Abstract
Detection of cancer cells at early stages could potentially increase survival rates in cancer patients. Aberrant promoter hypermethylation is a major mechanism for silencing tumor suppressor genes in many kinds of human cancers. A recent report from our laboratory described the use of quantitative methylation-specific PCR assays for discriminating patients with lung cancer from those without lung cancer using lung biopsies as well as sputum samples. TCF21 is known to be essential for differentiation of epithelial cells adjacent to mesenchyme. Using restriction landmark genomic scanning, a recent study identified TCF21 as candidate tumor suppressor at 6q23-q24 that is epigenetically inactivated in lung and head and neck cancers. Using DNA sequencing technique, we narrowed down a short CpG-rich segment (eight specific CpG sites in the CpG island within exon 1) of the TCF21 gene, which was unmethylated in normal lung epithelial cells but predominantly methylated in lung cancer cell lines. We specifically targeted this short CpG-rich sequence and developed a quantitative methylation-specific PCR assay suitable for high-throughput analysis. We showed the usefulness of this assay in discriminating patients with lung cancer from those without lung cancer using biopsies and sputum samples. We further showed similar applications with multiple other malignancies. Our assay might have important implications in early detection and surveillance of multiple malignancies.
Introduction
Transcriptional inactivation of CpG island-containing promoters of tumor suppressor genes by DNA hypermethylation has been well documented in many human cancers (1). Methylation of specific CpG residues within a CpG island of a tumor suppressor gene may reflect gene silencing and indicate, at least in part, the expression status of the gene. Gene promoter hypermethylation potentially provides a noninvasive screen for early cancer detection (2).
Methylation analyses have been conducted using conventional methodologies such as COBRA, direct sequencing, or methylation-specific PCR of the bisulfite-treated DNA. However, these methods are labor intensive, amenable to false-positive results, and not suitable for high-throughput analysis. Methyl Light assays are not only highly specific, sensitive, and reproducible but also are nonsubjective and allow for rapid analysis of many samples at multiple gene loci (3, 4). Recent publications have shown the presence of promoter hypermethylation of various genes in clinical specimens containing exfoliated tumor cells (such as malignant effusions, sputum, serum, etc.; refs. 5–8). Recently, we reported quantitative methylation-specific PCR analysis of sputum DNA based on a panel of methylated genes (9). The panel separated patients with lung cancer from those without lung cancer, showing the potential of the quantitative methylation-specific PCR analysis of sputum as an effective biomarker assay. Subsequently, we decided to explore additional novel markers that might further improve this assay.
TCF21 is known to be essential for differentiation of epithelial cells adjacent to mesenchyme (10). Using restriction landmark genomic scanning, Smith et al. (11) identified TCF21 as a candidate tumor suppressor at 6q23-q24 that is epigenetically inactivated in lung and head and neck cancers. In our article, using DNA sequencing, we examined lung cancer and bronchial epithelial cell lines for methylation of the CpG island within exon 1 of the TCF21 gene (GenBank accession no. AF047419). We identified a short CpG-rich segment that was unmethylated in normal bronchial cells but predominantly methylated in lung cancer cell lines. We decided to develop a high-throughput quantitative bioassay to determine prevalence of methylation in clinical samples and evaluate its potential as cancer biomarker assay applicable for multiple cancer types.
Materials and Methods
Surgically resected non-small lung and bladder cancers and their adjacent nonmalignant tissues were obtained from the University of Texas M. D. Anderson Cancer Center. Surgically resected breast cancers and their adjacent nonmalignant breast tissues were obtained from Parkland Memorial Hospital. Leukemia cases, all acute myeloid leukemia, were obtained from Parkland Memorial Hospital. Peripheral blood mononuclear cells were obtained from healthy individuals with a family history of cancer. We established all cell lines used in this study. Sputum samples were obtained from 13 patients with non–small cell lung cancer (NSCLC) and 25 individuals with chronic obstructive pulmonary disease; all were heavy smokers without lung cancer in the Canisius Wilhelmina Hospital. Three-day pooled early morning sputum samples were collected in Saccomanno’s fixative (2% polyethylene glycol in 50% ethanol). Informed consent and institutional review board permission were obtained at each site.
Gene Expression in Cell Lines
Gene expression studies were conducted as described previously (11) with some modifications. Semiquantitative real-time PCR was carried out by using QuantiTect SYBR Green PCR kit. The expression levels were quantitated using comparative Ct method. In case of both TCF21 and WNT4, means of expression values for the two human bronchial epithelial cells (HBEC) were considered to have a value of 1. 5-Aza-2-deoxcytidine treatment of lung cell lines (H1299, H2887, HCC95, and H661) was done using protocol as described previously (12).
DNA Extraction and Bisulfite Modification and DNA Sequencing
Genomic DNA was extracted from cell lines, primary tumors and nonmalignant cells, sputum samples as described previously (9). Sodium bisulfite treatment was done as described previously (13). The modified DNA was used as a template for quantitative PCR (qPCR) analysis. The DNA sequencing was carried out as described previously using Applied Biosystems prism dye terminator cycle sequencing method (Perkin-Elmer; ref. 14). Primers were designed to exclude CG sites, rendering DNA amplification independent of the methylation status: forward 5′-ATGTGGAGGATTTTTAAGAGGT-3′ and reverse 5′-CTAAAAAAAACCTTACTCAACACTC-3′. The sequences were confirmed by sequencing in both directions.
Quantitative Real-time PCR Analysis
qPCR analysis was done using the Chromo4 MJ Research Real-time PCR System. Sodium bisulfite–treated genomic DNA was amplified by fluorescence-based real-time methylation-specific PCR using TaqMan technology as described previously (9, 15, 16). In brief, primers (forward 5′-CGAGGAGAGTTTTAATTGCGAGA-3′ and reverse 5′-CCTAACTAACCCCGCTCAAAAAA-3′) and probe (5′-FAM-TAGAAGGGTCGCGGCGGTTTGGBHQ-1–3′) were designed to specifically amplify bisulfite-converted DNA within the region of the test genes that was differentially methylated between expression-positive and expression-negative cell lines (Fig. 1). The non-methylated form of MYOD1 was used as an internal reference standard (9). The sputum DNA samples were coded and shipped from The Netherlands and analyzed in Dallas in a blinded fashion. Some of the lung cancer DNA was also independently analyzed by three investigators (N.S., V.S., and C.P.) in a blinded fashion. The data from all the three investigators were in close agreement (data not shown).
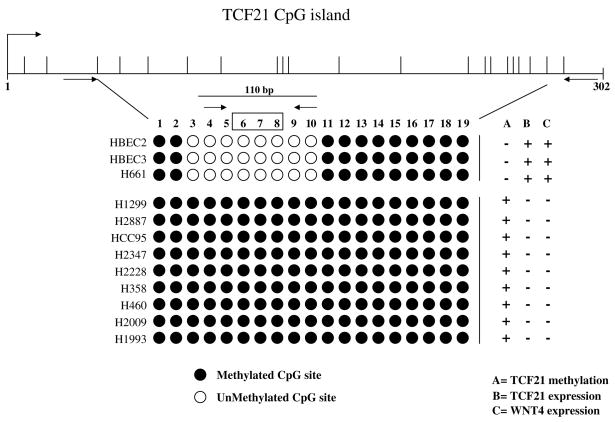
Figure 1.
Direct sequencing of the bisulfite PCR product from the CpG island within exon 1 of the TCF21 gene for 10 lung cancer cell lines and 2 normal lung epithelial cell lines (HBEC2 and HBEC3). Closed circles, methylated CpG sites; open circles, unmethylated CpG sites. The expression levels were quantitated using comparative Ct method. In both TCF21 and WNT4 expression analysis, means of expression values for the two HBECs were considered to have a value of 1. (expression positive, relative expression >1; expression negative, relative expression <0.50). Sequencing and expression analyses were conducted as described in Materials and Methods. Arrows, locations of primers; open box, location of the probe.
Nested qPCR protocol was accomplished by doing two rounds of PCR as reported previously (17). In the first round, external primers used for DNA sequencing were used. The successful amplification in the first round was confirmed through agarose gel electrophoresis. The second round was carried on the 500× dilution of the first round reaction using the probe and primer sets as in quantitative methylation-specific PCR protocol described above.
Statistical Analysis
The receiver operating characteristic curves, a plot of the sensitivity versus specificity across all possible cutoff values, were used to identify the accuracy of a marker in discriminating cancer from nonmalignant tissue. The quantitative methylation data for the gene were correlated with tumor stage using the Mann-Whitney U test, which does not require parametric assumption on the distribution of quantitative methylation. Statistical differences between groups were examined using Fisher’s exact test. P values <0.05 were considered significant.
Results
We sequenced the CpG island within exon 1 of the TCF21 gene from 10 lung cancer cell lines and 2 HBECs (18) and also analyzed them for gene expression. The results showed that all the 19 CpG sites were methylated in 9 of 10 lung cancer cell lines, whereas 8 specific CpG sites of the 19 were unmethylated only in H661 (a cancer cell line) and in the HBECs (Fig. 1). All methylation-positive cancer cell lines were negative for TCF21 expression (relative expression <0.50). H661 and HBECs were negative for methylation and positive for TCF21 expression (relative expression ≥1; Fig. 1). 5-Aza-2′-deoxycytidine treatment to four lung cell lines (H1299, H2887, HCC95, and H661) resulted into reactivation of TCF21 expression in all the cell lines, except H661, which was positive for TCF21 expression in untreated population (data not shown). The results suggest that methylation of the specific sites is related to gene silencing. However, it is possible that other factors in addition to methylation (19) might also contribute to gene silencing in these lung cancer cell lines. We examined WNT4 expression for two reasons. Our preliminary studies showed consistent loss of its expression in lung cancer cell lines compared with HBECs.8 Secondly, the gene was also shown to be induced significantly when the lung cancer cells were transfected with TCF21 (11). Interestingly, all the lung cancer cell lines (except H661) were negative for WNT4 expression (relative expression <0.50; Fig. 1). Based on all above observations, the specific CpG region was considered a potential target to develop an assay to discriminate between cancer cells and normal cells.
We tested the qPCR assay for methylation of TCF21 by analysis of 40 NSCLC tumors (22 adenocarcinoma and 18 squamous carcinomas) and their adjacent nonmalignant lung tissues. Additionally, peripheral blood mononuclear cells from 12 healthy subjects recruited for genetic epidemiology studies were also quantitatively analyzed. Figure 2A shows the quantitative methylation data for NSCLC and their adjacent nonmalignant lung tissue. We found aberrant methylation of TCF21 in 30 of 40 (75%) of primary lung tumors (QR, 0–129.67) and in 7 of 40 (18%) adjacent lung tissue (QR, 0–0.8). Based on the highest QR in nonmalignant tissue as the cutoff, 28 of 40 (70%) of primary NSCLC were found to have TCF21 methylated. Additionally, peripheral blood mononuclear cells from 18 healthy subjects recruited for genetic epidemiologic studies were analyzed for TCF21 methylation and found to be below the levels of detection. Figure 2B shows the receiver operating characteristic curve, which provides evidence for the excellent discriminatory capacity of TCF21 methylation in separating cancer from adjacent normal lung tissue. We also correlated quantitative methylation data for TCF21 with tumor stage using the Mann-Whitney P test. Increased methylation levels (QR) were observed with increase in stage (stage I–II, P = 0.008; stage I combined with stages II and III, P = 0.002).
Figure 2.
A. Methylation levels of mTCF21 in NSCLC (Tumor), adjacent nonmalignant lung, and peripheral blood mononuclear cells from cancer-free individuals. Methylation levels were quantitated by semiquantitative real-time PCR. Real-time analysis was done as described in Materials and Methods. Quantitative ratio is defined as the ratio of the fluorescence emission intensity values for the PCR products of the biomarker gene to those of PCR products of MYOD1 multiplied by 100. The ratio is a measure for the relative level of methylation in an individual sample. Because values are expressed on a log scale, completely negative values are expressed as values of 0.01. Solid horizontal bar, threshold above which the samples are considered positive for methylation. B. Receiver operating characteristic curves for mTCF21 at separating cancer from the adjacent nonmalignant lung. Receiver operating characteristic curves are plots of the true-positive rate (Y axis) against the false-positive rate (X axis) for the different possible cutoff points of a diagnostic test. The closer the curve follows the left-hand border and then the top border of the receiver operating characteristic space, the more accurate the test [area under the curve (AUC) = 0.85]. C. Methylation levels of mTCF21 in sputum from NSCLC patients and in sputa from patients without malignancy (chronic obstructive pulmonary diseases). Methylation levels were quantitated as described in Materials and Methods. The table shows the frequency of methylation in the sputum samples following the combination of nested PCR and qPCR. The two-round PCR protocol was carried out as described in Materials and Methods.
We further tested the qPCR assay for methylation of TCF21 in sputum DNA from patients with lung cancer (Fig. 2C). We analyzed 38 sputum DNA samples (13 from cancer and 25 noncancer patients) for methylation of TCF21. We found 7 of 13 (54%) cancer sputa (QR, 0–34.63) and 0 of 25 (0%) noncancer sputa showed methylation of TCF21 (P < 0.0001; Fig. 2C). Combination of nested PCR and qPCR led to increase in methylation frequencies from 7 of 13 (54%) to 9 of 13 (70%) in cancer cases and from 0 of 25 (0%) to 3 of 25 (12%) in chronic obstructive pulmonary diseases (table in Fig. 2C). Whereas no methylation was detected (Ct values of 50) in normal lung and normal lymphocytes, the methylated status of the newly observed five positive samples (all with Ct values <35) following two-round PCR protocol was confirmed through DNA sequencing (data not shown). We speculate that these five cases might be at higher risk to develop lung cancer than other cases, all of which (like normal lung and normal lymphocytes) showed Ct values of 50 even after two rounds of PCR. We have proven the feasibility of the nested qPCR combination approach for increasing the sensitivity without significantly compromising the specificity. Further development of the nested qPCR assay is in progress to assess real effect of this approach.
We further tested the qPCR assay for methylation of TCF21 by analysis of 30 breast tumors and their adjacent nonmalignant tissues. We found aberrant methylation of TCF21 in 26 of 30 (87%) of breast tumors (QR, 0–850.68) and in 13 of 30 (40%) adjacent breast tissue (QR, 0–2; Table 1). Based on the highest QR in nonmalignant tissue as the cutoff, 20 of 30 (67%) of primary breast tumors were found to have TCF21 methylated. The data show that TCF21 methylation has excellent discriminatory capacity at separating cancer from adjacent nonmalignant breast tissue (Table 1).
Table 1.
Quantitative data for methylation of TCF21 in multiple malignancies
| Tissue | Quantitative ratio (range) |
% Samples positive for methylation in tumors with QR above the highest in normal group | |
|---|---|---|---|
| Cancer | Normal | ||
| Breast | 0–850.68 (n = 30) | 0–2.0 (n = 30) | 20/30 (67) |
| Bladder | 0–52.60 (n = 41) | 0–0 (n = 6) | 18/41 (44) |
| Colon | 0–256.83 (n = 20) | 0–8.12 (n = 20) | 15/20 (75) |
| Leukemia | 0–74.80 (n = 27) | 0–0 (n = 12) | 11/27 (41) |
Additionally, we tested the qPCR assay for methylation of TCF21 in bladder cancers, colon cancers, and leukemias with corresponding nonmalignant tissues (Table 1). Also, in the case of bladder cancer cases, a strong trend was observed between methylation and invasiveness (52% in invasive and 27% in noninvasive cases). In all the malignancies, TCF21 methylation appears to have excellent discriminatory capacity at separating cancer from nonmalignant tissue.
Discussion
In this article, we identified a short CpG-rich sequence within exon 1 of the TCF21 gene that was differentially methylated in lung cancer cells compared with normal lung cells. We targeted this region to design a methylation-specific qPCR assay suitable for high-throughput analysis. We successfully showed the application of this assay, in discriminating patients with lung cancer from those without lung cancer, using lung cancer biopsies and sputa from cancer patients. We further showed similar application of this assay with multiple other malignancies, covering different organ systems.
Transition between epithelial cell to mesenchymal cell is known to occur during tumorigenesis (20, 21). Epithelial-mesenchymal transition has been described in many cancers and correlates with clinical outcome (20). Malignant lesions are often defined by their differentiation status, where benign tumors and low-grade cancers typically retain their epithelial phenotype and malignant cells acquire more fibroblastic mesenchymal phenotype (20). As mentioned in the results, Smith et al. (11) previously reported highly significant induction of WNT4 expression in lung cancer cells (TCF21 expression negative) when transfected with TCF21. WNT4 has been associated previously with epithelial-mesenchymal transition and epithelial phenotype (22). We observed significant expression of WNT4 in HBECs with down-regulation in all the lung cancer cells analyzed in our study, except H661 (TCF21 expression positive). Our results suggest the interesting possibility that WNT4 might be one of the proximate downstream targets of TCF21. This possibility should be further explored. Thus, our bioassay to quantitate the prevalence of methylation of TCF21 in clinical samples may help determine the risk of cancer progression in patients and have considerable clinical application.
It appears from our data that methylation of the short CpG-rich sequence in the TCF21 gene may have significant potential as a biomarker assay in lung cancer and multiple other cancers. As evident from sputum analysis, the feasibility of increasing the sensitivity without significantly compromising the specificity (through combination of nested and qPCR) might make the assay more appealing for clinical as well as epidemiologic applications. Studies are in progress to further evaluate the potential of the assay in noninvasive detection of lung and other cancers.
Acknowledgments
Grant support: Early Detection Research Network grant U01CA084971, National Cancer Institute University of Texas SPORE in Lung Cancer grant P50CA70907, and Department of Defense grant W81XWH-04-1-0142.
Footnotes
Unpublished data.
References
- 1.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163 – 7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 2.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210 – 219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 3.Eads CA, Nickel AE, Laird PW. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic mice. Cancer Res. 2002;62:1296 – 9. [PubMed] [Google Scholar]
- 4.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410 – 8. [PubMed] [Google Scholar]
- 5.Brabender J, Usadel H, Danenberg KD, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20:3528 – 32. doi: 10.1038/sj.onc.1204455. [DOI] [PubMed] [Google Scholar]
- 6.Usadel H, Brabender J, Danenberg KD, et al. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371 – 5. [PubMed] [Google Scholar]
- 7.Harden SV, Tokumaru Y, Westra WH, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370 – 5. [PubMed] [Google Scholar]
- 8.Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284 – 8. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 9.Shivapurkar N, Stastny V, Suzuki M, et al. Application of a methylation gene panel by quantitative PCR for lung cancers. Cancer Lett. 2007;247:56 – 71. doi: 10.1016/j.canlet.2006.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quaggin SE, Schwartz L, Cui S, et al. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771 – 83. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 11.Smith LT, Lin M, Brena RM, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23–24 in lung and head and neck cancer. Proc Natl Acad Sci U S A. 2006;103:982– 7. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shigematsu H, Suzuki M, Takahashi T, et al. Aberrant methylation of HIN-1 (high in normal-1) is a frequent event in many human malignancies. Int J Cancer. 2005;113:600 – 4. doi: 10.1002/ijc.20622. [DOI] [PubMed] [Google Scholar]
- 13.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821 – 6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642 – 6. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 15.Toyooka KO, Toyooka S, Maitra A, et al. Establishment and validation of real-time polymerase chain reaction method for CDH1 promoter methylation. Am J Pathol. 2002;161:629 – 34. doi: 10.1016/S0002-9440(10)64218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shivapurkar N, Stastny V, Takahashi T, et al. Novel real-time PCR assay using a universal molecular marker for diagnosis of hematologic cancers. Int J Cancer. 2005;116:656 – 60. doi: 10.1002/ijc.21070. [DOI] [PubMed] [Google Scholar]
- 17.Fackler MJ, McVeigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442 – 52. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027 – 34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 19.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490 – 5. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Hugo H, Ackland ML, Blick T, et al. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374 – 83. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 21.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305 – 18. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 22.Taki M, Kamata N, Yokoyama K, et al. Down-regulation of Wnt-4 and up-regulation of Wnt-5a expression by epithelial-mesenchymal transition in human squamous carcinoma cells. Cancer Sci. 2003;94:593 –7. doi: 10.1111/j.1349-7006.2003.tb01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]