Abstract
Yeast is a highly tractable model system that is used to study many different cellular processes. The common laboratory strain Saccharomyces cerevisiae exists in either a haploid or diploid state. The ability to combine alleles from two haploids and the ability to introduce modifications to the genome requires the production and dissection of asci. Asci production from haploid cells begins with the mating of two yeast haploid strains with compatible mating types to produce a diploid strain. This can be accomplished in a number of ways either on solid medium or in liquid. It is advantageous to select for the diploids in medium that selectively promotes their growth compared to either of the haploid strains. The diploids are then allowed to sporulate on nutrient-poor medium to form asci, a bundle of four haploid daughter cells resulting from meiotic reproduction of the diploid. A mixture of vegetative cells and asci is then treated with the enzyme zymolyase to digest away the membrane sac surrounding the ascospores of the asci. Using micromanipulation with a microneedle under a dissection microscope one can pick up individual asci and separate and relocate the four ascopores. Dissected asci are grown for several days and tested for the markers or alleles of interest by replica plating onto appropriate selective media.
Keywords: Cellular Biology, Issue 27, asci, ascospores, diploid, zygote, sporulation, yeast dissection, micromanipulator
Protocol
Construction of diploid strains Protocol for production of diploids by mating
The initial strains should be struck out on YPD, or selective medium if necessary, to produce single colonies. The two haploid strains should be of opposite mating type (ie. MATa and MATα).
Using a sterile inoculation loop pick a small portion of a colony from one of the two haploid strains. On a new YPD plate streak the loop once in a straight line across the plate. Mark the direction of the streak with an arrow on the bottom of the plate.
Repeat this for the second strain on the same YPD plate. This time streak the colony perpendicular to the first, crossing the first streak with the second. Mark the direction of the second streak and note the area where the second streak crosses the first. This area will be where diploids will be produced.
Incubate the YPD plate at 30°C overnight.
Replica plate the YPD plate onto selective media that will selectively permit growth of the newly-formed diploids.
Pick a single colony if possible, or a small portion of the diploid streak and streak onto the same selective medium to generate single colonies.
Sporulation and digestion of asci Protocol
Pick single colonies from the plate above and make small (1cm x 1cm) patches on sporulation (SPO) plates and incubate at 30°C for 3-7 days to sporulate.
Sporulation can be checked by looking at cells from each patch under a light microscope. Place cells on a microscope slide in 5-10 μl of water, place a coverslip on top of the droplet, and look for the presence of asci. If asci are not easily found then put back in the incubator.
If there is effective sporulation in one of the patches, prepare a sterile 15mL conical tube with 100μL of tetrad juice.
Pick cells up from this sporulation patch with a sterile 3mm inoculation loop (cells should cover ¼ of the loop) and place in the tetrad juice. Gently swirl the loop to allow the cells to fall off the loop and mix with the solution.
Leave in tetrad juice (containing zymolyase) for 4-10mins (depending on the concentration and activity of zymolyase and the strain itself).This allows for digestion of the cell wall surrounding the ascospores.
Mark the top and bottom of the inoculum region on a YPD plate.
Optional. Reaction can be stopped by adding 1mL of ice cold sterile dH2O. Allow the cells to settle for 5mins. Remove 1mL from the supernatant then mix lightly by hand and keep on ice.
Take 5μL of digested cells from step 5 and gently place drops in a line in the inoculum region of the pre-marked YPD plate.
Use a sterile 3mm inoculation loop to very gently spread the drops 1-3 times in a line while barely making contact with the agar.
Allow the liquid to absorb into the agar (If step 7 is omitted, this is when the digestion of the outer cell wall will be reduced or stop). Proceed to the dissection microscope.
Dissection of Asci Protocol
Note that the following protocol is described for a Singer MSM System 200 micromanipulation microscope. This protocol can be followed with slight modifications if using a manual micromanipulator such as a Zeiss micromanipulator MR.
Focus on a region of the plate where half of the field contains cells and half is empty. Slowly bring the needle into focus such that it touches the empty region of the field. This will ensure that the needle does not contain cells from a previous use. Contact between the needle and the agar is seen as a dark black circle that slowly comes into focus.
Go to the matrix at position A1 and make a mark by piercing the agar again. This is to help determine the orientation of the plate if it is taken off the microscope before completion.
Switch to the search mode. Locate an asci. Preferably choose asci which are in areas where the cells are well spread out. Avoid areas with cell clusters. The four ascospores from an asci should still be attached to each other. Two ascospores may be slightly larger than the other two. Avoid picking asci where one ascospore is loose and not connected to the rest of the ascospores.
Pick up an ascus with the needle. Slowly bring the needle up to the plate. When the shadow of the microneedle is seen, line this up with the ascus to be picked. Again approach the needle until the black outline of a circle (the tip of the needle) is seen. Touch the tetrad with this circle and then quickly pull the needle away. Make sure all four ascospores from an ascus have adhered to the needle and that no additional cells were gathered on the microneedle that are not part of this ascus.
Pull the microneedle far away from the plate and go to the matrix (for the first ascus, go to position A1). Take note of each position where an ascus is relocated. Place the ascus down by touching the plate with the microneedle and pulling away until all four ascospores come off the microneedle. Gentle tapping of the needle arm or the platform holding the YPD plate is often useful at this step.
Repeat steps 3-5 and place asci down at consecutive positions on the matrix until the desired number of asci have been picked.
Go back to each ascus and break the four cells apart by teasing with the needle, tapping the platform gently or a combination of both. Pick up the cells leaving one behind at a new position in the matrix until each ascospore in each ascus is separated in rows of four.
Place the YPD plate in an incubator at 30°C for several days to observe the growth pattern.
Replica plate the dissected cells to selective medium if necessary.
Relocation of Zygotes Protocol
Streak out two haploid strains with opposite mating types on the appropriate medium to obtain single colonies.
Mate these two strains as described above and allow to grow from 4 to 6 hours (or overnight if necessary).
Check for zygotes under a light microscope. Zygotes will appear as dumbbell-shaped cells either with or without a bud.
Pick cells up with a 3mm inoculation loop (cells should cover ¼ of the loop) and place gently in 100μL of sterile dH2O.
Mark the top and bottom of the inoculum region on a YPD plate.
Take 5μL and gently place drops in a line in the inoculum region of the pre-marked YPD plate.
Use a 3mm inoculation loop to spread the drops in a line very gently while barely making contact with the agar.
Allow the liquid to absorb into the agar. Proceed to the dissection microscope.
Focus on a region of the plate where half of the field contains cells and half is empty. Slowly bring the needle into focus such that it touches the empty region of the field. This will ensure that the needle does not contain cells from a previous use. Contact between the needle and the agar is seen as a dark black circle that slowly comes into focus.
Go to the matrix at position A1 and make a mark by piercing the agar again. This is to help determine the orientation of the plate if it is taken off the microscope before completion.
Switch to the search mode. Locate a zygote.
Pick up a zygote with the needle.
Place zygotes individually on the matrix.
Allow zygotes to grow for several days.
Replica plate the zygotes onto the appropriate selective medium.
Representative Results
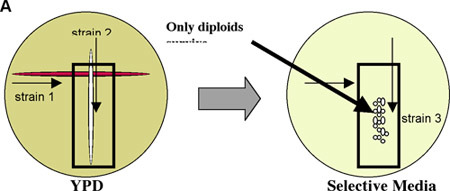 Figure 1: The mating of two haploid strains of opposite mating types generates a diploid strain.
Figure 1: The mating of two haploid strains of opposite mating types generates a diploid strain.
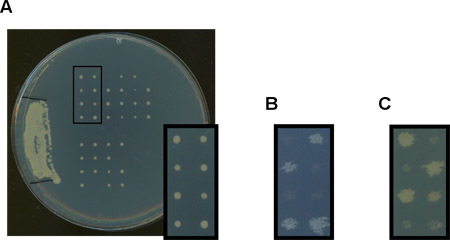 Figure 2: (A) A YPD plate with thre results of a mating between a temperature sensitive strain (with a leu2 auxotrophy) and a wild type strain (LEU2). Digested cells are spread in the inoculum region on the left where thick growth between the two black lines is seen. Asci that have been dissected grow evenly spaced in the matrix on the right. (B) The plate in (A) was replicated onto an SD -Leucine drop-out plate. Note the 2:2 growth for the LEU2 marker indicating a validated ascospore was dissected. (C) The plate in (A) was replicated onto a YPD plate and incubated at 37°C. Note the 2:2 growth that further validates that an ascospore was dissected.
Figure 2: (A) A YPD plate with thre results of a mating between a temperature sensitive strain (with a leu2 auxotrophy) and a wild type strain (LEU2). Digested cells are spread in the inoculum region on the left where thick growth between the two black lines is seen. Asci that have been dissected grow evenly spaced in the matrix on the right. (B) The plate in (A) was replicated onto an SD -Leucine drop-out plate. Note the 2:2 growth for the LEU2 marker indicating a validated ascospore was dissected. (C) The plate in (A) was replicated onto a YPD plate and incubated at 37°C. Note the 2:2 growth that further validates that an ascospore was dissected.
Discussion
Yeast dissection is a useful tool to select new strains with desired markers. When determining the theoretical growth pattern of four ascospores it is important to look at the genotype of the vegetative cell. If a plasmid is present, one must know if it is single, low or high copy as this will affect which ascospore(s) receive the plasmid and will affect predictions and conclusions regarding the strains being manipulated.
Certain strains can sporulate better than others producing many ascospores in a short period of time. Other strains may only yield a low percentage of ascospores. This does not have any affect on the experiment as long as enough true asci can be selected. Furthermore, strains respond differently during cell wall digestion and it is necessary to empirically adjust the incubation time in the tetrad juice.
When selecting cells with the desired markers, one should only choose those that come from an asci where the growth pattern of all ascospores correspond to the theoretical growth pattern and from a dissection where the vast majority of all ascospores lead to the expected growth pattern.
Disclosures
The authors have nothing to disclose.
Acknowledgments
Work in the author’s laboratory is funded by the Canadian Institutes of Health Research, the Canada Foundation for Innovation, the Natural Sciences and Engineering Research Council and Concordia University.
References
- Sherman F, Hicks J. Micromanipulation and Dissection of Asci. In: Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. pp. 21–37. [Google Scholar]


