lett 582:819–825 [DOI] [PubMed] [Google Scholar]
- Aytac U, Claret FX, Ho L, Sato K, Ohnuma K, Mills GB, Cabanillas F, et al. (2001) Expression of CD26 and its associated dipeptidyl peptidase IV enzyme activity enhances sensitivity to doxorubicin-induced cell cycle arrest at the G(2)/M checkpoint. Cancer Res 61:7204–7210 [PubMed] [Google Scholar]
- Baleato RM, Aitken RJ, Roman SD (2005) Vitamin A regulation of BMP4 expression in the male germ line. Dev Biol 286:78–90 [DOI] [PubMed] [Google Scholar]
- Birrell AM, Heffernan SJ, Kirwan P, McLennan S, Gillin AG, Yue DK (2002) The effects of aminoguanidine on renal changes in a baboon model of Type 1 diabetes. J Diabetes Complications 16:301–309 [DOI] [PubMed] [Google Scholar]
- Bjelke JR, Christensen J, Nielsen PF, Branner S, Kanstrup AB, Wagtmann N, Rasmussen HB (2006) Dipeptidyl peptidase 8 and 9 specificity and molecular characterization compared to dipeptidyl peptidase IV. Biochem J 396:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso N, Wagtmann N, Herling C, Chobaz-Peclat V, Bischof-Delaloye A, So A, Grouzmann E (2005) Circulating CD26 is negatively associated with inflammation in human and experimental arthritis. Am J Pathol 166:433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conarello S, Li Z, Ronan J, Roy R, Zhu L, Jiang G, Liu F, et al. (2003) Mice lacking dipeptidyl peptidase IV are protected against obesity and insulin resistance. Proc Natl Acad Sci USA 100:6825–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolznig H, Schweifer N, Puri C, Kraut N, Rettig WJ, Kerjaschki D, Garin-Chesa P (2005) Characterization of cancer stroma markers: in silico analysis of an mRNA expression database for fibroblast activation protein and endosialin. Cancer Immun 5:10–18 [PubMed] [Google Scholar]
- Dubois V, Ginneken CV, De Cock H, Lambeir A-M, Van der Veken P, Augustyns K, Chen X, et al. (2009) Enzyme activity and immunohistochemical localization of dipeptidyl peptidase 8 and 9 in male reproductive tissues. J Histochem Cytochem 57:531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentke GR, Munoz E, Huber BT, Plaut AG, Kettner CA, Bachovchin WW (1991) Inhibition of dipeptidyl aminopeptidase IV (DP-IV) by Xaa-boroPro dipeptides and use of these inhibitors to examine the role of DP-IV in T-cell function. Proc Natl Acad Sci USA 88:1556–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerker N, Wagner L, Wolf R, Heiser U, Hoffmann T, Rahfeld J-U, Schade J, et al. (2007) Neuropeptide Y (NPY) cleaving enzymes: Structural and functional homologues of dipeptidyl peptidase 4. Peptides 28:257–268 [DOI] [PubMed] [Google Scholar]
- Gilbert DC, Chandler I, McIntyre A, Goddard NC, Gabe R, Huddart RA, Shipley J (2009) Clinical and biological significance of CXCL12 and CXCR4 expression in adult testes and germ cell tumours of adults and adolescents. J Pathol 217:94–102 [DOI] [PubMed] [Google Scholar]
- Gorrell MD (2005) Dipeptidyl peptidase IV and related enzymes in cell biology and liver disorders. Clin Sci 108:277–292 [DOI] [PubMed] [Google Scholar]
- Gorrell MD, Gysbers V, McCaughan GW (2001) CD26: a multifunctional integral membrane and secreted protein of activated lymphocytes. Scand J Immunol 54:249–264 [DOI] [PubMed] [Google Scholar]
- Gorrell MD, Wickson J, McCaughan GW (1991) Expression of the rat CD26 antigen (dipeptidyl peptidase IV) on subpopulations of rat lymphocytes. Cell Immunol 134:205–215 [DOI] [PubMed] [Google Scholar]
- Gorrell MD, Yu DMT (2005) Diverse functions in a conserved structure: The dipeptidyl peptidase IV gene family. In Robinson JW, ed. Trends in Protein Research. New York, Nova Science Publishers, Inc., 1–78
- Gossrau R (1979) Histochemical and biochemical distribution of dipeptidyl peptidase IV (DP IV). Histochemistry 60:231–248 [DOI] [PubMed] [Google Scholar]
- Hartel S, Gossrau R, Hanski C, Reutter W (1988) Dipeptidyl peptidase (DPP) IV in rat organs. Comparison of immunohistochemistry and activity histochemistry. Histochemistry 89:151–161 [DOI] [PubMed] [Google Scholar]
- Hegen M, Camerini D, Fleischer B (1993) Function of dipeptidyl peptidase IV (CD26, Tp103) in transfected human T cells. Cell Immunol 146:249–260 [DOI] [PubMed] [Google Scholar]
- Hegen M, Niedobitek G, Klein CE, Stein H, Fleischer B (1990) The T cell triggering molecule Tp103 is associated with dipeptidyl aminopeptidase IV activity. J Immunol 144:2908–2914 [PubMed] [Google Scholar]
- Heike M, Mobius U, Knuth A, Meuer S, Meyer-zum-Buschenfelde KH (1988) Tissue distribution of the T cell activation antigen Ta1. Serological, immunohistochemical and biochemical investigations. Clin Exp Immunol 74:431–434 [PMC free article] [PubMed] [Google Scholar]
- Holt JE, Jackson A, Roman SD, Aitken RJ, Koopman P, McLaughlin EA (2006) CXCR4/SDF1 interaction inhibits the primordial to primary follicle transition in the neonatal mouse ovary. Dev Biol 293:449–460 [DOI] [PubMed] [Google Scholar]
- Kim D, Wang L, Beconi M, Eiermann GJ, Fisher MH, He H, Hickey GJ, et al. (2005) (2R)-4-oxo-4-[3-(trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: a potent, orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. J Med Chem 48:141–151 [DOI] [PubMed] [Google Scholar]
- Kirby M, Yu DMT, O'Connor S, Gorrell MD (In Press) Inhibitor selectivity in the clinical application of dipeptidyl peptidase-4 inhibition. Clin Sci [DOI] [PubMed]
- Kirby M, Yu DMT, O'Connor S, Gorrell MD (In Press) Inhibitor selectivity in the clinical application of dipeptidyl peptidase-4 inhibition. Clin Sci. (DOI: 10.1042/CS20090047) [DOI] [PubMed]
- Lankas G, Leiting B, Roy R, Eiermann G, Beconi M, Biftu T, Chan C, et al. (2005) Dipeptidyl peptidase IV inhibition for the treatment of type 2 diabetes: potential importance of selectivity over dipeptidyl peptidases 8 and 9. Diabetes 54:2988–2994 [DOI] [PubMed] [Google Scholar]
- LeBeau AM, Brennen WN, Aggarwal S, Denmeade SR (2009) Targeting cancer stroma with a fibroblast activation protein-activated promelittin protoxin. Mol Cancer Ther 8:1378–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch GW, Slaytor EK, Elliott FD, Saurajen A, Turville SG, Sloane AJ, Cameron PU, et al. (2003) CD4 is expressed by epidermal Langerhans' cells predominantly as covalent dimers. Exp Dermatol 12:700–711 [DOI] [PubMed] [Google Scholar]
- Maes MB, Dubois V, Brandt I, Lambeir AM, Van der Veken P, Augustyns K, Cheng JD, et al. (2007) Dipeptidyl peptidase 8/9-like activity in human leukocytes. J Leukoc Biol 81:1252–1257 [DOI] [PubMed] [Google Scholar]
- Maes MB, Lambeir AM, Gilany K, Senten K, Van der Veken P, Leiting B, Augustyns K, et al. (2005) Kinetic investigation of human dipeptidyl peptidase II (DPPII)-mediated hydrolysis of dipeptide derivatives and its identification as quiescent cell proline dipeptidase (QPP)/dipeptidyl peptidase 7 (DPP7). Biochem J 386:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguet D, Baggio L, Kobayashi T, Bernard AM, Pierres M, Nielsen PF, Ribel U, et al. (2000) Enhanced insulin secretion and improved glucose tolerance in mice lacking CD26. Proc Natl Acad Sci USA 97:6874–6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan GW, Wickson JE, Creswick PF, Gorrell MD (1990) Identification of the bile canalicular cell surface molecule GP110 as the ectopeptidase dipeptidyl peptidase IV: an analysis by tissue distribution, purification and N-terminal amino acid sequence. Hepatology 11:534–544 [DOI] [PubMed] [Google Scholar]
- Park J, Knott HM, Nadvi NA, Collyer CA, Wang XM, Church WB, Gorrell MD (2008) Reversible inactivation of human dipeptidyl peptidases 8 and 9 by oxidation. The Open Enzyme Inhibition Journal 1:52–61 [Google Scholar]
- Pitman MR, Sulda ML, Kuss B, Abbott CA (2009) Dipeptidyl peptidase 8 and 9—guilty by association? Front Biosci 14:3619–3633 [DOI] [PubMed] [Google Scholar]
- Preller V, Gerber A, Wrenger S, Togni M, Marguet D, Tadje J, Lendeckel U, et al. (2007) TGF-beta1-mediated control of central nervous system inflammation and autoimmunity through the inhibitory receptor CD26. J Immunol 178:4632–4640 [DOI] [PubMed] [Google Scholar]
- Qi SY, Riviere PJ, Trojnar J, Junien JL, Akinsanya KO (2003) Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine proteases. Biochem J 373:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold D, Goihl A, Wrenger S, Reinhold A, Kühlmann UC, Faust J, Neubert K, et al. (In Press) Review: Role of dipeptidyl peptidase IV (DP IV)-like enzymes in T lymphocyte activation: investigations in DP IV/CD26-knockout mice. Clin Chem Lab Med. Published online February 10, 2009 (DOI: 10.1515/CCLM.2009.1062) [DOI] [PubMed]
- Rettig WJ, Su SL, Fortunato SR, Scanlan MJ, Raj BK, Garin-Chesa P, Healey JH, et al. (1994) Fibroblast activation protein: purification, epitope mapping and induction by growth factors. Int J Cancer 58:385–392 [DOI] [PubMed] [Google Scholar]
- Schade J, Stephan M, Schmiedl A, Wagner L, Niestroj AJ, Demuth HU, Frerker N, et al. (2008) Regulation of expression and function of dipeptidyl peptidase 4 (DP4), DP8/9, and DP10 in allergic responses of the lung in rats. J Histochem Cytochem 56:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Reynolds CJ, Elder EA (1992) The evolution of proteinase substrates with special reference to dipeptidylpeptidase IV. Histochem J 24:637–647 [DOI] [PubMed] [Google Scholar]
- Stremenova J, Krepela E, Mares V, Trim J, Dbaly V, Marek J, Vanickova Z, et al. (2007) Expression and enzymatic activity of dipeptidyl peptidase-IV in human astrocytic tumours are associated with tumour grade. Int J Oncol 31:785–792 [PubMed] [Google Scholar]
- Tanaka S, Murakami T, Horikawa H, Sugiura M, Kawashima K, Sugita T (1997) Suppression of arthritis by the inhibitors of dipeptidyl peptidase IV. Int J Immunopharmacol 19:15–24 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Camerini D, Seed B, Torimoto Y, Dang NH, Kameoka J, Dahlberg HN, et al. (1992) Cloning and functional expression of the T cell activation antigen CD26. J Immunol 149:481–486 [PubMed] [Google Scholar]
- Tanaka T, Kameoka J, Yaron A, Schlossman SF, Morimoto C (1993) The costimulatory activity of the CD26 antigen requires dipeptidyl peptidase IV enzymatic activity. Proc Natl Acad Sci USA 90:4586–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bonin A, Hühn J, Fleischer B (1998) Dipeptidyl-peptidase IV/CD26 on T cells: analysis of an alternative T cell activation pathway. Immunol Rev 161:43–53 [DOI] [PubMed] [Google Scholar]
- Wang M, Gorrell MD, Abbott CA, Jaggi R, Marguet D, Dickinson RG (2002) Hepatic covalent adduct formation with zomepirac in the CD26-deficient mouse. J Gastroenterol Hepatol 17:66–71 [DOI] [PubMed] [Google Scholar]
- Wang XM, Yao T-W, Nadvi NA, Osborne B, McCaughan GW, Gorrell MD (2008) Fibroblast activation protein and chronic liver disease. Front Biosci 13:3168–3180 [DOI] [PubMed] [Google Scholar]
- Wang XM, Yu DMT, McCaughan GW, Gorrell MD (2005) Fibroblast activation protein increases apoptosis, cell adhesion and migration by the LX-2 human stellate cell line. Hepatology 42:935–945 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Kuppuswamy D, Li Y, Livanos EM, Hixon M, White A, Beach D, et al. (1996) Alteration of cell cycle kinase complexes in human papillomavirus E6- and E7-expressing fibroblasts precedes neoplastic transformation. J Virol 70:999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DMT, Wang XM, McCaughan GW, Gorrell MD (2006) Extra-enzymatic functions of the dipeptidyl peptidase (DP) IV related proteins DP8 and DP9 in cell adhesion, migration and apoptosis. FEBS J 273:2447–2461 [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhou ZM, Lu L, Xu M, Wang H, Li JM, Sha JH (2005) Expression of a novel dipeptidyl peptidase 8 (DPP8) transcript variant, DPP8-v3, in human testis. Asian J Androl 7:245–255 [DOI] [PubMed] [Google Scholar]