Summary
Transcription factor movement is well established in plants. Since the initial report of KNOTTED movement, more than a dozen transcription factors have been shown to move in plants. However, the developmental significance of movement is not known. Using the SHORT-ROOT (SHR) transcription factor as a tool for studying cell-to-cell trafficking, we show that movement of SHR from its site of synthesis is necessary for normal development of the Arabidopsis root. We identify multiple regions of SHR that are required for intra-and intercellular movement of SHR, including a region that is necessary for movement but not activity. We made the surprising discovery that the capacity for intercellular movement may be conserved among other GRAS family proteins. Finally, we provide evidence that movement requires both cytoplasmic and nuclear localization, strongly suggesting a mechanistic link between nuclear transport and cell-to-cell movement.
Keywords: SHORT ROOT, Arabidopsis root development, intercellular protein movement, plasmodesmata, protein trafficking
Introduction
In plants, cell-to-cell communication is possible by the direct trafficking of transcription factors between cells. Transcription factor movement was initially described by Lucas et al. (1995), who showed that the maize KNOTTED1 (KN1) protein is found beyond the domain of KN1 RNA transcription. Since then, additional transcription factors have been shown to traffic between cells, including KNAT1/BREVIPEDICELLUS (KNAT1/BP), SHOOT MERISTEMLESS (STM) (Kim et al., 2003), DEFICIENS (DEF) (Perbal et al., 1996), CAPRICE (CPC) (Wada et al., 2002), LEAFY (LFY) (Sessions et al., 2000) and SHORT-ROOT (SHR) (Nakajima et al., 2001).
Transcription factor movement is possible in plants due to the presence of plasmodesmata (Lucas, 1995; Lucas et al., 1995; Maizel, 2006; Perbal et al., 1996; Ruiz-Medrano et al., 2004; Wu et al., 2003; Zambryski and Crawford, 2000). Plasmodesmata are membrane-lined channels that connect nearly all plant cells (Gunning and Overall, 1983). Functionally they are similar to gap junctions (or ring canals), in that they allow passage of small molecules between cells. However, unlike gap junctions, plasmodesmata selectively allow passage of macromolecules between cells. When tested using GFP, the size-exclusion limit of plasmodesmata in most growing tissues was found to range from approximately 30–50 kDa (Crawford and Zambryski, 2001; Kim et al., 2005a). In theory, transcription factors could either diffuse through open plasmodesmata or actively dilate plasmodesmata to facilitate their own movement (Crawford and Zambryski, 2001). However, recent results with KNOTTED1, in which Tassetto et al. (2005) showed that the KNOTTED1 homeodomain can traffic between animal cells in culture, suggest that at least some transcription factors may selectively move between cells by a plasmodesmata-independent pathway.
While the precise mechanisms regulating intercellular trafficking are not known, structure/function analysis of several of these transcription factors as well as other plant-derived non-cell-autonomous proteins revealed two primary modes of cell-to-cell movement (Aoki et al., 2002; Kim et al., 2005b; Kurata et al., 2005; Lucas et al., 1995; Wu et al., 2003). The first, which is referred to as selective movement, relies upon sequences within the transported protein for movement (Lucas et al., 1995). For example, within CPC, two motifs are necessary for transport: one in the N-terminus and the other in the MYB domain (Kurata et al., 2005). Likewise, the KN1 homeodomain has been shown to mediate KNI movement (Kim et al., 2005b). In SHR, substitution of a single amino acid in the VHIID domain inhibits movement (Gallagher et al., 2004). In contrast, there is no requirement for movement sequences in proteins that move non-selectively between cells (i.e. by diffusion), as has been shown for LFY (Wu et al., 2003). For all of the selectively transported proteins, the sequences relevant to protein movement appear to function only within the context of the protein as a whole. No short autonomous movement domain akin to the 9–11 amino acid HIV Tat transduction domain, which is able to cause movement to unrelated proteins (Chauhan et al., 2007), has been found in plants. Recently, however, it was shown that 38 amino acids from the non-cell-autonomous Cm-PP16-1 phloem protein appear to confer movement to the normally cell-autonomous glutathione-S-transferase protein (Taoka et al., 2007).
It is generally believed that cell-to-cell transport of transcription factors is developmentally relevant, because zones of symplastic connectivity change over developmental time (Gisel et al., 1999; Kim et al., 2002a,b; Rinne and van der Schoot, 1998) and because movement of specific transcription factors correlates with specific developmental events. However, direct evidence showing that movement is required for developmental patterning is lacking.
Here we examine SHR movement in the Arabidopsis root with respect to both the relevance of movement to development, and the mechanisms regulating cell-to-cell trafficking. SHR is a member of the plant-specific family of GRAS transcription factors. It is required for proper root development, moving from the stele into the adjacent ground tissue where it is required for an asymmetric cell division that creates the two distinct ground tissue layers, the cortex and endodermis, which are located between the pericycle and epidermal cell layers (Figure 1) (Nakajima et al., 2001). Recent results have shown that SHR occurs directly upstream of SCARECROW (SCR) in a pathway that leads to this patterning of the ground tissue and maintenance of quiescent center (QC) cells (Nakajima et al., 2001). However, SHR and SCR are expressed in non-overlapping domains. SHR is expressed only in the stele, whereas SCR is confined to the ground tissue. This raises the question of how SHR regulates SCR expression.
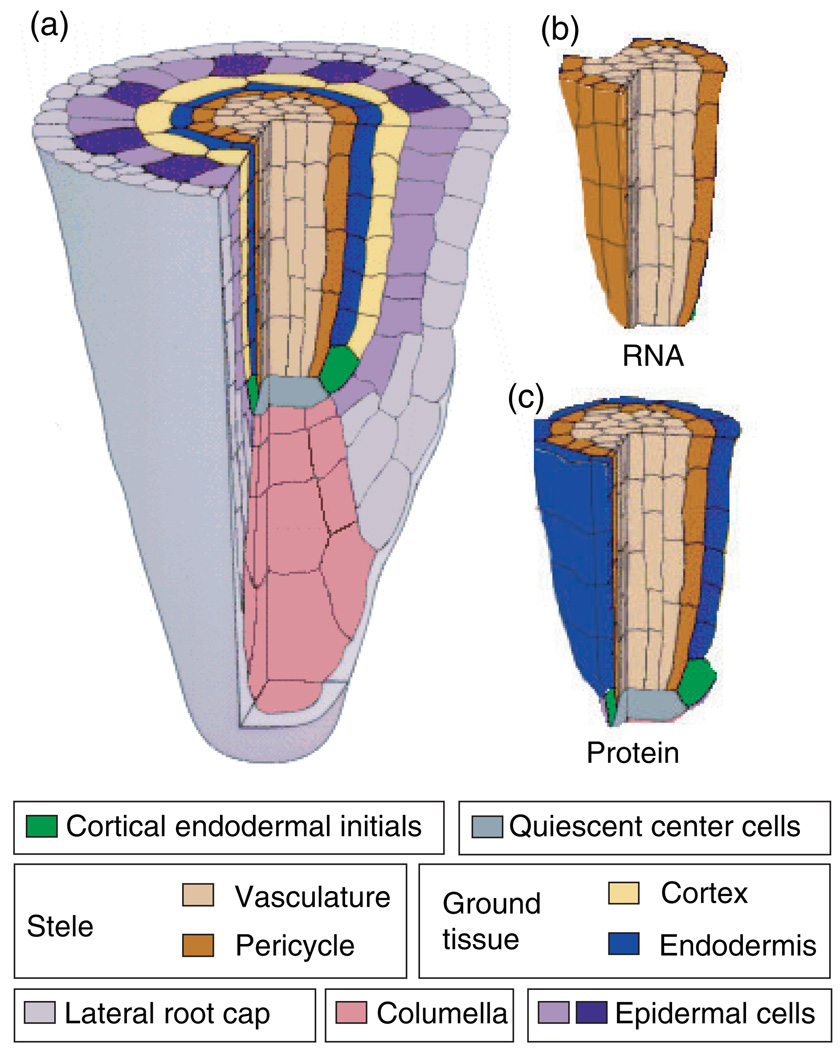
Figure 1. Diagram illustrating SHR RNA and protein localization in the Arabidopsis root meristem.
(a) Cut-away view of the Arabidopsis root tip.
(b) The cells in which SHR RNA is detected.
(c) The cells in which SHR protein is detected.
This figure is modified from Benfey and Scheres (2000).
In the present study, we show that SHR movement from the stele is required for proper patterning of the root. In addition, we add to our earlier findings that SHR movement is selective and requires cytoplasmic localization (Gallagher et al., 2004), identifying multiple domains that are required for SHR movement and for proper sub-cellular localization, and suggesting that both nuclear and cytoplasmic localization are required for SHR movement. These data expand upon our previous results (Gallagher et al., 2004) and the results of Cui et al. (2007) who showed that SCR is required in endodermal cells to prevent movement of SHR into the cortical cell layer and expansion of the ground tissue. Collectively, these data allow us to propose a model for SHR trafficking in which a balance between nuclear localization and nuclear export promotes SHR movement into the ground tissue, where further movement is blocked by SCR (Cui et al., 2007).
Results
The demonstration that SHR moves from the stele into the endodermis (Figure 1) and is responsible for up-regulating SCR expression (Nakajima et al., 2001) strongly suggested that SHR movement is required for normal ground-tissue patterning. However, this has not been directly demonstrated. To address this issue, we determined whether SHR could rescue the radial patterning and root length of shr-2 plants (shown in Figure 2a) without moving into the endodermis or QC.
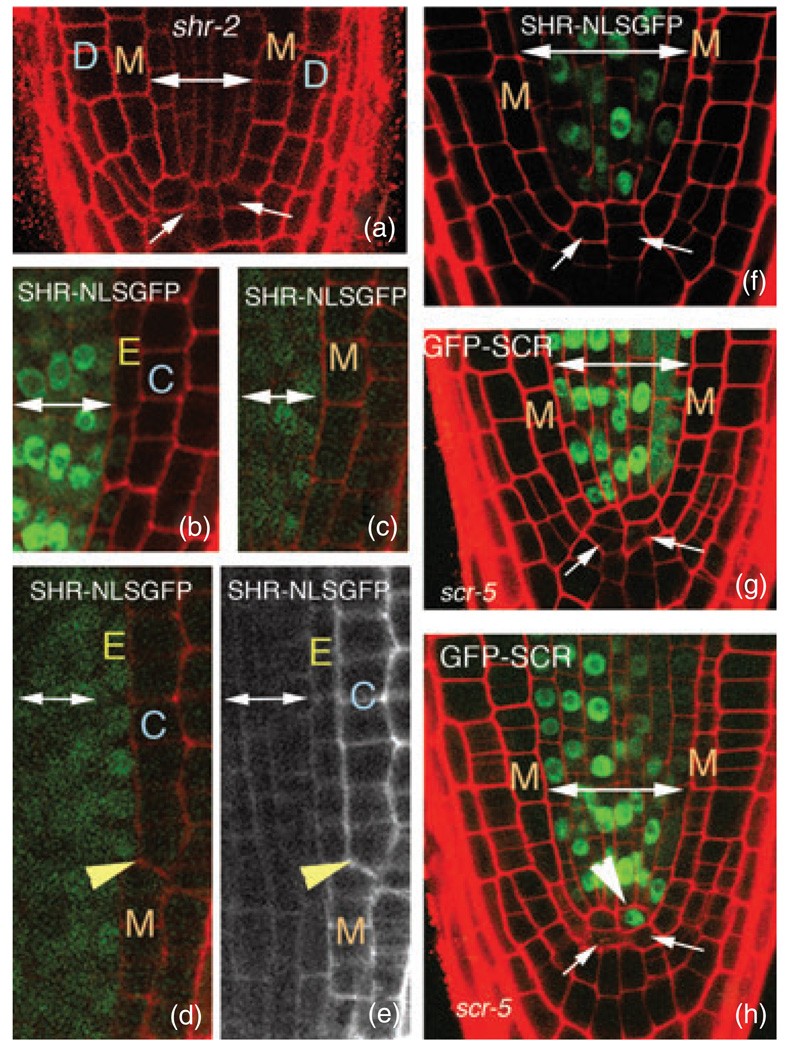
Figure 2. Cell-autonomous SHR does not rescue shr-2.
(a) Longitudinal confocal section through the tip of a shr-2 root. Arrows indicate abnormal QC morphology.
(b) Longitudinal confocal section through the tip of a wild-type root expressing SHRpro–SHR–NLSGFP.
(c–e) Longitudinal confocal micrographs of shr-2 plants expressing SHRpro–SHR–NLSGFP. (c) The cell-autonomous SHR–NLSGFP protein is unable to rescue either the radial patterning or QC morphology. However, in (d), rescue of radial patterning is coincident with movement of the SHR–NLSGFP protein into the ground tissue. The yellow arrowheads in (d) and (e) indicate the wall between the endodermis and cortex. Notice that above the yellow arrowhead there are two distinct ground-tissue layers, and SHR–NLSGFP is clearly detected in the endodermal layer. (e) The red-channel image of the root in (d) is shown in gray scale to allow easier visualization of the rescue of the endodermal layer.
(f) shr-2 root expressing SHRpro–SHR–NLSGFP. This root has aberrant radial patterning and QC morphology, but normal root length despite a lack of detectable GFP in the QC cells.
(g,h) scr-5 roots expressing SHRpro–GFP–SCR. Expression of SCR in the stele does not rescue the scr-5 radial patterning defects, but these roots do have normal lengths. In (h), there is an indication of divisions at the base of the stele (arrowhead) above the abnormal QC (arrows) forming a population of cells that morphologically resemble a new QC.
Double-headed arrows delineate stele cells throughout. M, mutant ground-tissue layer; D, epidermis; E, endodermis; C, cortex.
Previously we have shown that efficient nuclear localization of SHR by addition of a canonical nuclear localization signal (NLS) (SHR–NLSGFP) inhibited movement into the endodermis when expressed in the stele of wild-type roots (Figure 2b), but did not affect the activity of SHR when expressed directly in the endodermis (Gallagher et al., 2004). We used SHR–NLSGFP to test whether movement of SHR is required to rescue the phenotype of shr-2. shr-2 plants have only one ground-tissue layer between the pericycle and epidermal cell layers, and have a disorganized QC (Figure 2a). The QC cells are thought to serve as a signaling center that maintains the stem-cell population (initial cells) in the root meristem (Helariutta et al., 2000; Sabatini et al., 1999, 2003). Loss of the QC population results in a shortened root due to exhaustion of the initial cells. Thus SHR is required both for correct radial patterning and maintenance of root growth.
The SHR–NLSGFP construct expressed from the SHR promoter (SHRpro) was transformed into plants heterozygous for the shr-2 mutation, and T2 generation seedlings with the shr-2 phenotype were identified. In approximately 70% of the shr-2 seedlings with SHR-NLSGFP expression, fluorescence due to GFP was restricted to the stele, and the radial pattern resembled that of shr-2, with one ground-tissue layer and a disorganized QC (Figure 2c–e). Approximately 30% of seedlings with short roots showed sporadic rescue of the ground tissue (Figure 2d,e). In all of these plants, SHR–GFP was detectable in the inner layer of ground tissue, indicating that the protein had moved. We did not observe rescue of the radial pattern without the presence of SHR in the inner ground-tissue layer. These results show that addition of an exogenous nuclear localization signal does not entirely abrogate movement in the shr-2 background. More importantly, they indicate that movement of SHR out of the stele is consistent with normal radial patterning of the root.
In addition to being required for proper radial patterning, both SHR and SCR are required to prevent differentiation of the initial cells and maintain root growth, presumably due to their role in preventing loss of the QC (Sabatini et al., 2003). In all of the T2 lines expressing SHRpro–SHR–NLSGFP and segregating for shr-2, there was an increase in the expected ratio of long to short roots (from 3:1 to approximately 10:1), indicating that the SHRpro–SHR–NLSGFP transgene was often able to rescue shr-2 root length. We examined seedlings with long roots and found two classes: those with aberrant root radial patterning, which were genotyped as shr-2 homozygotes, and wild-type plants. In shr-2 plants with a normal root length, the SHR–NLSGFP protein was often not detected in the QC cells (Figure 2f). These results suggest that expression of nuclear-localized SHR in the stele is sufficient to maintain continued divisions of the root initials in a shr-2 background.
Like shr-2 plants, scr plants also have aberrant QC cells and fail to maintain continued division of the root initial cells. As SCR is also required for maintenance of the QC, we tested whether expression of GFP–SCR in the stele could rescue scr root length, by expressing it from the SHR promoter in scr-5 (a null allele) plants (Paquette and Benfey, 2005). Analogous to the results with SHR, expression of GFP–SCR in the stele is able to rescue scr-5 root length without detection of GFP in the QC cells (Figure 2g,h).
The above results suggest that SHR movement from the stele is required for correct radial patterning of the Arabidopsis root, as expression of largely cell-autonomous SHR in the stele tissue is not sufficient for rescue of the division that produces the separate cortex and endodermal cell layers. However, rescue of root length may not require movement of SHR. Instead, it appears that expression of cell-autonomous SHR or SCR in the stele is able to rescue the root length defect in shr-2 or scr-5 plants respectively.
Identification of domains within SHR required for movement
We previously showed that threonine at position 289 (T289) is required for proper SHR localization and movement, and that, independent of localization and movement, T289 is also required for SHR’s ability to function (Gallagher et al., 2004). Here we determined whether T289 is the critical residue for SHR movement, or whether other regions of the SHR protein are also required to promote cell-to-cell transport. In addition, we determined whether there is complete overlap between the sequences that are required for SHR movement and SHR activity.
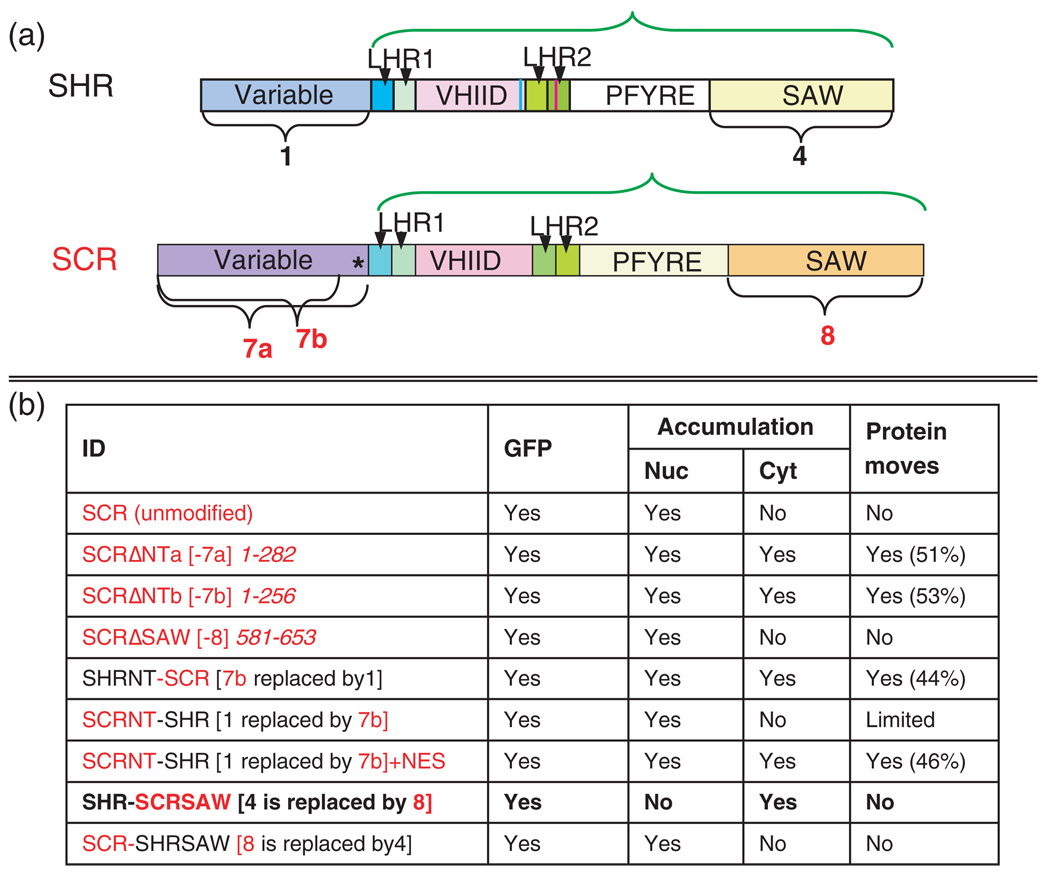
The SHR protein can be divided into eight distinct domains based upon sequence homology with the other GRAS proteins (Figure 3a) (Pysh et al., 1999). The functional significance of these regions of the GRAS domain is not known except for SLENDER RICE, in which the first set of leucine heptad repeats has been shown to be required for homodimerization (Itoh et al., 2002). To determine the role of these sequences in SHR movement, we produced deletions within the SHR protein corresponding to the domain structure and then assayed the effects of these mutations on movement and activity in a wild-type Columbia background. Expression of the mutated proteins as GFP fusions from the SHR promoter allowed us to assess the ability of the protein to properly localize and move. To determine whether the mutated proteins can function independently of movement, we expressed them directly in the endodermis, using the SCR promoter (SCRpro) (Gallagher et al., 2004). Expression of SHR from SCRpro in wild-type plants results in an increase in the number of ground-tissue layers (Gallagher et al., 2004; Nakajima et al., 2001). Mutated SHR proteins were considered to retain SHR activity if they caused an increase in the number of ground-tissue layers when expressed from SCRpro.
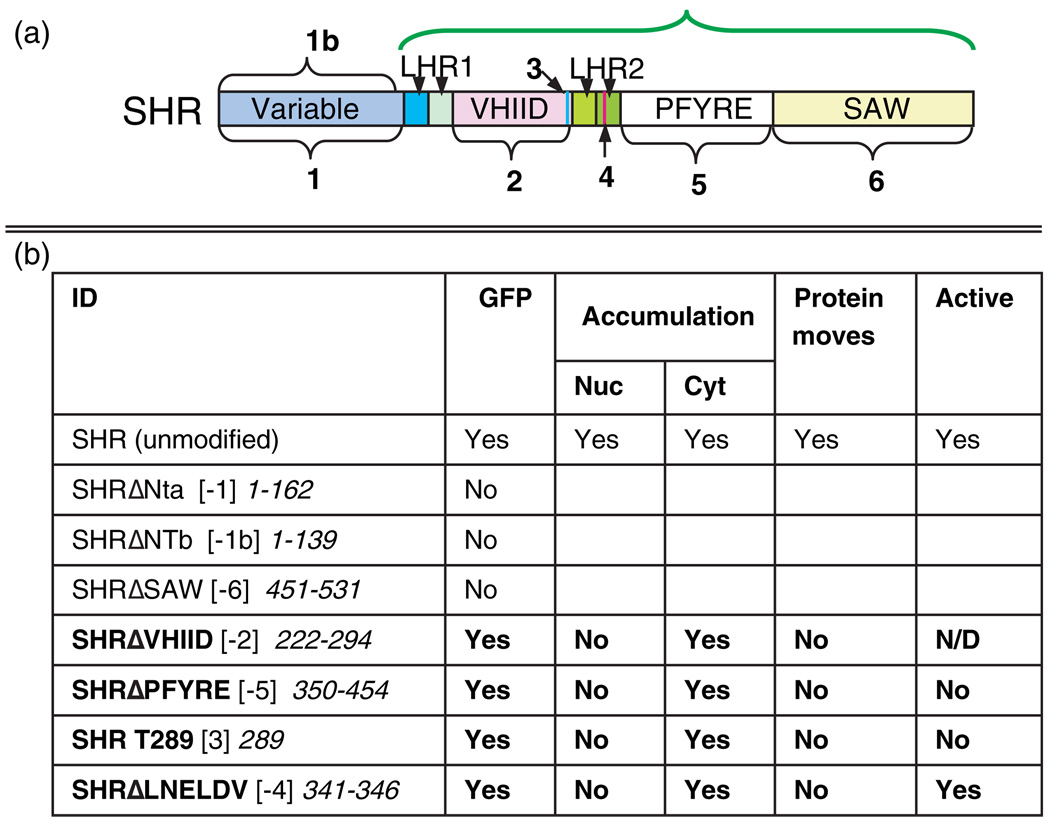
Figure 3. SHR deletion constructs.
(a) Diagram of the domains deleted in SHR for analysis of SHR movement and activity. Arrow 3 indicates the single conserved threonine (T289) in the VHIID domain, which we previously showed was required for movement. Arrow 4 indicates the LNELDV motif. The green bracket shows the region designated as the SHR GRAS domain.
(b) Summary of the results of the deletion analysis. The bracketed numbers in the ID column refer to the numbered domains in (a). The numbers in italics refer to the amino acids deleted or mutated in each construct. Entries in bold show disruptions in both nuclear accumulation and movement.
The results of the structure/function analysis are summarized in Figure 3b. In general, the deletion constructs fell into three distinct classes: (1) those that were undetectable and thus required other approaches to test their function, (2) those that lost the ability to move and function, and (3) one that did not move, but retained activity.
Class 1: The SHR N-terminus and SAW domain appear to be required for protein stability
Two different N-terminal (NT) deletions of SHR were produced (Figure 3). No GFP was detected in any of the lines expressing either construct (n > 10). This was also the case for deletion of the SAW domain (n > 10). These results suggest that the N-terminus and SAW domains are required for protein stability (Waldo et al., 1999), and that other methods are required to assay their function.
Class 2: Deletion of the VHIID or PFYRE domains disrupts movement and activity
Deletion of either the VHIID domain (Figure 4) or the PFYRE domain affected subcellular localization and disrupted movement from the stele, with neither protein detected in the endodermis. As we had previously demonstrated that T289, within the VHIID domain, is required for SHR activity (Gallagher et al., 2004), we did not express the SHRDVHIID construct from SCRpro to test for activity, as deletion of the VHIID domain would eliminate this critical residue. Expression of the SHRΔPFYRE protein from the SCR promoter did not cause an increase in the number of ground-tissue layers (data not shown, Figure 5a–d). These results indicate that both the VHIID and PFYRE domains are necessary for movement and activity of the SHR protein. It is noteworthy that nuclear localization was reduced by these mutations.
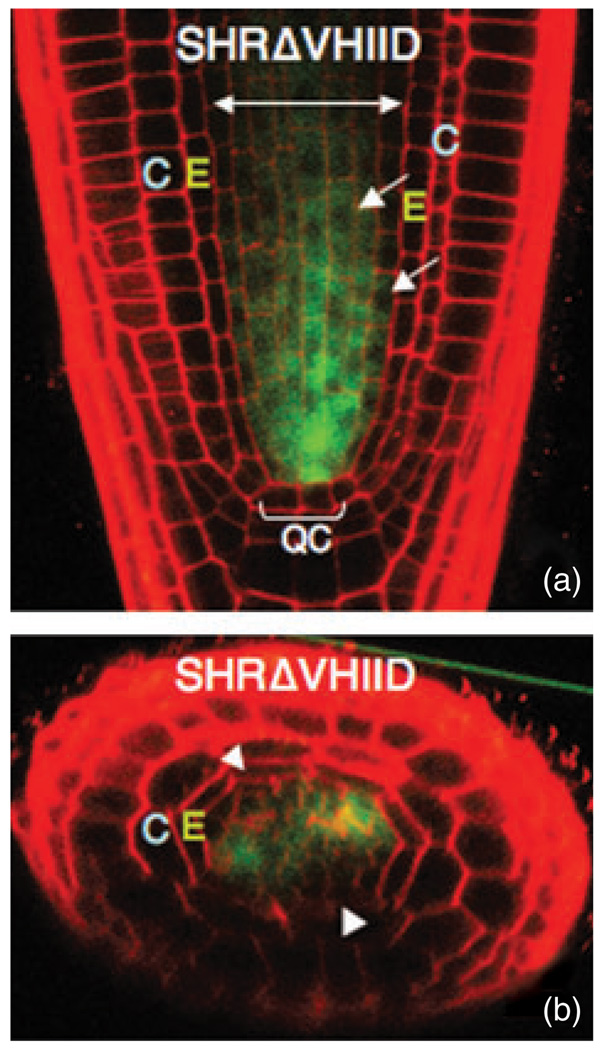
Figure 4. The SHR VHIID domain is required for SHR movement.
(a) Longitudinal and (b) transverse confocal sections through a wild-type root expressing SHRpro–SHRΔVHIID–GFP. The mutant protein is not excluded but does not accumulate in the nuclei of stele cells (arrows). No protein is detected in the endodermis. Arrowheads in (b) indicate the endodermal cells. E, endodermis; C, cortex; QC, quiescent center.
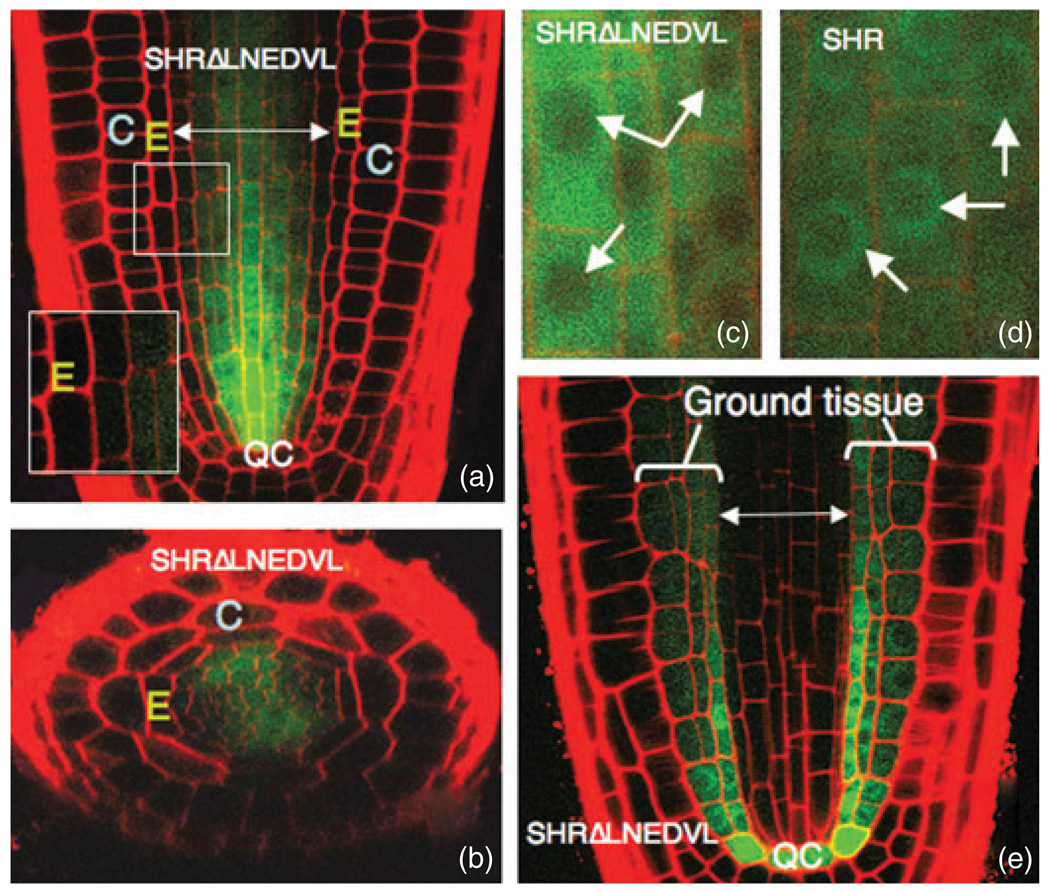
Figure 5. Mutation of the LNELDV residues within LHR2 (arrow 4 in Figure 3a) results in loss of SHR movement, but not SHR function.
(a) Longitudinal and (b) transverse confocal sections through a wild-type root expressing SHRpro–SHRΔLNELDV–GFP. No protein is detected in the endodermis.
(c,d) Compared to wild-type SHR in (d), the mutant SHRΔLNELDV protein in (c) does not accumulate in the nuclei (indicated by arrows) of stele cells.
(e) When expressed directly in the endodermis from SCRpro, the SHRΔLNELDV protein increases the number of ground-tissue layers in a manner consistent with wild-type SHR function.
E, endodermis; C, cortex; QC, quiescent center.
Class 3: Mutation of the leucine heptad repeat II disrupts movement, but not activity
To assess the function of the second leucine heptad repeat in SHR movement, we disrupted the heptad repeat structure, substituting an LNELDV motif with a polyalanine (AAA) stretch. In the stele, there was a reduction in nuclear accumulation compared to wild-type SHR, with SHRΔLNELDV distributed throughout the cells (compare Figure 5c,d). The mutant protein was not detected in the endodermis, quiescent center cells or the root initials.
To test for activity of the SHRΔLNELDV protein, we expressed it directly in the ground tissue from SCRpro (Figure 5e). In this tissue, the protein localized to both the cytoplasm and the nucleus. We observed a multi-layered ground-tissue phenotype indistinguishable from that of wild-type SHR expression from SCRpro. Thus, in contrast to the other mutations that affected SHR movement, SHRΔLNELDV is a functional SHR protein. These results suggest that the LNELDV motif is required for SHR movement, but not its activity.
The SCR GRAS domain has the capacity to move
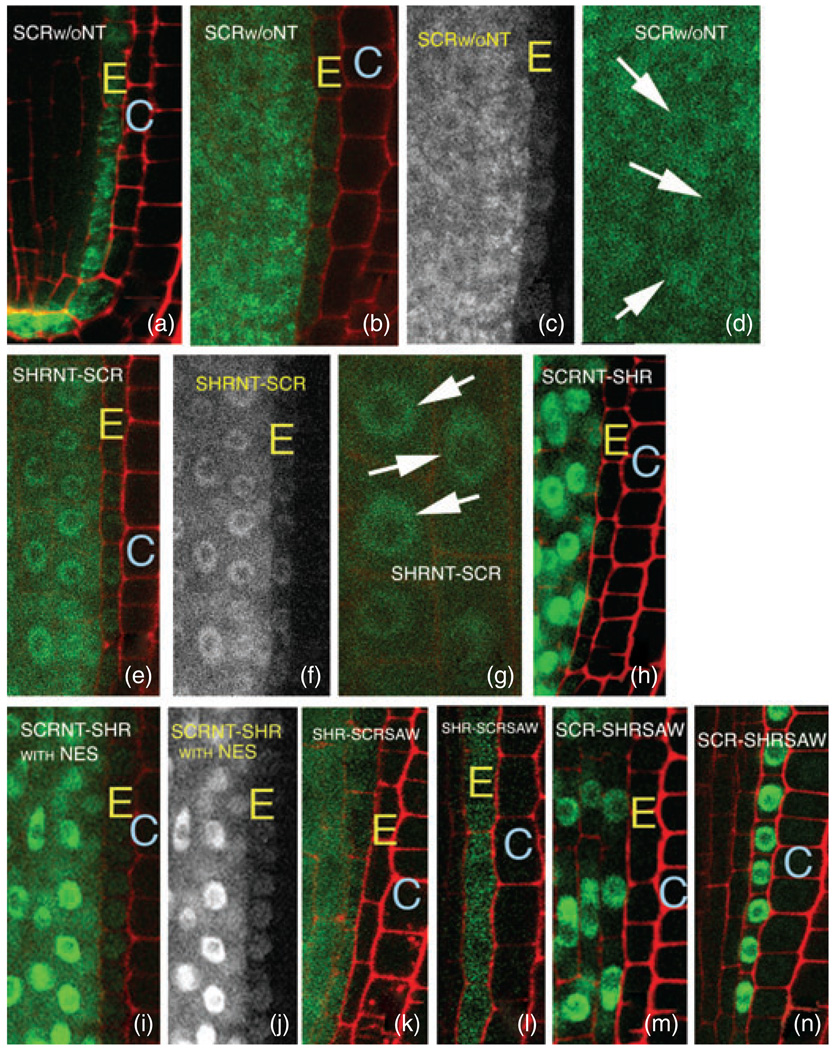
As multiple regions within the conserved GRAS domain are required for trafficking of SHR, we hypothesized that the GRAS domains from other family members may also have the capacity for cell-to-cell movement. We tested this hypothesis using the SCR protein. The SCR protein is normally expressed in the endodermis, with low-level expression in the cortex. In these tissues, the SCR protein is nuclear-localized and cell-autonomous. When expressed in the stele, SCR–GFP is also nuclear-localized, and does not move into the endodermis, quiescent center cells or root initials (Gallagher et al., 2004). To test whether the SCR GRAS domain alone (i.e. SCR without the N-terminus) has the capacity for movement, we expressed it as a GFP fusion from the SCR and SHR promoters. Neither SCR GRAS protein (SCRΔNTa or SCRΔNTb; see Figure 6) showed exclusive nuclear localization in the endodermis or the stele (Figure 7a–d) as is seen for wild-type SCR, indicating that the SCR N-terminus confers proper subcellular localization to the SCR GRAS domain. Localization is independent of the predicted NLS, as the SCRΔNTa construct lacks this region. The SCR GRAS protein was found throughout stele and endodermal cells, with some indication of weak protein aggregation. Significantly, the SCR GRAS domain was able to move from the stele into the endodermis, quiescent center cells and the cortical endodermal root initials, indicating that the SCR GRAS domain has the capacity to move (Figure 7b,c).
Figure 6. Key to the domain swaps.
(a) Diagrams of SHR and SCR proteins indicating relevant domains. In both, the GRAS domains are indicated by a green bracket and include the entire protein minus the N-terminus. The asterisk in SCR represents the predicted NLS.
(b) Summary of the results of the SCR deletions and domain swaps between SHR and SCR. The bracketed ID numbers refer to the numbered domains in (a). The numbers in italics refer to the amino acids deleted or mutated in each construct. SCR is indicated in red and SHR in black. The entry in bold shows disruption in both nuclear accumulation and movement. The percentages listed in the column ‘Protein moves’ are relative to SHR–GFP movement in the wild-type background in the root meristem.
Figure 7. The SHR and SCR GRAS domains are necessary for movement.
(a–d) Expression of the SCR GRAS domain in the endodermis (a) and the stele (b–d). Loss of the SCR N-terminus independent of the predicted NLS results in localization of the SCR GRAS protein throughout the cell and movement of the protein from the stele into the endodermis when expressed from SHRpro (as in b–d). (c) GFP channel only from (b) to facilitate visualization of the GFP signal in the endodermis. (d) Higher-resolution image of localization of the SCR GRAS protein in stele cells. Arrows indicate nuclei.
(e–g) Replacement of the SCR N-terminus with the SHR N-terminus (designated 7b/1 in Figure 6) results in both nuclear and cytoplasmic localization in the stele without increasing the efficiency of movement of the SCR GRAS domain [see (b) for comparison]. (f) GFP channel only from (e) to facilitate visualization of the GFP signal in the endodermis. (g) Higher-resolution image of localization of the SHRNT–SCR protein in stele cells. Arrows indicate nuclei.
(h–j) Exchange of the SHR N-terminus with the SCR N-terminus (designated 1/7b in Figure 6) independent of the predicted NLS leads to nuclear localization of the chimeric protein (h). Addition of an NES (i,j) allows movement of the SCR–SHR chimera. (j) GFP channel only from (i) to facilitate visualization of the GFP signal in the endodermis.
(k,l) Expression of the SHR–SCRSAW protein (designated 8/4 in Figure 6) from the SHR (k) and SCR promoters (l). Replacement of the SHR SAW domain with the SCR SAW domain results in mislocalization of SHR in the stele (k) and the endodermis (l), and loss of movement from the stele (k).
(m,n) Replacement of the SCR SAW domain with the SHR SAW domain (designated 4/8 in Figure 6) does not affect subcellular localization in either the stele (m) or the endodermis (n), indicating that the SHR SAW domain does not represent an autonomous movement domain.
E, endodermis; C, cortex. For all images, expression in the stele is achieved using the SHR promoter and expression in the endodermis is achieved using the SCR promoter.
To quantify movement of the SCR GRAS domain, we measured GFP intensities in the stele and endodermis. The ratio of endodermal fluorescence to stele fluorescence was used as an indicator of the efficiency of transport from the stele into the endodermis (Figure 6b). Using such calculations, we found that movement of the SCR GRAS domain was approximately 50% of that observed for SHR–GFP. This suggests that the SCR GRAS domain is not as efficient in promoting movement as SHR, perhaps due to sequences within the SHR N-terminus that promote movement. In addition, the fact that SCR can move when the SCR N-terminus is removed suggests that the SCR N-terminus inhibits movement. To test these hypotheses, we performed domain swaps between the SHR and SCR N-termini.
The N-terminus of SHR was fused to the C-terminus of SCR (the SCR GRAS domain) to produce SHRNT–SCR–GFP (see Figure 6). This construct was then expressed from the SHR promoter (Figure 7e–g) or the SCR promoter (data not shown). In the stele, the SHRNT–SCR–GFP protein localized similarly to SHR, with GFP detected in both the nucleus and cytoplasm of individual stele cells (Figure 7e–g), suggesting that the SHR N-terminus is, at least in part, responsible for SHR subcellular localization. There was no evidence, however, that the SHR N-terminus increased the ability of the SCR GRAS domain to move. In fact, movement of the SHRNT–SCR–GFP construct was 80% of that of the SCR GRAS domain alone, indicating that, if anything, the SHR N-terminus reduces the efficiency with which the SCR GRAS domain moves.
To test the ability of the SCR N-terminus to inhibit movement, we performed the reciprocal swap (SCRNT–SHR), removing the N-terminus of SHR and replacing it with the N-terminus of SCR, which does not include the predicted NLS. The chimeric protein was fused to GFP and expressed from the SHR and SCR promoters. In both the stele (Figure 7h) and endodermis (data not shown), the SCRNT–SHR protein was predominantly nuclear-localized. When expressed from SHRpro, we rarely detected the chimeric SCRNT–SHR protein in the endodermis, indicating very limited movement of the nuclear-localized SCRNT–SHR protein. However, addition of a nuclear export signal (NES) allowed the SCRNT–SHR protein to accumulate in the cytoplasm of stele cells, and, more importantly, to move into the endodermis (Figure 7i,j). Movement of the SCRNT–SHR chimera with the NES was approximately 50% of that of wild-type SHR. These results suggest that the role of the SHR N-terminus in potentiating SHR movement may derive primarily from its ability to stabilize the SHR GRAS domain and provide nuclear export activity. For SCR, the N-terminus also controls subcellular localization, which is likely to play a role in inhibiting movement of the intact SCR protein.
The SHR SAW domain is required for proper localization and movement
As removal of the SAW domain from SHR–GFP appeared to result in an unstable protein, swaps with the SCR SAW domain were performed to assess the role of this region in SHR movement. Removal of the SCR SAW domain has no affect on SCR localization and does not allow SCR to move (data not shown). The SHR SAW domain was removed and replaced with the equivalent region from SCR to produce SHR–SCRSAW–GFP (Figure 7k,l). Localization of the chimeric protein was similar to the localization observed for the class 2 and class 3 mutations. In addition, there was no indication that the protein moved from the stele into the endodermis (Figure 7k), suggesting that the SHR SAW domain is required for SHR movement, and that the SCR domain cannot substitute for the SHR domain in this context.
For the reciprocal swap, the SCR SAW domain was replaced with the equivalent domain from SHR (SCR–SHRSAW–GFP) (Figure 7m,n). No effect on SCR localization was seen when expressed in either the stele (Figure 7m) or the endodermis (Figure 7n). The protein was detected only in nuclei, and there was no movement into neighboring cells.
Collectively, these results indicate that the SHR SAW domain is required for stability of the SHR protein, and, independent of stability, for nuclear accumulation and movement. The SHR SAW domain is not, however, sufficient for movement, as it did not confer movement to SCR.
Both cytoplasmic and nuclear localization are required for SHR movement
Previous results have shown that cytoplasmic localization of SHR is required for cell-to-cell movement. The results of the structure/function analysis, however, show a positive correlation between nuclear accumulation and the ability of SHR to move between cells. All mutations that significantly reduced nuclear accumulation also prevented cell-to-cell transport (indicated in bold in Figure 3 and Figure 6). However, for modifications that resulted in a decrease in the cytoplasmic localization of SHR, addition of an NES promoted movement. Together, these results suggest that both nuclear and cytoplasmic localization are required for cell-to-cell transport of SHR. To further explore the relationship between subcellular localization and SHR movement, we attempted to rescue movement of one of the class 2 mutations by restoring nuclear localization.
Increasing nuclear localization can enhance movement
If nuclear localization is mechanistically required for SHR movement, then at least some of the mutations that disrupted both nuclear localization and movement of SHR might be primary defects in nuclear localization that indirectly affect protein movement. For example, conversion of T289 to an isoleucine (I) resulted in reduction of nuclear accumulation of SHRand a lack ofSHRmovement (Gallagher et al., 2004). These results suggest that T289 is independently required for both nuclear localization and movement of SHR. However, the alternative is that T289 is necessary for nuclear localization, which is in turn necessary for cell-to-cell trafficking. If the latter scenario were correct, then one would expect that restoration of nuclear localization could (at least partially) restore movement. We tested this by adding NLSGFP to the mutated SHRTΔI protein (T289 mutation) and expressing the resulting SHRTΔI–NLSGFP construct from the SHR promoter in wild-type plants (Figure 8a,b). In these lines (SHRpro–SHRTΔI–NLSGFP), the modified protein was found in both the nucleus and cytoplasm of stele cells, and we consistently detected SHRTΔI–NLSGFP in the endodermis, indicating that protein movement was restored. These results indicate that T289 is not specifically required for SHR movement, and that instead this residue is required for nuclear accumulation, which is in turn required for SHR movement. These data indicate a mechanistic link between nuclear localization of SHR and movement.
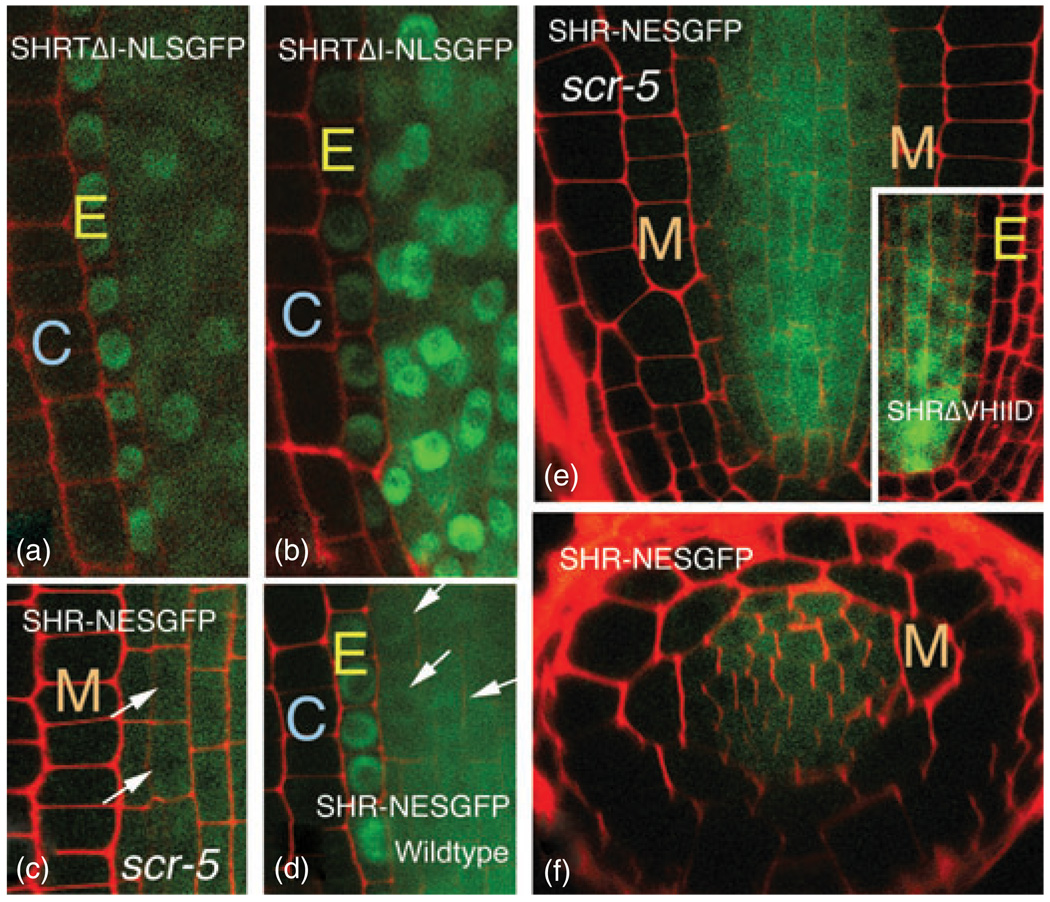
Figure 8. Nuclear accumulation correlates with the ability to move.
(a,b) Movement of the T289 mutation is rescued by addition of an NLSGFP construct. Two different roots expressing the SHRpro–SHRΔT289–NLSGFP construct are shown. GFP is detected in the endodermis in both roots.
(c–f) Expression of SHRpro–SHR–NESGFP in scr-5 resulted in SHR localization similar to that for SHRpro–SHRΔVHIID [for comparison, see inset in (e)] and a reduction in SHR movement from the stele into the endodermis. The effect of the NES on SHR localization is more pronounced in scr-5 (c,e,f) than in wild-type plants (d).
M, mutant ground-tissue layer; E, endodermis; C, cortex. Arrows indicate nuclei.
Decreasing nuclear localization can inhibit SHR movement
If nuclear accumulation is a prerequisite for cell-to-cell movement of SHR, then reducing that ability of SHR to localize to the nucleus should inhibit movement. It was previously shown that SCR is required for nuclear accumulation of SHR both in the endodermis and when ectopically expressed in the epidermis (Sena et al., 2004). In an effort to dramatically reduce nuclear accumulation of SHR in stele cells (essentially phenocopy the class 2 or 3 mutations), we expressed SHR fused to NESGFP (SHRpro–SHR–NESGFP) in the stele in the scr-5 null background (Paquette and Benfey, 2005) (Figure 8c,e,f). In these plants, there was a reduction in nuclear accumulation of SHR–NESGFP in both stele and in the mutant layer of cells compared to unmodified SHR or SHR–NESGFP in the wild-type background (Figure 8d). As predicted if nuclear accumulation is required for cell-to-cell transport, there was a strong reduction in the amount of SHR–NESGFP detected in the mutant layer (Figure 8e,f), indicating a reduction in movement. These results provide further evidence that nuclear accumulation is necessary for optimal SHR movement, and that the nuclear localization signal in SHR does not directly serve as a movement sequence, but that the sequences required for nuclear localization promote movement as a consequence of nuclear accumulation of the SHR protein. These results also extend the results of Sena et al. (2004) who showed a non-cell-autonomous role for SCR (that is not dependent upon SCR movement) in promoting nuclear accumulation (or inhibiting nuclear export) of SHR in epidermal tissues.
Discussion
Recent evidence indicates that there are many transcription factors that traffic between plant cells. Estimates by Lee et al. (2006) suggest that as many as 17% of transcription factors move in the root. To date, very little is known about the developmental significance of movement, or the sequences and mechanisms controlling cell-to-cell trafficking. Using SHR as a model for transcription factor movement, we have shown that SHR movement from the stele is essential for radial patterning of the Arabidopsis root. Through a structure/function analysis, we have identified a strong correlation between SHR nuclear localization and movement, and show that the GRAS domain is the important region for intercellular trafficking. Further, we provide evidence that SHR movement is controlled in part by nuclear/cytoplasmic partitioning.
SHR movement is required for normal root development
Many groups have shown that cell-to-cell movement of transcription factors, as well as other macromolecules, is developmentally regulated (Crawford and Zambryski, 2001; Imlau et al., 1999; Kim et al., 2003; Kurata et al., 2005; Sena et al., 2004; Sessions et al., 2000; Wu et al., 2003; Zambryski, 2004). However, conclusive evidence that movement is required for normal development has been lacking. Our data indicate that SHR movement from the stele is necessary for normal patterning of the Arabidopsis root. In shr-2 roots, in which SHR–NLSGFP was expressed and only detected in the stele, the ground tissue failed to divide and the QC was often disorganized. Rescue of shr-2 radial patterning correlated with the cell-to-cell movement of SHR–NLSGFP, indicating that movement of SHR is developmentally relevant.
Surprisingly, we observed rescue of root length in shr-2 plants with abnormal patterning of the ground tissue and no detectable GFP signal in the presumptive QC. These results raise the possibility that SHR movement is not required to maintain the root meristem. Based upon the results of Sabatini et al. (2003), we had anticipated that SHR would be required in the QC for maintenance of the stem-cell niche. However, our results suggest that expression of nuclear-localized SHR or SCR in the stele is sufficient to maintain continued divisions of the root initials in shr-2 and scr-5 mutant backgrounds, respectively. SCR expression in the stele correlates with ectopic divisions above the QC in the stele cells, perhaps resulting in reorganization of the QC signaling center to a more basal region of the root. This is not unlike what was found when the QC was laser-ablated (Xu et al., 2006). Initially, SHR–GFP became nuclear-localized and SCR was expressed in the provascular cells immediately above the ablated QC, then a new QC signaling center was organized in what was the apical region of the stele (Sabatini et al., 1999; Xu et al., 2006).
An alternative explanation for the above results is that there was low-level movement of SHR or SCR into the QC cells that was below the level of detection. As addition of the NLS to SHR–GFP did not completely abrogate movement in shr-2, and as our data indicate that the SCR GRAS domain does have the capacity to move, it is possible that the GFP-tagged proteins moved into the QC cells of the rescued roots. If this is the case, then the levels of SHR and SCR required for QC maintenance are likely to be lower than those needed for radial patterning. In support of this second hypothesis is the finding by Mace et al. (2006) that low levels of GFP expression are difficult to detect.
Previously, Wu et al. (2003) presented data suggesting that movement of LFY, a transcription factor required for the transition from vegetative to floral development, is required for LFY function. Movement-compromised GLFY (LFY with a GFP insertion at amino acid 31) does not completely rescue the lfy-11 phenotype. This is in contrast to constructs with GFP at either the N- or C-terminus, which are movement-competent, and rescue lfy-11. While these results strongly suggest that LFY movement is required for its function, the possibility cannot be excluded that GLFY is compromised in its ability to fully function as a transcription factor independently of movement. If movement of GLFY were restored, and GLFY was shown to rescue the lfy-11 phenotype, this would provide conclusive evidence that LFY movement is required for function.
A significant distinction between LFY and SHR movement is that LFY is expressed throughout the entire region in which the protein moves. Thus cell-to-cell trafficking of LFY does not increase its effective domain, whereas SHR movement allows SHR to function in cells in which it is not normally expressed. The LFY results suggest that movement per se is required for LFY function. This is not the case for SHR, for which we have no evidence that movement in and of itself is required for function (Gallagher et al., 2004; Levesque et al., 2006).
Multiple regions within the SHR GRAS domain are necessary for movement
Our data indicate that multiple regions within the GRAS domain are required for nuclear accumulation and cell-to-cell movement of SHR. Experiments in which the N-termini of SHR and SCR were swapped (or deleted in the case of SCR) suggest that the conserved GRAS domain, which defines the family, is the important region for cell-to-cell transport. This is analogous to what has been shown for the KN1 and ENGRAILED2 (EN2) proteins (Tassetto et al., 2005), of which the conserved homeodomain has the capacity to move. It has been suggested that intercellular transfer may be an evolutionarily conserved feature of many DNA-binding homeoproteins (Tassetto et al., 2005). Sequences outside the conserved homeodomain either promote or inhibit movement. Likewise, for SCR, sequences outside the GRAS domain probably regulate movement. Further analysis of the GRAS domains from additional GRAS proteins will allow us to determine how widespread the potential for cell-to-cell movement is for this family.
There is not complete overlap between domains required for movement and activity
Previously we identified a single amino acid in the VHIID domain as being necessary for SHR movement and activity (Gallagher et al., 2004). In the present study, most of the mutations that inhibited movement also disrupted activity. Significantly, the LNELDV motif in LHR2 is not required for SHR activity but is required for movement, indicating that there is not complete overlap in the sequences that are required for movement and activity. This is in contrast to KN1 and CPC, for which all of the sequences that have been identified to date as being involved in movement (when examined) are also required for activity (Kim et al., 2005b; Kurata et al., 2005; Lucas et al., 1995). The overlap between movement and activity has made determination of the mechanism of cell-to-cell trafficking problematic. Loss of both movement and activity could be attributed to improper protein folding rather than a specific movement sequence. However, for some of these proteins, such as KN1, the capacity for specific sequences to confer movement has been directly demonstrated (Kim et al., 2005b; Tassetto et al., 2005). This has not been possible for SHR (Gallagher et al., 2004). However, the ability to separate movement and activity in SHR allows us to confidently conclude that specific sequences within SHR are required for movement, and that loss of movement is not a consequence of improper folding.
Correlation between nuclear localization and movement
An interesting similarity between CPC, KN1 and SHR movement is that all mutations that diminish nuclear localization also reduce movement (Gallagher et al., 2004; Kim et al., 2005b; Kurata et al., 2005; Lucas et al., 1995; Prochiantz and Joliot, 2003; Tassetto et al., 2005). Mutation of the Myb domain in CPC inhibited both nuclear localization and movement (Kurata et al., 2005). Mutation of the predicted NLS in KN1 blocked movement (Lucas et al., 1995; Prochiantz and Joliot, 2003; Tassetto et al., 2005). Likewise, for SHR, all of the mutations within the GRAS domain that diminished nuclear accumulation also prevented movement. These results suggest that either there is broad overlap between the sequences required for nuclear localization and those required for movement, or that nuclear localization mechanistically promotes movement. The latter hypothesis is supported by the results showing that reduction of nuclear localization of SHR–NESGFP in scr-5 decreased movement, and, more importantly, that movement of the SHRT289I mutant could be rescued by restoration of nuclear localization.
Interestingly, for the KN1 homeodomain, the correlation between nuclear localization and movement has been extended to transport between animal cells. Tassetto et al. (2005) showed that the KN1 homeodomain can traffic between animal cells in culture. The mutation within the NLS that blocked trafficking in planta also inhibited nuclear localization in and transport between animal cells. Movement of the mutant KN1 homeodomain could be restored by addition of a heterologous NLS.
Both nuclear and cytoplasmic localization are required for SHR movement. The finding that promoting either nuclear or cytoplasmic localization at the expense of the other inhibits movement suggests that a balance between nuclear import and export must be maintained for continued movement. The movement of many endogenous animal proteins that traffic by non-conventional secretion, including thioredoxin, galectin, HMGB1 and homeodomain proteins is also correlated with nuclear localization (Prochiantz and Joliot, 2003; Tassetto et al., 2005). Secretion of the EN2 homeodomain protein requires both nuclear import and nuclear export. The significance of subcellular localization for EN2 protein movement is not known. However, Prochiantz and Joliot (2003) have speculated that specific modifications occur in the nucleus, which confer competence to move. They have also suggested that nuclear localization could promote intercellular trafficking by bringing the cargo proteins into proximity with factors that facilitate movement or by allowing access to the secretory compartment. These are possible explanations for the correlation that we see between SHR subcellular localization and movement.
While these data collectively suggest parallels between SHR movement and secretion of homeodomain proteins (such as KN1 and EN2), we have no evidence to suggest that SHR moves through an unconventional secretory pathway. Similarities between transport via plasmodesmata and nuclear–cytoplasmic transport have long been suggested (Lee et al., 2000; Lucas, 1995; Lucas et al., 1995), as has a possible mechanistic link between nuclear localization and movement (Oparka, 2004). This is not to suggest that the NLS equates to a plasmodesmata-targeting signal, as nuclear-localized proteins clearly do not traffic between cells and we were able to diminish SHR movement by reducing nuclear localization without disrupting the nuclear localization sequences. However, the correlation between nuclear localization and cell-to-cell movement may provide clues as to the proteins and processes involved in intercellular transport of non-cell-autonomous proteins in planta, and perhaps movement through plasmodesmata.
Model for SHR movement
Our previous data suggested a model for SHR movement in which cytoplasmically localized SHR protein was selectively trafficked between cells, based upon sequences within the SHR protein. These data further suggested that cytoplasmic localization of SHR was necessary but not sufficient for SHR movement (Gallagher et al., 2004). Our current data, together with recent findings by Cui et al. (2007), allow us to expand upon this model to reflect a requirement for nuclear accumulation of SHR in cell-to-cell movement. However, it appears that there needs to be a balance between nuclear import and nuclear export, as movement is inhibited by either a strong NLS (nuclear trapping) (Cui et al., 2007; Gallagher et al., 2004) or loss of nuclear accumulation. These results suggest that nuclear accumulation of SHR may in some way confer onto SHR a movement-promoting modification that is then recognized in the cytoplasm. The requirement for multiple domains in the nuclear localization of SHR indicates the likelihood that protein–protein interaction is required for nuclear localization of SHR. In the endodermis, SCR is involved in nuclear accumulation of SHR, and therefore plays a role in regulating SHR movement (Cui et al., 2007). Thus nuclear localization of SHR appears to play both positive and negative roles in SHR trafficking. As we discover the proteins that regulate the subcellular localization of SHR, and perhaps the downstream targets of SCR, we will learn more about the factors that control intercellular protein movement.
Experimental procedures
Construction of transgenes
Standard molecular biology techniques were used for all cloning procedures. The SHR promoter, SCR promoter and NLS–GFP constructs used were as previously described (Gallagher et al., 2004; Nakajima et al., 2001). mGFP5 was used for translational fusions not requiring an additional NLS (Haseloff et al., 1997). All GFP fusions are to the N-terminus of mGFP5. To aid in cloning, a BamHI site was added to the 5′ end of mGFP5 that encodes an aspartate–proline–glycine linker. The NES is identical to that used by Haasen et al. (1999). For all constructs containing SHR, a modified coding sequence was used that silently eliminates the internal SpeI and SacI sites. In addition, an SpeI site was created just upstream of the ATG, and the stop codon was replaced in-frame with a BamHI site (Nakajima et al., 2001). The junctions of the deletion and fusion constructs are as shown in Figure 3 and Figure 6. Specific information about the primer sequences used is available upon request.
Plant materials, transformation and growth conditions
The shr and scr lines used have been described previously (Nakajima et al., 2001; Paquette and Benfey, 2005; Sena et al., 2004). All lines are in the Columbia background. Plants used for confocal imaging were grown vertically in Percival growth chambers (http://www.percival-scientific.com) on MS medium containing 1.0% sucrose and 1.0% granulated agar (BBL, http://www.bd.com). All plant transformations were performed using standard dipping protocols.
Confocal microscopy
All confocal images were obtained on 4–7-day-old roots using either a Zeiss LSM-510 microscope (http://www.zeiss.com/) or a Leica SP2 microscope (http://www.leica-microsystems.com) equipped with an argon–krypton ion laser with the appropriate filter sets for visualizing GFP and propidium iodide. Prior to visualization, the seedlings were incubated for 0.5–2 min in 0.01 µg ml−1 propidium iodine in H2O. Images were collected using a 60× water immersion lens. Root cross-sections were obtained in line-imaging mode on the Zeiss LSM-510.
Percentage movement was calculated by comparing the ratio of GFP levels in the endodermis to levels in the stele, and then comparing these ratios between modified and unmodified SHR. These calculations were performed using root images with less than 5% saturated pixels in the GFP channel. In some cases, roots with more than 5% saturated pixels in the GFP channel are shown to make visualizing the signal in the endodermis easier. However, these images were not used to calculate the percentage movement.
Acknowledgements
We thank Jose Dinneny, Doris Wagner (Department of Biology, University of Pennsylvania) and Dolf Weijers (Laboratory of Biochemistry, Wageningen University) for helpful comments on the manuscript prior to publication. Myra Hu, Betty Kelley and Heather Belcher provided valuable technical assistance with cloning and plant maintenance. K.L.G. was partially supported by National Institutes of Health post-doctoral fellowship GM065722. This work was supported by a grant to P.N.B. from the National Institutes of Health (RO1GM043778).
References
- Aoki K, Kragler F, Xoconostle-Cazares B, Lucas WJ. A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl Acad. Sci. USA. 2002;99:16342–16347. doi: 10.1073/pnas.252427999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Scheres B. Root development. Curr. Biol. 2000;10:R813–R815. doi: 10.1016/s0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Tikoo A, Kapur AK, Singh M. The taming of the cell penetrating domain of the HIV Tat: myths and realities. J. Control. Release. 2007;117:148–162. doi: 10.1016/j.jconrel.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford KM, Zambryski PC. Non-targeted and targeted protein movement through plasmodesmata in leaves in different developmental and physiological states. Plant Physiol. 2001;125:1802–1812. doi: 10.1104/pp.125.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- Gisel A, Barella S, Hempel FD, Zambryski PC. Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development. 1999;126:1879–1889. doi: 10.1242/dev.126.9.1879. [DOI] [PubMed] [Google Scholar]
- Gunning BES, Overall RL. Plasmodesmata and cell-to-cell transport in plants. Bioscience. 1983;33:260–265. [Google Scholar]
- Haasen D, Kohler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl Acad. Sci. USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Hempel FD, Sha K, Pfluger J, Zambryski PC. Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development. 2002a;129:1261–1272. doi: 10.1242/dev.129.5.1261. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl Acad. Sci. USA. 2002b;99:4103–4108. doi: 10.1073/pnas.052484099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005a;102:2227–2231. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Rim Y, Wang J, Jackson D. A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 2005b;19:788–793. doi: 10.1101/gad.332805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, et al. Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development. 2005;132:5387–5398. doi: 10.1242/dev.02139. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yoo BC, Lucas WJ. Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta. 2000;210:177–187. doi: 10.1007/PL00008124. [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl Acad. Sci. USA. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ. Plasmodesmata: intercellular channels for macromolecular transport in plants. Curr. Opin. Cell Biol. 1995;7:673–680. doi: 10.1016/0955-0674(95)80109-x. [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Mace DL, Lee JY, Twigg RW, Colinas J, Benfey PN, Ohler U. Quantification of transcription factor expression from Arabidopsis images. Bioinformatics. 2006;22:e323–e331. doi: 10.1093/bioinformatics/btl228. [DOI] [PubMed] [Google Scholar]
- Maizel A. Transcription factor movement in plants. J. Soc. Biol. 2006;200:221–227. doi: 10.1051/jbio:2006025. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Oparka KJ. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 2004;9:33–41. doi: 10.1016/j.tplants.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Paquette AJ, Benfey PN. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005;138:636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal MC, Haughn G, Saedler H, Schwarz-Sommer Z. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development. 1996;122:3433–3441. doi: 10.1242/dev.122.11.3433. [DOI] [PubMed] [Google Scholar]
- Prochiantz A, Joliot A. Can transcription factors function as cell-cell signalling molecules? Nat. Rev. Mol. Cell Biol. 2003;4:814–819. doi: 10.1038/nrm1227. [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Rinne PL, van der Schoot C. Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development. 1998;125:1477–1485. doi: 10.1242/dev.125.8.1477. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cazares B, Kragler F. The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 2004;7:641–650. doi: 10.1016/j.pbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D. Cell–cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science. 2000;289:779–782. doi: 10.1126/science.289.5480.779. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ham BK, Xoconostle-Cazares B, Rojas MR, Lucas WJ. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassetto M, Maizel A, Osorio J, Joliot A. Plant and animal homeodomains use convergent mechanisms for intercellular transfer. EMBO Rep. 2005;6:885–890. doi: 10.1038/sj.embor.7400487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- Wu X, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, Weigel D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- Zambryski P. Cell-to-cell transport of proteins and fluorescent tracers via plasmodesmata during plant development. J. Cell Biol. 2004;164:165–168. doi: 10.1083/jcb.200310048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski P, Crawford K. Plasmodesmata: gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 2000;16:393–421. doi: 10.1146/annurev.cellbio.16.1.393. [DOI] [PubMed] [Google Scholar]