SYNOPSIS
Assays that measure minimal residual disease (MRD) can determine the response to treatment in patients with acute lymphoblastic leukemia (ALL) much more precisely than morphological screening of bone marrow smears. The clinical significance of MRD detected by flow cytometry or polymerase chain reaction-based methods in childhood ALL has been conclusively established. Hence, MRD is being used in several clinical trials to adjust treatment intensity. Similar findings have been gathered in adult patients with ALL, making MRD one of the most powerful and informative parameters to guide clinical management. This article discusses practical issues related to MRD methodologies and the evidence supporting the use of MRD for risk assignment in clinical trials.
Keywords: acute lymphoblastic leukemia, minimal residual disease, flow cytometry, polymerase chain reaction
INTRODUCTION
The response to treatment in patients with leukemia has traditionally been assessed by counting cells in blood while attempting to identify residual leukemic blasts in blood and bone marrow by microscopic analysis. The latter task is challenging when leukemic calls are present in small numbers. This is particularly true in patients with acute lymphoblastic leukemia (ALL) because the morphology of ALL blast cells is often indistinguishable from that of lymphoid precursors (the progenitors of B lymphocytes, often called “hematogones” by hemopathologists) and that of activated mature lymphocytes. The distinction between leukemic and normal cells is exceedingly difficult in bone marrow samples recovering after cessation of chemotherapy or after transplant, where the percentage of hematogones may surpass 10% of the total cellular population. Hence, the morphologic assessment of remission by morphology in patients with ALL can be imprecise, especially if samples are examined when normal hematopoiesis is reconstituting. With contemporary chemotherapy regimens, only a minority of patients with ALL have unusually high percentages of marrow leukemic lymphoblasts persisting during remission induction therapy (1); most patients achieve morphologic remission and the amount of residual disease not detectable by morphology, that is minimal residual disease (MRD), has remained unknown until relatively recently.
Janossy and colleagues were among the first to examine the remission status of patients with ALL with a method more sensitive and specific than morphology. While performing the initial attempts to immunophenotype ALL nearly 30 years ago, these investigators noticed that T-lineage ALL cells simultaneously expressed nuclear terminal deoxynucleotidil transferase (TdT) and T-cell markers whereas peripheral blood and bone marrow cells of healthy individuals did not (2). Logically, they used this cell marker combination to investigate whether patients with T-lineage ALL in morphologic remission had measurable MRD and found cells with the immunophenotype of T-lineage ALL in the bone marrow of some patients (3). The usefulness of immunologic markers to identify residual leukemic cells was corroborated and expanded by the development of monoclonal antibodies and clinical flow cytometers, which allowed the detection of MRD not only in T-lineage ALL but also in B-lineage ALL.(4-6) In parallel to these developments, other investigators took advantage of the development of the polymerase chain reaction (PCR) technique to amplify fusion transcripts in ALL cells (7;8), and to use antigen-receptor genes as a PCR target to detect MRD (9-12). This groundbreaking work was enriched by the subsequent research of numerous laboratories resulting in methods for objective MRD detection whose sensitivity is much higher than that of morphology.(13;14).
In addition to developing and refining MRD assays, early studies had to define the value of MRD testing to assess response to treatment and predict relapse. Although the clinical significance of MRD is now clear, initial efforts to systematically study MRD in patients were often met with skepticism about the clinical value of MRD testing. This often stemmed from the belief, supported by animal studies and clinical observations,(15;16) that leukemia distribution might be extremely heterogeneous, rendering MRD testing uninformative in regards to residual leukemic burden and treatment response. Others thought that MRD studies might not provide any additional information over established clinicobiologic prognostic features of ALL. As discussed in this article, numerous studies have now conclusively demonstrated that MRD is a powerful prognostic indicator in childhood ALL and there is mounting evidence that this is also the case in adult ALL patients. Therefore, an increasing number of treatment protocols use MRD measurements for ALL risk assignment.
MRD ASSAYS
Targets for flow cytometric studies
ALL cells express immunophenotypic features that can be used to distinguish them from normal hematopoietic cells, including hematogones and activated lymphocytes (14). These leukemia-associated immunophenotypes can be grouped into three main categories. First, are immunophenotypes that are expressed during normal development but are limited to cells in certain tissues. This group is exemplified by the immunophenotypic features of T-lineage ALL cells, which are only expressed by a subset of thymocytes and never found outside the thymus. These leukemia-associated immunophenotypes are those that were used by Janossy and colleagues in their early studies of MRD (3), and can now be effectively used with flow cytometry to monitor MRD in T-lineage ALL, and also to detect disease dissemination in T-cell lymphoblastic lymphoma.(17)
The second group of leukemia-associated immunophenotypes is constituted by the expression of fusion proteins derived from chromosomal breakpoints, such as BCR-ABL1, ETV6-RUNX1, or TCF3-PBX1. Within this category, we can also include the ectopic expression of proteins promoted by gene translocations, such as expression of PBX1 in lymphoblasts (PBX1 expression is normally confined to non-lymphoid cells),(18) and the high molecular weight melanoma associated antigen, the human homolog of the rat NG2, on the surface of 11q23-positive ALL (1, 2). This set of markers is attractive because of its leukemia specificity. However, its use has been limited by the lack of suitable antibodies for reliable flow cytometric analysis of these proteins.
The third type of leukemia-associated immunophenotype is represented by markers normally expressed during lymphohematopoiesis but found in abnormal combinations in leukemic cells. These phenotypes are, at times, termed “asynchronous” or “aberrant”(4;19-21). They offer the most widely available option to monitor MRD in ALL by flow cytometry, and the only one that can currently be used to track MRD in B-lineage ALL. The use of this approach requires a deep understanding of the immunophenotypes expressed by normal hematopoietic cells not only during steady-state conditions but also during chemotherapy and during active regeneration.
Targets for PCR studies
Two main categories of targets can be used to distinguish leukemic cells from normal cells with PCR. One is represented by gene fusions, such as BCR-ABL1, MLL-AFF1, TCF3-PBX1, and ETV6-RUNX1, which result in the expression of aberrant mRNA transcripts in leukemic cells (22;23). Recurrent abnormalities suitable for amplification in clinical samples are present in approximately 40% of children and 50% of adults with ALL (23;24). However, with the uncovering of genetic abnormalities afforded by the application of whole-genome screening technologies (25;26), it is very likely that additional genetic targets will enrich the available gamut.
The second category of PCR targets for MRD studies in ALL is composed by the clonal rearrangement of immunoglobulin (IG) and T-cell receptor (TCR) genes whose junctional regions are unique to the leukemic clone, forming a molecular signature of sorts. The most commonly used way to target these rearrangements for MRD detection requires identification of the various IG and/or TCR gene rearrangements in each lymphoid malignancy at diagnosis (27). Thus, the presence of rearranged genes is typically screened by using PCR primers matched to opposite sides of the junctions, to the V and J regions of various IG and TCR genes. If an apparently clonal rearrangement is found, one must ensure that it derives from ALL cells and not from contaminating normal cells by analyzing the PCR product for their clonal origin, e.g., by heteroduplex analysis. The ALL-derived PCR products are then used for direct sequencing of the junctional regions of the IG/TCR gene rearrangements which, in turn, is used to design junctional region-specific oligonucleotides, also called allele-specific oligonucleotides (27). Clonal IG/TCR gene rearrangements can also be detected with high-resolution electrophoresis systems, such as radioactive fingerprinting or fluorescent gene scanning, without the need for patient-specific oligonucleotides, but this approach has a considerably lower sensitivity, usually not better than 0.1% (28;29).
The majority (>95%) of B-lineage ALL cases have IG gene rearrangements (30;31). Cross-lineage TCR gene rearrangements also occur in up to 90% of B-lineage ALL cases (32). TCR genes are rearranged in most cases of T-lineage ALL (33-35). Cross-lineage IG gene rearrangements occur in approximately 20% of T-ALL (30).
Quantitation of MRD by PCR using either fusion transcripts or IG/TCR gene rearrangements is most frequently performed by using “real-time” quantitative PCR (RQ-PCR) (36).
Strengths and weaknesses of various MRD assays
In virtually all patients with ALL, leukemia-associated immunophenotypes can be defined at diagnosis and then used to monitor MRD during treatment. Immunophenotypes sufficiently dissimilar from those of normal cells to allow a sensitivity of detection of 0.01% are expressed by the majority of cells in approximately 95% of cases (20;37). In addition to their potential for accurate quantification of MRD, flow cytometric analysis can also be used to examine the status of normal hematopoietic cell maturation at the same time. This gives information about the degree of hemodilution in the sample studied and on the degree of lympho-hematopoietic recovery.
The reliability of flow cytometric MRD assays depends on several factors. First, the immunophenotypes used to distinguish leukemic cells must not overlap with those of normal lymphoid cells. Certain immunophenotypes apparently absent among cells from a bone marrow sample from a healthy donor may become apparent if the bone marrow is actively proliferating after chemotherapy. The label “leukemia-associated immunophenotype” should be reserved to those marker combinations that are truly never expressed by normal hematopoietic cells, regardless of their proliferative and developmental status. A second factor that affects the reliability of flow cytometric MRD assays is the number of cells available for study. If one wants to detect 1 leukemic cell in 10,000, at least 100,000 mononuclear cells must be examined, because 10 leukemic “events” are the minimum required to interpret flow cytometry results (20). Third, the markers used need to be stably expressed on leukemic cells, and the investigator must be aware of fluctuations that may occur during chemotherapy (38). Finally, and perhaps most importantly, the laboratory performing the studies must have specific expertise in MRD assays. Simple availability of a flow cytometer and experience in leukemia immunophenotyping is not sufficient to perform MRD studies proficiently.
A strength of PCR amplification of fusion transcripts is the stable association between the molecular abnormality and the leukemic clone, regardless of cellular changes caused by therapy or clonal selection. Moreover, a positive MRD result with this technique may alert to the presence of pre-leukemic or “leukemia-initiating” cells (39), which might be missed by other methods. There are, however, some disadvantages in using fusion transcripts as a target for MRD studies. Perhaps the most worrysome is the imprecise estimate of the percentage of leukemic cells present. This is due to the fact that the amount of transcripts per leukemic cell may vary from patient to patient with the same genetic subtype of ALL, and could be affected by chemotherapy and by the cell integrity when the sample is examined.(23) Because of these variables, it is practically impossible to establish a precise relation between quantity of PCR product and number of leukemic cells.
PCR amplification of antigen-receptor genes is a reliable and accurate method for monitoring MRD and can be used in the majority of cases of childhood and adult ALL (27;36;40). In contrast to fusion transcripts, rearranged IG and TCR genes are present in one copy per cell which; with the use of RQ-PCR, a very precise quantitation of MRD can be achived. IG and TCR genes in ALL might undergo continuing or secondary rearrangements (41), resulting in oligoclonality, i.e., the presence of subclones having distinct clonal IG/TCR gene rearrangements. IGH genes in B-lineage ALL are prone to subclone formation, with multiple gene rearrangements found in 30% to 40% of cases at diagnosis (42). Minor clones which might be undetected at diagnosis but may become predominant during the course of the disease (43;44), an observation that has lead to recommending targeting two or more different rearrangements (27). Indeed, multiple targets are identifiable in the majority of ALL cases but multiple targets that allow detection of MRD with a high sensitivity (e.g., 0.01%) are not identifiable in approximately 30% of cases (45;46).
When applied in parallel to study MRD in the same samples, flow cytometry and PCR amplification of IG/TCR genes yield remarkably similar measurements, if MRD is present at a ≥ 0.01% level (Figure 1)(47-49).
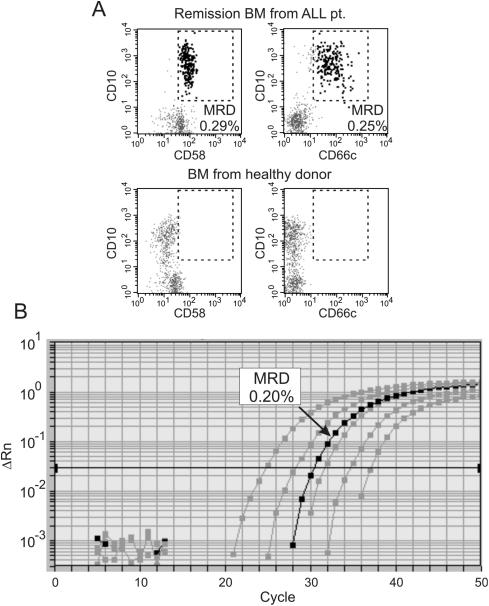
Figure 1. Detection of MRD by flow cytometry and PCR.
A bone marrow samples collected at the end of remission induction therapy from a patient with B-lineage ALL in morphologic remission was examined for MRD. (A) Flow cytometry studies of mononuclear cells using two different leukemia-associated markers (CD58 and CD66c) indicated the presence of 0.29% and 0.25% ALL cells, respectively (area within the dashed line in top panels). No cells (0.01%) within the equivalent areas of the dot plots were seen in the bone marrow of a healthy donor. (B) Molecular analysis of MRD was performed using RQ-PCR amplification of a clonal IG gene rearrangement determined at diagnosis. The black line corresponds to the amplification signal obtained in the patient sample; grey lines are the signal from serial dilutions of DNA extracted from the leukemic cell at diagnosis with that of peripheral blood from healthy donors. The estimated MRD levels was 0.20%, similar to the estimates by flow cytometry
Feasibility of MRD testing in prospective studies
MRD assays have been incorporated into clinical trials for children with ALL and their feasibility for routine analysis of treatment response is now well established. Of the 2143 patients with B-lineage ALL enrolled on 9900 series treatment protocols of the Children's Oncology Group, day 29 samples were submitted from 2086 patients (97.3%) to be studied for MRD by flow cytometry.(50) In only 4% of cases, sample cellularity was too low or the immunophenotype of the leukemic cells (determined at diagnosis) was not sufficiently distinct to allow a sensitivity of detection of 0.01%. MRD results were classified as indeterminate in 1.4% of cases. Overall, a test with the sensitivity of at leass 0.01% was performed in 92% of patients (50).
MRD was prospectively studied by PCR amplification of IG/TCR genes in pediatric patients enrolled in the AIEOP-BFM ALL 2000 trial (46). Bone marrow samples were obtained at initial diagnosis and on days 33 and 78 of therapy. Of the 3341 diagnostic samples examined, 88 (3%) lacked suitable gene rearrangements targets for PCR analysis, and an additional 217 (7%) had a target but not sufficient to reach a sensitivity of 0.01%. In 671 patients (20%) there was only one sensitive target, whereas in the remaining 71% of patients at least two sensitive targets were available for MRD analysis. At least one IG or TCR target could be identified in 98% of B-lineage ALL patients, with two or more targets detected in 93%. In T-lineage ALL patients, these proportions were 93% and 88%, respectively. Overall, adequate data for MRD-based stratification were obtained in 2594 (78%) of the 3341 patients.
In the St Jude Total XV trial for children with newly diagnosed ALL, our laboratory monitored MRD by using flow cytometric detection of aberrant immunophenotypes and/or PCR amplification of antigen-receptor genes. Flow cytometry was applied in patients with B-lineage ALL and T-ALL whereas PCR studies were done only in patients with B-lineage ALL. Overall, 482 of 492 patients (98%) were monitored by flow cytometry, and 403 of 492 (82%) by PCR. As previously shown (47-49), both methods yielded virtually identical results above the threshold level of 0.01% used to defined MRD positivity. In the few cases with discrepant results, we used the highest MRD value. The two methods in combination could be applied to study 491 of 492 (99.8%) patients. The single patient with no available immunophenotypic or antigen-receptor gene rearrangements had a MLL-AF9 fusion transcript and was monitored by RQ-PCR using that marker.
In adult ALL, 23% of B-ALL lacked a clonal marker for PCR analysis by one group (3) and 10% in another (4) of whom 7% had no clonal marker detected and in 3% the clonal markers were not suitable for MRD quantification.
CLINICAL SIGNIFICANCE OF MRD
Prognostic significance of MRD in childhood ALL
One of the most immediately obvious applications of MRD testing is its use in measuring early treatment response and identify patients who achieve morphologic remission but still harbor considerable levels of disease. The prognostic value of such tests in childhood ALL was most convincingly demonstrated by 3 large prospective studies reported in the late 90 by the EORTC,(51) St Jude (52) and BFM groups (53), which together many other reports (reviewed in (14;54) unequivocally demonstrated that MRD detected during the first 2-3 months of therapy is as the strongest predictor of relapse. MRD can also help identify patients with a higher risk of relapse among those with specific ALL subtypes (55-58), as well as among patients with first-relapse ALL who achieve a second remission (59-61), and patients with “isolated” extramedullary relapse (62). Detection of MRD before allogeneic hematopoietic stem cell transplantation (HSCT) is associated with an increase risk of relapse post-HSCT (29;63-67).
A commonly used cut-off level to define MRD positivity is 0.01% of bone marrow mononuclear cells. The selection of this level is due to the fact that this is the typical limit of detection for routine flow cytometric and molecular assays and it has been shown to discriminate patients with different risks of relapse. To this end, patients who had MRD of 0.01% or higher in bone marrow at any treatment interval monitored had a significantly higher risk of relapse in earlier St Jude studies (37;52;55). Likewise, MRD ≥ 0.01% on day 29 was the strongest prognostic indicator in studies of the Children's Oncology Group (50). Investigators of the EORTC, however, found that a cut-off level of 0.1% at the end of remission induction and at subsequent timepoints was very informative (51), as did those of the Austrian BFM group (68), and the Dana-Farber Cancer Institute ALL Consortium (69).
During the early phases of treatment, levels of MRD are directly proportional to the risk of subsequent relapse. For example, MRD ≥ 1% at the end of remission induction therapy was associated with an extremely high rate of relapse in St Jude Studies (55). Investigators of the I-BFM Study Group reported that patients whose bone marrow had MRD ≥0.1% on both day 33 and day 78 of treatment had a relapse rate of 75% (46;53).
An unexpected observation made while sequentially testing MRD in children with ALL was that, in a substantial proportion of patients, remission induction therapy induces a remarkable reduction in the leukemic cell burden, resulting in undetectable (<0.01%) MRD after only 2-3 weeks of therapy. In a recent analysis of 402 patients with B-lineage ALL, we found that 183 (45.5%) had such excellent response to treatment and were MRD <0.01% after 19 days of treatment (70). The vast majority of patients who become MRD negative at early stage of therapy had an excellent prognosis overall (37;71). This led us to develop a simplified flow cytometric MRD test that can detect residual leukemia on day 15-26 of treatment with a minimum panel of antibodies (72). Although this test cannot be used beyond this treatment interval (owing to the high risk of false-positive results in recovering marrow samples) it is well suited for the identification of patients whose leukemic cells have the highest sensitivity to early treatment and are predicted to have good treatment responses overall.
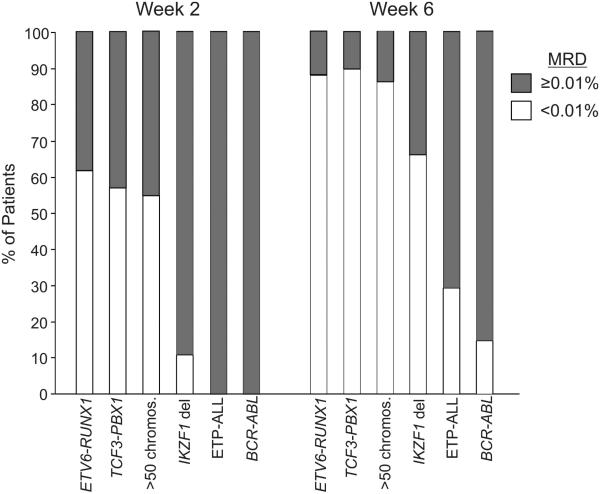
It is known that genetic abnormalities in childhood ALL are associated with a different prevalence of MRD during remission induction therapy (73;74). We found that among patients with B-lineage ALL MRD on days 19 and 43 of treatment was much more prevalent in those with BCR-ABL1 ALL, and less prevalent overall in patients with ETV6-RUNX1, hyperdiploid (>50 chromosomes) and TCF3-PBX1 ALL (75). Recent studies have identified novel subtypes of ALL with a significantly higher prevalence of MRD. Thus, patients with B-lineage ALL and mutations or deletions of Ikaros (IKZF1) were significantly more likely to have MRD detected during remission induction therapy than those without this abnormalitiy (26). Among patients with T-lineage ALL, those classified as early thymic precursor (ETP)-ALL, had significantly higher levels of MRD during remission induction therapy than patients with typical T-ALL (76). Figure 2 illustrates the prevalence of MRD among patients with various genetic subgroups of childhood ALL.
Figure 2. Prevalence of MRD in different subtypes of childhood ALL.
The percentage of MRD positive (≥ 0.01% of bone marrow mononuclear cells) and negative as determined by flow cytometry 2 and 6 weeks from diagnosis is shown. The number of patients studied in each subgroups was 89 for ETV6-RUNX1, 21 for TCF3-PBX1, 115 for hyperdiploidy with >50 chromosomes, 100 for IKZF1 deletions/mutations, 14 for ETP-ALL and 13 for BCR-ABL. For more details, please see references 26, 70 and 76.
Prognostic significance of MRD in adult ALL
Although the clinical significance of MRD has been studied less extensively in adult patients with ALL, there is considerable evidence supporting its potential usefulness. In an early study, Mortuza et al. (77) used PCR amplification of antigen-receptor genes to study MRD in a group of 85 patients with Philadelphia chromosome-negative B-lineage ALL and found that its presence 3-5 months after induction therapy correlated with a poorer outcome. Subsequently, Bruggeman et al. (40) used PCR amplification of antigen-receptor genes in 196 standard-risk patients to define three risk groups: a low-risk group including 10% of patients who had <0.01% MRD on day 11 and day 24 and a 3-year relapse rate of 0%; a high-risk group comprising 23% of patients defined by MRD ≥ 0.01% until week 16 with a relapse rate of 94%; and an intermediate-risk group including all remaining patients with a relapse rate of 47%. These team of investigators also reported results of a prospective analysis of post-consolidation samples in 105 patients enrolled in the German Multicenter GMALL trial. Inclusion criteria were hematologic remission, completion of the first-year chemotherapy, and MRD-negativity prior to enrollment in the study. Conversion to MRD positivity was observed in 28 patients, 17 of whom relapsed (median time from MRD test to clinical relapse, 9.5 months); of the 77 patients who remained MRD-negative, only 5 had relapsed at the time of the report (78). Holowiecki et al. (79) used flow cytometry to estimate MRD in 116 patients with Philadelphia-negative ALL enrolled in the Polish Adult leukemia Group ALL 4-2002 MRD study and found that MRD ≥ 0.1% after remission induction therapy was an independent predictor for relapse in both standard- and high-risk groups. Bassan et al. (80) studied MRD using fusion transcripts and/or IG/TCR gene rearrangements as targets. At the end of consolidation, 58 were MRD negative (0.01%) and 54 were MRD positive. Five-year overall disease-free survival estimates were 72% in the MRD negative group versus 14% in MRD positive group, regardless of clinical risk factors. MRD was the most significant risk factor for relapse.
Monitoring of MRD in adult patients with Philadelphia-positive ALL receiving HSCT and/or imatinib therapy has been shown to predict treatment outcome (81-83). Similarly, in Philadelphia-negative ALL, MRD detected by flow cytometry in bone marrow samples of patients with ALL before initiation of conditioning for HSCT was a significant predictor of failure post-HSCT (84). Finally, in a study of 43 adult patients with ALL undergoing HSCT, the relapse rate at 36 months was 0% for the 12 patients who were MRD-negative before HSCT versus 46% for those who were MRD-positive (85).
Uses of MRD testing for risk classification
There are numerous ways to include MRD studies in clinical trials, depending on treatment schedule, intensity and previous experience. For example, the AIEOP-BFM group used MRD to classify patients into three risk groups: standard risk (MRD negative on days 33 and 78), intermediate risk (any MRD positivity on days 33 and 78 but <0.1% on day 78), and high risk (MRD ≥ 0.1% on day 78); treatment intensity was regulated accordingly (46). At St Jude Children's Research Hospital, we use MRD levels on day 15 and day 42 for treatment assignment. Patients with MRD of ≥ 1% on day 15 receive intensified remission induction therapy; further intensification is reserved for patients with ≥ 5% leukemic cells. On the other hand, patients with MRD <0.01% on day 15 receive a slightly less intensive reinduction therapy and lower cumulative doses of anthracyclin. Patients with standard-risk ALL who have MRD of ≥ 0.01% on day 42 are reclassified as high-risk; patient with MRD ≥ 1% are eligible for HSCT in first remission.
Our team also uses MRD to guide treatment for patients with first-relapse ALL who achieve a second remission. Those with persistent MRD are candidates for HSCT whereas those who achieve MRD negativity (in the context of other favorable clinical features) are eligible for continuing chemotherapy. For patients who undergo HSCT, MRD is used to aid the timing of transplant. Thus, additional courses of chemotherapy may be administered in efforts to reduce MRD levels before HSCT. Monitoring MRD post-HSCT may be helpful to make informed decisions about modulation of immunosuppressive therapy and administrations of donor lymphocyte infusions.
All the MRD methods described here are being used to monitor MRD by various groups, the selection depending primarily on existing expertise and strength of preclinical studies withing the group. Flow cytometry results can be obtained with a few hours of sample collection. The development of a patient-specific PCR assay is time consuming but, once the assay is developed, MRD estimates can also be obtained quite rapidly. Because of the time required to develop a patient-specific PCR assay (often more than 2 weeks), flow cytometry may be preferable for studies at very early timepoints during therapy while PCR might be best for studies at the end of therapy or post-HSCT because of its higher sensitivity. Our current strategy is to use preferentially flow cytometry to monitor MRD during remisison induction therapy, and develop a PCR assay for IG/TCR genes only if a suitable immunophenotype is not identified at diagnosis. After studying MRD with either one or the other method on days 15 and 42, MRD monitoring is stopped in patients with B-lineage ALL who are MRD negative on day 42. Sequential MRD monitoring continues in patients with B-lineage ALL who are MRD positive and in any patient with T-lineage ALL. Although it has been estimated that PCR is more expensive than flow cytometry (86), in our experience the two methods have comparable costs.
In patients with B-lineage ALL, MRD is usually present at higher levels in bone marrow than in peripheral blood (87-89). This is not the case in T-lineage ALL, where MRD levels in peripheral blood are similar to those in bone marrow (88;89). Based on this observations, it is our current practice to use blood instead of marrow to monitor MRD after day 42 in patients with T-lineage ALL.
Concluding remarks
It is unquestionable that MRD tests allow to define leukemia “remission” in a way that is much more accurate and rigorous than the one afforded by conventional morphologic techniques. In addition to their capacity to predict outcome on the basis of early response to therapy, MRD methods can also be used to recognize leukemia relapse before it is morphologically overt, to determine the leukemia burden before HSCT, and to measure the efficacy of a treatment regimen in relation to that of its predecessor. Current studies incorporating MRD to guide treatment decisions should clarify whether this approach produces significantly higher cure rates and/or lower toxicities. Beyond its direct clinical application, MRD measurements can also be used to reveal new molecular determinants of treatment response, as shown by correlative studies with the gene expression profiles of leukemic lymphoblasts (90-92), as well as germline or leukemia-associated gene polymorphisms (93;94).
MRD assays are complex and require expertise to be performed well. Also, immunophenotype change at relapse was observed in 46% of cases (both pediatric and adult cases) in one study (5); specifically, the myeloid antigens were frequently lost or acquired. Therefore, simplification of the methodologies should be an objective for MRD researchers. Although MRD testing is relatively expensive compared to other routine laboratory assays performed at diagnosis in patients with ALL, it provides unique and powerful information which should not only improve treatment but, in the long run, also reduce overall clinical management costs (86).
ACKNOWEDGEMENTS
I thank Elaine Coustan-Smith, Pat Stow and Laura Key for providing Figure 1, and all the staff, patients and families at St Jude Children's Research Hospital for their continuous support.
This work was supported by grants CA60419 and CA21765 from the National Cancer Institute, and by the American Lebanese Syrian Associated Charities (ALSAC)
REFERENCES
- 1.Sandlund JT, Harrison PL, Rivera G, et al. Persistence of lymphoblasts in bone marrow on day 15 and days 22 to 25 of remission induction predicts a dismal treatment outcome in children with acute lymphoblastic leukemia. Blood. 2002;100(1):43–47. doi: 10.1182/blood.v100.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Janossy G, Bollum FJ, Bradstock KF, et al. Cellular phenotypes of normal and leukemic hemopoietic cells determined by analysis with selected antibody combinations. Blood. 1980;56(3):430–441. [PubMed] [Google Scholar]
- 3.Bradstock KF, Janossy G, Tidman N, et al. Immunological monitoring of residual disease in treated thymic acute lymphoblastic leukaemia. Leuk Res. 1981;5(45):301–309. doi: 10.1016/0145-2126(81)90002-3. [DOI] [PubMed] [Google Scholar]
- 4.Hurwitz CA, Loken MR, Graham ML, et al. Asynchronous antigen expression in B lineage acute lymphoblastic leukemia. Blood. 1988;72(1):299–307. [PubMed] [Google Scholar]
- 5.Terstappen LW, Loken MR. Myeloid cell differentiation in normal bone marrow and acute myeloid leukemia assessed by multi-dimensional flow cytometry. Anal Cell Pathol. 1990;2(4):229–240. [PubMed] [Google Scholar]
- 6.Campana D, Coustan-Smith E, Janossy G. The immunologic detection of minimal residual disease in acute leukemia. Blood. 1990;76(1):163–171. [PubMed] [Google Scholar]
- 7.Kawasaki ES, Clark SS, Coyne MY, et al. Diagnosis of chronic myeloid and acute lymphocytic leukemias by detection of leukemia-specific mRNA sequences amplified in vitro. Proc Natl Acad Sci U S A. 1988;85(15):5698–5702. doi: 10.1073/pnas.85.15.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermans A, Gow J, Selleri L, et al. bcr-abl oncogene activation in Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia. 1988;2(10):628–633. [PubMed] [Google Scholar]
- 9.d'Auriol L, MacIntyre E, Galibert F, et al. In vitro amplification of T cell gamma gene rearrangements: a new tool for the assessment of minimal residual disease in acute lymphoblastic leukemias. Leukemia. 1989;3(2):155–158. [PubMed] [Google Scholar]
- 10.Hansen-Hagge TE, Yokota S, Bartram CR. Detection of minimal residual disease in acute lymphoblastic leukemia by in vitro amplification of rearranged T-cell receptor delta chain sequences. Blood. 1989;74(5):1762–1767. [PubMed] [Google Scholar]
- 11.Yamada M, Hudson S, Tournay O, et al. Detection of minimal disease in hematopoietic malignancies of the B-cell lineage by using third-complementarity-determining region (CDR-III)-specific probes. Proc Natl Acad Sci U S A. 1989;86(13):5123–5127. doi: 10.1073/pnas.86.13.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brisco MJ, Condon J, Hughes E, et al. Outcome prediction in childhood acute lymphoblastic leukaemia by molecular quantification of residual disease at the end of induction. Lancet. 1994;343(8891):196–200. doi: 10.1016/s0140-6736(94)90988-1. [DOI] [PubMed] [Google Scholar]
- 13.Szczepanski T, Orfao A, van der Velden VH, et al. Minimal residual disease in leukaemia patients. Lancet Oncology. 2001:2409–417. doi: 10.1016/s1470-2045(00)00418-6. [DOI] [PubMed] [Google Scholar]
- 14.Campana D. Determination of minimal residual disease in leukemia patients. Br J Haematol. 2003;121:823–838. doi: 10.1046/j.1365-2141.2003.04393.x. [DOI] [PubMed] [Google Scholar]
- 15.Martens AC, Schultz FW, Hagenbeek A. Nonhomogeneous distribution of leukemia in the bone marrow during minimal residual disease. Blood. 1987;70(4):1073–1078. [PubMed] [Google Scholar]
- 16.Mathe G, Schwarzenberg L, Mery AM, et al. Extensive histological and cytological survey of patients with acute leukaemia in “complete remission”. Br Med J. 1966;5488:640–642. doi: 10.1136/bmj.1.5488.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coustan-Smith E, Sandlund JT, Perkins SL, et al. Minimal disseminated disease in childhood T-cell lymphoblastic lymphoma: a report from the Children's Oncology Group. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.21.1318. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Dijk MA, Voorhoeve PM, Murre C. Pbx1 is converted into a transcriptional activator upon acquiring the N-terminal region of E2A in pre-B-cell acute lymphoblastoid leukemia. Proc Natl Acad Sci U S A. 1993;90(13):6061–6065. doi: 10.1073/pnas.90.13.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucio P, Parreira A, van den Beemd MW, et al. Flow cytometric analysis of normal B cell differentiation: a frame of reference for the detection of minimal residual disease in precursor-B- ALL. Leukemia. 1999;13(3):419–427. doi: 10.1038/sj.leu.2401279. [DOI] [PubMed] [Google Scholar]
- 20.Campana D, Coustan-Smith E. Detection of minimal residual disease in acute leukemia by flow cytometry. Cytometry. 1999:38139–152. doi: 10.1002/(sici)1097-0320(19990815)38:4<139::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 21.Ciudad J, San Miguel JF, Lopez-Berges MC, et al. Prognostic value of immunophenotypic detection of minimal residual disease in acute lymphoblastic leukemia. J Clin Oncol. 1998;16(12):3774–3781. doi: 10.1200/JCO.1998.16.12.3774. [DOI] [PubMed] [Google Scholar]
- 22.van Dongen JJ, Macintyre EA, Gabert JA, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13(12):1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 23.Gabert J, Beillard E, van der Velden V, et al. Standardization and quality control studies of `real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia. 2003;17(12):2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- 24.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23(26):6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 25.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 26.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Velden V, Cazzaniga G, Schrauder A, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21(4):604–611. doi: 10.1038/sj.leu.2404586. [DOI] [PubMed] [Google Scholar]
- 28.Delabesse E, Burtin ML, Millien C, et al. Rapid, multifluorescent TCRG Vgamma and Jgamma typing: application to T cell acute lymphoblastic leukemia and to the detection of minor clonal populations. Leukemia. 2000;14(6):1143–1152. doi: 10.1038/sj.leu.2401750. [DOI] [PubMed] [Google Scholar]
- 29.Knechtli CJC, Goulden NJ, Hancock JP, et al. Minimal residual disease status before allogeneic bone marrow transplantation is an important determinant of successful outcome for children and adolescents with acute lymphoblastic leukemia. Blood. 1998;92(11):4072–4079. [PubMed] [Google Scholar]
- 30.van Dongen JJ, Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part II: Possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta. 1991;198(12):93–174. doi: 10.1016/0009-8981(91)90247-a. [DOI] [PubMed] [Google Scholar]
- 31.Beishuizen A, Verhoeven MA, Mol EJ, et al. Detection of immunoglobulin heavy-chain gene rearrangements by Southern blot analysis: recommendations for optimal results. Leukemia. 1993;7(12):2045–2053. [PubMed] [Google Scholar]
- 32.Szczepanski T, Beishuizen A, Pongers-Willemse MJ, et al. Cross-lineage T cell receptor gene rearrangements occur in more than ninety percent of childhood precursor-B acute lymphoblastic leukemias: alternative PCR targets for detection of minimal residual disease. Leukemia. 1999;13(2):196–205. doi: 10.1038/sj.leu.2401277. [DOI] [PubMed] [Google Scholar]
- 33.Breit TM, Wolvers-Tettero IL, Beishuizen A, et al. Southern blot patterns, frequencies, and junctional diversity of T-cell receptor-delta gene rearrangements in acute lymphoblastic leukemia. Blood. 1993;82(10):3063–3074. [PubMed] [Google Scholar]
- 34.Szczepanski T, Langerak AW, Willemse MJ, et al. T cell receptor gamma (TCRG) gene rearrangements in T cell acute lymphoblastic leukemia refelct `end-stage' recombinations: implications for minimal residual disease monitoring. Leukemia. 2000;14(7):1208–1214. doi: 10.1038/sj.leu.2401765. [DOI] [PubMed] [Google Scholar]
- 35.Kneba M, Bolz I, Linke B, et al. Analysis of rearranged T-cell receptor beta-chain genes by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood. 1995;86(10):3930–3937. [PubMed] [Google Scholar]
- 36.van der Velden V, Hochhaus A, Cazzaniga G, et al. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia. 2003;17(6):1013–1034. doi: 10.1038/sj.leu.2402922. [DOI] [PubMed] [Google Scholar]
- 37.Coustan-Smith E, Sancho J, Behm FG, et al. Prognostic importance of measuring early clearance of leukemic cells by flow cytometry in childhood acute lymphoblastic leukemia. Blood. 2002;100(1):52–58. doi: 10.1182/blood-2002-01-0006. [DOI] [PubMed] [Google Scholar]
- 38.Gaipa G, Basso G, Maglia O, et al. Drug-induced immunophenotypic modulation in childhood ALL: implications for minimal residual disease detection. Leukemia. 2005;19(1):49–56. doi: 10.1038/sj.leu.2403559. [DOI] [PubMed] [Google Scholar]
- 39.Hong D, Gupta R, Ancliff P, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319(5861):336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 40.Bruggemann M, Raff T, Flohr T, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood. 2006;107(3):1116–1123. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- 41.Szczepanski T, Pongers-Willemse MJ, Langerak AW, et al. Unusual immunoglobulin and T-cell receptor gene rearrangement patterns in acute lymphoblastic leukemias. Curr Top Microbiol Immunol. 1999;246:205–13. doi: 10.1007/978-3-642-60162-0_26. [DOI] [PubMed] [Google Scholar]
- 42.Beishuizen A, Hahlen K, Hagemeijer A, et al. Multiple rearranged immunoglobulin genes in childhood acute lymphoblastic leukemia of precursor B-cell origin. Leukemia. 1991;5(8):657–667. [PubMed] [Google Scholar]
- 43.Szczepanski T, Willemse MJ, Brinkhof B, et al. Comparative analysis of Ig and TCR gene rearrangements at diagnosis and at relapse of childhood precursor-B-ALL provides improved strategies for selection of stable PCR targets for monitoring of minimal residual disease. Blood. 2002;99(7):2315–2323. doi: 10.1182/blood.v99.7.2315. [DOI] [PubMed] [Google Scholar]
- 44.van der Velden V, Bruggemann M, Hoogeveen PG, et al. TCRB gene rearrangements in childhood and adult precursor-B-ALL: frequency, applicability as MRD-PCR target, and stability between diagnosis and relapse. Leukemia. 2004;18(12):1971–1980. doi: 10.1038/sj.leu.2403505. [DOI] [PubMed] [Google Scholar]
- 45.Pongers-Willemse MJ, Seriu T, Stolz F, et al. Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets. Leukemia. 1999:13110–118. doi: 10.1038/sj.leu.2401245. [DOI] [PubMed] [Google Scholar]
- 46.Flohr T, Schrauder A, Cazzaniga G, et al. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22:771–782. doi: 10.1038/leu.2008.5. [DOI] [PubMed] [Google Scholar]
- 47.Neale GA, Coustan-Smith E, Pan Q, et al. Tandem application of flow cytometry and polymerase chain reaction for comprehensive detection of minimal residual disease in childhood acute lymphoblastic leukemia. Leukemia. 1999;13(8):1221–1226. doi: 10.1038/sj.leu.2401459. [DOI] [PubMed] [Google Scholar]
- 48.Neale GA, Coustan-Smith E, Stow P, et al. Comparative analysis of flow cytometry and polymerase chain reaction for the detection of minimal residual disease in childhood acute lymphoblastic leukemia. Leukemia. 2004;18:934–938. doi: 10.1038/sj.leu.2403348. [DOI] [PubMed] [Google Scholar]
- 49.Kerst G, Kreyenberg H, Roth C, et al. Concurrent detection of minimal residual disease (MRD) in childhood acute lymphoblastic leukaemia by flow cytometry and real-time PCR. Br J Haematol. 2005;128(6):774–782. doi: 10.1111/j.1365-2141.2005.05401.x. [DOI] [PubMed] [Google Scholar]
- 50.Borowitz MJ, Devidas M, Hunger SP, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors. a Children's Oncology Group study. Blood. 2008 doi: 10.1182/blood-2008-01-132837. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cave H, van der Werfften Bosch J, Suciu S, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer--Childhood Leukemia Cooperative Group. N Engl J Med. 1998;339(9):591–598. doi: 10.1056/NEJM199808273390904. [DOI] [PubMed] [Google Scholar]
- 52.Coustan-Smith E, Behm FG, Sanchez J, et al. Immunological detection of minimal residual disease in children with acute lymphoblastic leukaemia. Lancet. 1998;351(9102):550–554. doi: 10.1016/S0140-6736(97)10295-1. [DOI] [PubMed] [Google Scholar]
- 53.van Dongen JJ, Seriu T, Panzer-Grumayer ER, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet. 1998;352:1731–1738. doi: 10.1016/S0140-6736(98)04058-6. [DOI] [PubMed] [Google Scholar]
- 54.Campana D. Minimal residual disease in acute lymphoblastic leukemia. Semin Hematol. 2009;46(1):100–106. doi: 10.1053/j.seminhematol.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coustan-Smith E, Sancho J, Hancock ML, et al. Clinical importance of minimal residual disease in childhood acute lymphoblastic leukemia. Blood. 2000;96:2691–2696. [PubMed] [Google Scholar]
- 56.Biondi A, Valsecchi MG, Seriu T, et al. Molecular detection of minimal residual disease is a strong predictive factor of relapse in childhood B-lineage acute lymphoblastic leukemia with medium risk features. A case control study of the International BFM study group. Leukemia. 2000;14(11):1939–1943. doi: 10.1038/sj.leu.2401922. [DOI] [PubMed] [Google Scholar]
- 57.Attarbaschi A, Mann G, Panzer-Grumayer R, et al. Minimal residual disease values discriminate between low and high relapse risk in children with B-cell precursor acute lymphoblastic leukemia and an intrachromosomal amplification of chromosome 21: the Austrian and German acute lymphoblastic leukemia Berlin-Frankfurt-Munster (ALL-BFM) trials. J Clin Oncol. 2008;26(18):3046–3050. doi: 10.1200/JCO.2008.16.1117. [DOI] [PubMed] [Google Scholar]
- 58.van der Velden V, Corral L, Valsecchi MG, et al. Prognostic significance of minimal residual disease in infants with acute lymphoblastic leukemia treated within the Interfant-99 protocol. Leukemia. 2009 doi: 10.1038/leu.2009.17. in press. [DOI] [PubMed] [Google Scholar]
- 59.Eckert C, Biondi A, Seeger K, et al. Prognostic value of minimal residual disease in relapsed childhood acute lymphoblastic leukaemia. Lancet. 2001;358(9289):1239–1241. doi: 10.1016/S0140-6736(01)06355-3. [DOI] [PubMed] [Google Scholar]
- 60.Coustan-Smith E, Gajjar A, Hijiha N, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia after first relapse. Leukemia. 2004:18499–504. doi: 10.1038/sj.leu.2403283. [DOI] [PubMed] [Google Scholar]
- 61.Paganin M, Zecca M, Fabbri G, et al. Minimal residual disease is an important predictive factor of outcome in children with relapsed `high-risk' acute lymphoblastic leukemia. Leukemia. 2008;22(12):2193–2200. doi: 10.1038/leu.2008.227. [DOI] [PubMed] [Google Scholar]
- 62.Hagedorn N, Acquaviva C, Fronkova E, et al. Submicroscopic bone marrow involvement in isolated extramedullary relapses in childhood acute lymphoblastic leukemia: a more precise definition of “isolated” and its possible clinical implications, a collaborative study of the Resistant Disease Committee of the International BFM study group. Blood. 2007;110(12):4022–4029. doi: 10.1182/blood-2007-04-082040. [DOI] [PubMed] [Google Scholar]
- 63.van der Velden V, Joosten SA, Willemse MJ, et al. Real-time quantitative PCR for detection of minimal residual disease before allogeneic stem cell transplantation predicts outcome in children with acute lymphoblastic leukemia. Leukemia. 2001;15(9):1485–1487. doi: 10.1038/sj.leu.2402198. [DOI] [PubMed] [Google Scholar]
- 64.Bader P, Hancock J, Kreyenberg H, et al. Minimal residual disease (MRD) status prior to allogeneic stem cell transplantation is a powerful predictor for post-transplant outcome in children with ALL. Leukemia. 2002;16(9):1668–1672. doi: 10.1038/sj.leu.2402552. [DOI] [PubMed] [Google Scholar]
- 65.Uzunel M, Mattsson J, Jaksch M, et al. The significance of graft-versus-host disease and pretransplantation minimal residual disease status to outcome after allogeneic stem cell transplantation in patients with acute lymphoblastic leukemia. Blood. 2001;98(6):1982–1984. doi: 10.1182/blood.v98.6.1982. [DOI] [PubMed] [Google Scholar]
- 66.Krejci O, van der Velden V, Bader P, et al. Level of minimal residual disease prior to haematopoietic stem cell transplantation predicts prognosis in paediatric patients with acute lymphoblastic leukaemia: a report of the Pre-BMT MRD Study Group. Bone Marrow Transplant. 2003;32(8):849–851. doi: 10.1038/sj.bmt.1704241. [DOI] [PubMed] [Google Scholar]
- 67.Goulden N, Bader P, van der Velden V, et al. Minimal residual disease prior to stem cell transplant for childhood acute lymphoblastic leukaemia. Br J Haematol. 2003;122(1):24–29. doi: 10.1046/j.1365-2141.2003.04394.x. [DOI] [PubMed] [Google Scholar]
- 68.Dworzak MN, Froschl G, Printz D, et al. Prognostic significance and modalities of flow cytometric minimal residual disease detection in childhood acute lymphoblastic leukemia. Blood. 2002;99(6):1952–1958. doi: 10.1182/blood.v99.6.1952. [DOI] [PubMed] [Google Scholar]
- 69.Zhou J, Goldwasser MA, Li A, et al. Quantitative analysis of minimal residual disease predicts relapse in children with B-lineage acute lymphoblastic leukemia in DFCI ALL Consortium Protocol 95-01. Blood. 2007;110(5):1607–1611. doi: 10.1182/blood-2006-09-045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campana D. Molecular determinants of treatment response in acute lymphoblastic leukemia. Hematology (Am Soc Hematol Educ Program) 2008:366–373. doi: 10.1182/asheducation-2008.1.366. [DOI] [PubMed] [Google Scholar]
- 71.Panzer-Grumayer ER, Schneider M, Panzer S, et al. Rapid molecular response during early induction chemotherapy predicts a good outcome in childhood acute lymphoblastic leukemia. Blood. 2000;95(3):790–794. [PubMed] [Google Scholar]
- 72.Coustan-Smith E, Ribeiro RC, Stow P, et al. A simplified flow cytometric assay identifies children with acute lymphoblastic leukemia who have a superior clinical outcome. Blood. 2006;108(1):97–102. doi: 10.1182/blood-2006-01-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pui CH, Campana D, Evans WE. Childhood acute lymphoblastic leukemia - Current status and future perspectives. Lancet Oncology. 2001:2597–607. doi: 10.1016/S1470-2045(01)00516-2. [DOI] [PubMed] [Google Scholar]
- 74.Borowitz MJ, Pullen DJ, Shuster JJ, et al. Minimal residual disease detection in childhood precursor-B-cell acute lymphoblastic leukemia: relation to other risk factors. A Children's Oncology Group study. Leukemia. 2003;17(8):1566–1572. doi: 10.1038/sj.leu.2403001. [DOI] [PubMed] [Google Scholar]
- 75.Campana D. Molecular determinants of treatment response in acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program. 2008:366–73. doi: 10.1182/asheducation-2008.1.366. [DOI] [PubMed] [Google Scholar]
- 76.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mortuza FY, Papaioannou M, Moreira IM, et al. Minimal residual disease tests provide an independent predictor of clinical outcome in adult acute lymphoblastic leukemia. J Clin Oncol. 2002;20(4):1094–1104. doi: 10.1200/JCO.2002.20.4.1094. [DOI] [PubMed] [Google Scholar]
- 78.Raff T, Gokbuget N, Luschen S, et al. Molecular relapse in adult standard-risk ALL patients detected by prospective MRD monitoring during and after maintenance treatment: data from the GMALL 06/99 and 07/03 trials. Blood. 2007;109(3):910–915. doi: 10.1182/blood-2006-07-037093. [DOI] [PubMed] [Google Scholar]
- 79.Holowiecki J, Krawczyk-Kulis M, Giebel S, et al. Status of minimal residual disease after induction predicts outcome in both standard and high-risk Ph-negative adult acute lymphoblastic leukaemia. The Polish Adult Leukemia Group ALL 4-2002 MRD Study. Br J Haematol. 2008;142(2):227–237. doi: 10.1111/j.1365-2141.2008.07185.x. [DOI] [PubMed] [Google Scholar]
- 80.Bassan R, Spinelli O, Oldani E, et al. Improved risk classification for risk-specific therapy based on the molecular study of MRD in adult ALL. Blood. 2009 doi: 10.1182/blood-2008-11-185132. [DOI] [PubMed] [Google Scholar]
- 81.Radich J, Gehly G, Lee A, et al. Detection of bcr-abl transcripts in Philadelphia chromosome-positive acute lymphoblastic leukemia after marrow transplantation. Blood. 1997;89(7):2602–2609. [PubMed] [Google Scholar]
- 82.Wassmann B, Pfeifer H, Stadler M, et al. Early molecular response to posttransplantation imatinib determines outcome in MRD+ Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL) Blood. 2005;106(2):458–463. doi: 10.1182/blood-2004-05-1746. [DOI] [PubMed] [Google Scholar]
- 83.Pane F, Cimino G, Izzo B, et al. Significant reduction of the hybrid BCR/ABL transcripts after induction and consolidation therapy is a powerful predictor of treatment response in adult Philadelphia-positive acute lymphoblastic leukemia. Leukemia. 2005;19(4):628–635. doi: 10.1038/sj.leu.2403683. [DOI] [PubMed] [Google Scholar]
- 84.Sanchez J, Serrano J, Gomez P, et al. Clinical value of immunological monitoring of minimal residual disease in acute lymphoblastic leukaemia after allogeneic transplantation. Br J Haematol. 2002;116(3):686–694. doi: 10.1111/j.1365-2141.2002.3311a.x. [DOI] [PubMed] [Google Scholar]
- 85.Spinelli O, Peruta B, Tosi M, et al. Clearance of minimal residual disease after allogeneic stem cell transplantation and the prediction of the clinical outcome of adult patients with high-risk acute lymphoblastic leukemia. Haematologica. 2007;92(5):612–618. doi: 10.3324/haematol.10965. [DOI] [PubMed] [Google Scholar]
- 86.Goulden N, Oakhill A, Steward C. Practical application of minimal residual disease assessment in childhood acute lymphoblastic leukaemia annotation. Br J Haematol. 2001;112(2):275–281. doi: 10.1046/j.1365-2141.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- 87.Brisco MJ, Sykes PJ, Hughes E, et al. Monitoring minimal residual disease in peripheral blood in B-lineage acute lymphoblastic leukaemia. Br J Haematol. 1997;99(2):314–319. doi: 10.1046/j.1365-2141.1997.3723186.x. [DOI] [PubMed] [Google Scholar]
- 88.Coustan-Smith E, Sancho J, Hancock ML, et al. Use of peripheral blood instead of bone marrow to monitor residual disease in children with acute lymphoblastic leukemia. Blood. 2002:1002399–2402. doi: 10.1182/blood-2002-04-1130. [DOI] [PubMed] [Google Scholar]
- 89.van der Velden V, Jacobs DC, Wijkhuijs AJ, et al. Minimal residual disease levels in bone marrow and peripheral blood are comparable in children with T cell acute lymphoblastic leukemia (ALL), but not in precursor-B-ALL. Leukemia. 2002;16(8):1432–1436. doi: 10.1038/sj.leu.2402636. [DOI] [PubMed] [Google Scholar]
- 90.Cario G, Stanulla M, Fine BM, et al. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105(2):821–826. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- 91.Flotho C, Coustan-Smith E, Pei D, et al. Genes contributing to minimal residual disease in childhood acute lymphoblastic leukemia: prognostic significance of CASP8AP2. Blood. 2006;108:1050–1057. doi: 10.1182/blood-2006-01-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Flotho C, Coustan-Smith E, Pei D, et al. A set of genes that regulate cell proliferation predicts treatment outcome in childhood acute lymphoblastic leukemia. Blood. 2007;110(4):1271–1277. doi: 10.1182/blood-2007-01-068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rocha JC, Cheng C, Liu W, et al. Pharmacogenetics of outcome in children with acute lymphoblastic leukemia. Blood. 2005;105(12):4752–4758. doi: 10.1182/blood-2004-11-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301(4):393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]