Abstract
The human actin-binding protein filamin-A (also known as ABP-280) cross-links actin into a dynamic three-dimensional structure. It interacts with more than 45 proteins of diverse functions, serving as the scaffold in various signaling networks. BRCA2 is a protein that regulates RAD51 dependent recombinational repair of DNA double strand breaks (DSB). Proximate to the C-terminus of the BRCA2 protein, a conserved and DNA binding domain (BRCA2-DBD) interacts with filamin-A and BCCIP. In this study, we sought to test the hypothesis that filamin-A influences homologous recombinational repair of DSB and the maintenance of genomic stability. We used three pairs of cell lines with normal and reduced filamin-A expression, including breast cancer and melanoma cells. We found that lack or reduction of filamin-A sensitizes cells to ionizing radiation, slows the removal of DNA damage induced γH2AX nuclear foci, reduces RAD51 nuclear focus formation and recruitment to chromatin in response to irradiation, and results in a 2-fold reduction of homologous recombinational repair of DSB. Furthermore, filamin-A deficient cells have increased frequencies of micronucleus formation after irradiation. Our data illustrate the importance of the cytoskeleton structure in supporting the homologous recombinational DNA repair machinery and genome integrity, and further implicate a potential of filamin-A as a marker for prognosis in DNA damage based cancer therapy.
Keywords: filamin-A, ABP-280, BRCA2, homologous recombination, chromosome aberrations, DSB repair
Introduction
Homologous recombination (HR) plays a critical role in the repair of DNA double strand breaks (DSB). Due to the complexity of HR reactions, a large number of proteins are involved in HR, including enzymes that catalyze the chemical reactions, and accessory proteins that either regulate the activity of the enzymatic proteins or act at the interface between HR and other cellular processes. The key reactions in HR are the search for homology and strand exchange, which are catalyzed by the RecA protein in Escherichia coli and its homolog RAD51 in eukaryotes. The enzymes activities of these proteins are tightly regulated so that HR is coordinated with the cell cycle and other repair processes. In mammalian cells, RAD51 activity is regulated by several accessory proteins such as BRCA2, which was originally implicated as a HR-regulatory protein by its interaction with RAD51 (1–6). Subsequent studies have confirmed a direct role for BRCA2 and its interaction with RAD51 in HR (7–10). Mutations affecting the BRCA2-RAD51 interaction result in HR defects (6, 11). The human BRCA2 encodes a large protein of 3418 amino acids. Proximate to the BRCA2 C-terminus, a conserved region encoded by exons 14–24 is sometimes called the BRCA2-DNA binding domain (BRCA2-DBD) (12, 13). We previously reported that this BRCA2-DBD interacts with filamin-A and BCCIP (14, 15).
Filamin-A, also known as human actin-binding protein 280 (ABP-280) or filamin-1, encoded by the X-linked gene FLNa (16, 17). The human filamin-A gene encodes a protein of 2647 amino acids (18). It cross-links cortical actin filaments into a dynamic three-dimensional structure (19). Filamin-A has an elongated homodimeric and Y-shaped structure (18–22). At the N-terminus of the monomer, there is an actin-binding domain, followed by 24 tandem repeats of ~96 amino acids in length. Between repeats 15 and 16, there is a hinge domain, and repeat 24 is separated from repeat 23 by a second hinge domain. The last 65 amino acids of repeat 24 allow filamin-A dimerization to form a Y-shaped structure (18). In addition to filamentous actin, filamin-A interacts with more than 45 functionally diverse proteins including nuclear proteins, serving as the scaffold in various signaling networks (23, 24). These diverse interactions suggest that filamin-A is a key component of a versatile signaling scaffold.
The Filamin-A and BRCA2 interaction was originally identified by a yeast two-hybrid screen using the conserved BRCA2-DBD as the bait, and then confirmed by an in vitro binding assay using purified recombinant proteins, and an in vivo co-immunoprecipitation (14). Considering that filamin-A is a cytoskeletal protein that mostly associates with cell matrix, and BRCA2 is a critical protein in DNA HR (14), it is possible that filamin-A serves as a scaffold to anchor BRCA2 and/or to assist the assembly of the repair complex, and thus may play a role in DNA repair by HR. Although previous reports have showed that lack of filamin-A sensitizes cells to some DNA damage agents and delays the recovery of G2-cell cycle arrest (14, 25), no direct evidence has been documented for a role of filamin-A in DNA repair. In this report, we test the hypothesis that filamin-A plays a role in recombinational DNA repair and maintenance of genomic stability. We found that lack of filamin-A impairs HR, delays DSB repair, and promotes genomic instability.
Materials and Methods
Cell lines and Cell Cultures
Human A7, M2 and C8161 melanoma cells were cultured as described previously [18]. MDA-MB-231 cell was grown and routinely maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf bovine serum, 2 M glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. HT1080 cells were grown in a-MEM medium with 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were grown in a 37°C incubator supplied with 5% CO2 and 95% air. The same strategy as previously described (25) was used to knockdown filamin-A in C8161 melanoma cells, MDA-MB-231 breast cancer cells, and HT1080 fibrosarcoma cells.
Construction and Expression of Filamin-A Fragments
Several FLNa fragments (see Results for details) were amplified from pREP4 vector (kindly provided by Dr. TP Stossel) that contains the full length filamin-A cDNA by PCR. After digesting with restrict endonulease, fragments were inserted into pEYFP-C1 between the Hind III and XbaI sites. This results in the N-terminus fusion of the EYFP protein with the filamin-A fragments, stabilizing the expression of short filamin-A fragments. These vectors were transfected into the HT1080–1885 or M2 cells, and stable clones were selected with 500μg/ml of G418.
Western Blots and Extraction of Chromatin-bound Proteins
Cells were lysed in lysis buffer (50mM HEPES, pH7.6, 250mM NaCl, 5mM EDTA, 0.1% Nonidet P-40). The samples were then sonicated and boiled before loading onto 10% SDS-PAGE gel. Filamin-A was detected with an anti-filamin-A antibody (Chemicon International, Inc., CA). The β-actin was used as a loading control to confirm that an equal amount of proteins was loaded in each sample by probing with anti-β-actin monoclonal antibody (Sigma). RAD51 antibodies (Oncogene Research Products, Cambridge, MA), γH2AX (Serine 139) antibodies (Upstate Biotechnology, Lake Placid, NY), BRCA2 Antibodies (Calbiochem, San Diego, CA) were purchased. To extract chromatin-bound proteins, cells were suspended in hypotonic buffer (10mM HEPES pH 7.4, 10mM KCl, 0.05% NP-40, and protease inhibitors) and incubated on ice for 20 min to disrupt cell membranes. Thereafter, nuclei were recovered and treated with 0.2M HCl on ice for 10 min to extract chromatin-bound protein. Equal volumes of 1 M tris-HCl pH 8.0 were used to neutralize the acid-extracted protein solutions.
Radiation Survival Assay
Log-phase cells were plated on 100 mm culture dishes. The number of cells to be plated for each assay was determined by a pilot experiment so as to yield 50–150 surviving colonies per 100 mm dish. The cells were irradiated with Cs-137 γ-rays (dose rate: 0.893 Gy/min) 18 hours after the cells were plated. Colonies were grown for 12–14 days, after which the colonies were fixed with methanol and stained with 1% crystal violet. The number of colonies was normalized to the number of cells plated to calculate the surviving fraction. Each experiment was performed in triplicate and repeated at least twice.
Immunofluorescent Detection of γH2AX and RAD51 Nuclear Foci
Cells (5×104) were plated and grown on glass coverslips for 16 hours, then treated or not with γ-rays. The procedures to stain for γH2AX and RAD51 foci have been described previously (26, 27). Immunofluorescent signals were recorded using a Zeiss upright microscope with a UV laser source. RAD51 foci were visualized by the same procedure except that anti-RAD51 (1:1,000) primary antibody (Oncogene Research Products, Cambridge, MA) was used.
Measuring DSB-induced HR in HT1080–1885 Cells
To assess the role of filamin-A in DSB-induced HR, the HT1080–1885 cell line with a copy of installed HR substrate was used (28). The expression of endogenous filamin-A in HT1080–1885 was suppressed by RNA interference, using methods reported previously (25). To minimize clonal variation, multiple control and knockdown clones were selected and evaluated for the efficacy of filamin-A knock down (see Results). We then electroporated or transfected with an equal amount of pCMV-3NLS-I-SceI plasmid that expresses NLS-tagged I-SceI enzyme into each of the clones. Forty-eight hours after transfection, the cells were replated onto 100 mm culture dishes at ~100,000 viable cells per dish, and selected with puromycin. Seven to ten days after selection in puromycin (1 μg/ml), the cells that underwent I-SceI-induced HR formed colonies and were counted after staining. To determine plating efficiency, a parallel set of plates with appropriate cell dilution was plated to yield ~100 colonies in the absence of G418. The HR frequency was measured as the number of puromycin resistant colonies per 106 viable cells. The spontaneous HR frequency was measured in cells transfected with an empty pCMV vector. An average of 4–6 experiments was analyzed by Student's t-test to determine statistical significance.
Micronucleus Assay
Log-phase A7 and M2 cells were irradiated with 2 or 8 Gy Cs-137 γ-rays and harvested at indicated times. To assay micronucleus formation, cytochalasin B at 6 μg/mL final concentration was added into culture 24 hours after Cs-137 γ-irradiation and maintained for 24 hours. Cells were then trypsinized, treated with hypotonic saline and fixed. Air-dried slides were stained with Giemsa stain. Binucleated cells (~1000 cells) were scored under light microscopy for the presence or absence of micronuclei. Student's t-test was used for statistical analysis.
Chromosome Break and Aberration Assays
To prepare metaphase chromosome spreads, at 15 min, 8 hours, or 24 hours after irradiation, cells were blocked at mitosis with colchicine (at 0.08 μg/mL of final concentration) for four hours, washed with PBS, re-suspended in 75mM KCl for 30 min at 37°C and fixed twice with freshly made fixing solution (3:1, methanol:acetic acid, v/v) 20 min for each time. Then chromosome spreads were made on glass slides. After staining with Giemsa, forty metaphase cells were analyzed for each group. Unstable chromosome aberrations (dicentric, acentric ring, chromatid breaks and acentric fragments) were scored. The formation of dicentric and accentric rings reflects the mis-joining of DSB, and the existence of acentric fragments and breaks reflect un-repaired DSB. Statistical analyses for frequency of aberrations were performed using the Chi-Square Test and p<0.05 was considered significant.
Results
Down-regulation of Filamin-A Enhances Radiation Sensitivity
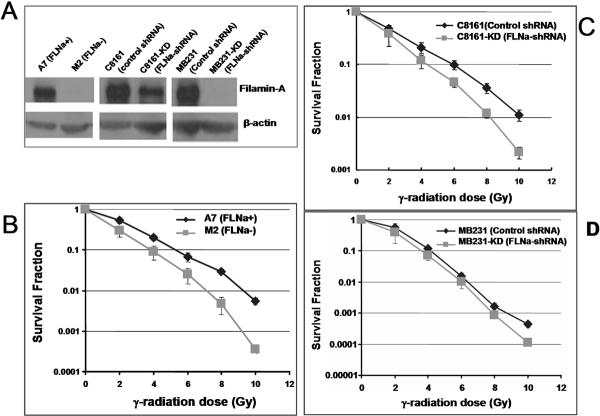
In a previous report, we showed that the M2 (filamin-A deficient) cells are more sensitive to γ-radiation than the A7 (filamin-A proficient) cells (14). M2 is a melanoma cell line that has spontaneously lost filamin-A expression, and A7 is a cell line derived from M2 by transfection with a filamin-A expressing vector (29). These data raised the question of whether depletion of filamin-A in filamin-A positive cells would sensitize the cells to radiation damage. To answer this question, we constructed filamin-A knockdown cell lines with C8161 melanoma and MDA-MB-231 breast cancer cells (referred to hereafter as MB231). The expression level of filamin-A in these cells was evaluated by Western blot (Figure 1A). The filamin-A deficiency in these cells did not affect their growth (see Supplement Figure S1). Using these cells, we measured their sensitivities to ionizing radiation by colony formation assays (see Materials and Methods for details). As shown in Figure 1, depletion of filamin-A by RNAi sensitizes cells to γ-rays, to a greater extent for the C8161 cells than the MB231 cells. The less extent of sensitization to radiation in filamin-A deficient MB231 cells may reflect a cell line specific dependence on ffilamin-A to DNA damage. Nevertheless, these data indicate that filamin-A is important for cellular resistance to radiation damage.
FIGURE 1.
Radiation sensitivity of filamin-A proficient and deficient cells. Panel A shows the anti-filamin-A western blot for the following three pairs of filamin-A proficient and deficient cell lines. 1) M2 (FLNa−) and A7 (FLNa+): M2 is a melanoma cell line that spontaneously lost filamin-A expression, and A7 is a M2-derived cell line that was transfected with filamin-A expressing vector (29); 2) C8161 (control shRNA) and C8161-KD (FLNa-shRNA): C8161 is filamin-A positive metastatic melanoma cell line expressing a control shRNA, and C8161-KD is a C8161 sub-clone expressing a filamin-A specific shRNA; 3) MB231 (control shRNA) and MB231-KD (FLNa-shRNA): MB231 is the MDA-MB-231 breast cancer cell line expressing a control shRNA, and MB231-KD is a MDA-MB-231 sub-clone expressing a filamin-A specific shRNA. Beta-actin was used as a loading control. Colony formation assays (see Materials and Methods) were used to determine the survival fractions after irradiation. Panel B-C are the radiation survival curves for the three pairs of filamin-A proficient and deficient cells. Error bars represent the standard deviation of three independent experiments.
Impaired DSB Repair in Filamin-A Deficient cells
Among the many factors that may affect cell sensitivity to DNA damage, DNA repair is the most critical one. The presence of γH2AX nuclear foci is widely regarded as a surrogate and sensitive marker for the presence of DSB in chromosomal DNA (30–33), and the number of γH2AX foci is believed to be closely related to the number of DSB in the cells (30, 31, 34). Therefore, a kinetic analysis of the appearance and disappearance of γH2AX foci after irradiation may reveal the DSB repair efficiency in the cells. To assess the role of filamin-A in DSB repair, we performed immunofluorescent staining and counted the number of γH2AX foci in M2 and A7 cells at time points after γ–irradiation. As shown in Figure 2A, the number of γH2AX foci per cell increased substantially after γ–irradiation. The focus numbers reached similar peak values at 0.5–1.0 hours after irradiation in M2 and A7 cells, suggesting that irradiation induces similar levels of initial DNA damage in the two cell lines. Afterwards, γH2AX foci decreased with time as DSBs were repaired. However, the numbers of γH2AX foci in A7 cells decreased significantly faster than those in M2 cells (p<0.01), especially within the first 4 hours after irradiation. This implies a faster DSB repair in A7 (filamin-A proficient) cells than in M2 (filamin-A deficient). To further confirm the role of filamin-A in DSB repair, we evaluated the effects of filamin-A knockdown on γH2AX protein levels after irradiation in C8161 melanoma and MB231 breast cancer cells. As shown in Figure 2B, the filamin-A deficient C8161 and MB231 cells retained higher levels of γH2AX than controls for up to 8 hours after γ-irradiation. These data strongly suggest that lack or reduction of filamin-A slows the repair of DSB. To confirm that filamin-A deficient impairs DSB repair, two additional approaches were used (Figures 2C and 2D).
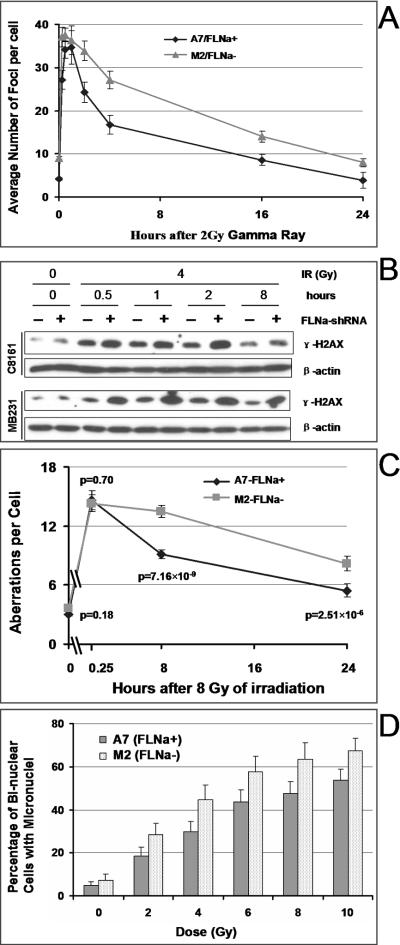
FIGURE 2.
Filamin-A deficiency impairs the DNA double strand break repair. Panel A shows the numbers of γH2AX foci per cell in A7 and M2 cells. At time points after the cells were treated with γ-radiation (panel A), cells were fixed and stained by immunoflurescent techniques with an anti-γH2AX antibody. The number of γH2AX foci was counted in >400 cells per time point in each experiment. Data shown are averages and standard errors of three experiments. Panel B shows the total γH2AX protein level measured by anti-γH2AX western blot at time points after irradiation (4 Gy) in C8161 and MB231 cells. The dose and times after irradiation are shown on the top of the panel. “FLNa-shRNA” indicates the whether the cells express filamin-A-specific shRNA. Beta-actin was used as a loading control. Panel C shows the total number of unstable chromosome aberrations per metaphase cell in A7 and M2. At 15 min, 8 hours, and 24 hours after 8 Gy of irradiation, mitotic chromosome spreads were prepared, and chromosome aberrations were scored from 40 independent metaphase cells as described in Materials and Methods. Data shown are averages ± SE (Standard Error). The Chi-Square Test was used to calculate the p-values, which are shown in the figure. Panel D shows the rate of micronuclear formations in A7 and M2 cells. Cytokinesis was blocked by treatment with cytochalasin B to form binuclear cells 24 h after γ-irradiation. Micronuclei in binuclear cells were visualized by Giemsa staining and scored under light microscopy. About 1000 binuclear cells in each experiment were analyzed and the data are averages of 5–6 independent experiments. Shown is the percentage of binuclear cells that have at least one micronucleus. The p-values indicate statistic significance based on t-tests.
First, incomplete repair of DSB before the cell enters mitosis would increase the level of chromosome fragments and chromatid breaks that are visible on mitotic chromosome spreads. Mis-joining of the DSB would increase the appearance of both dicentric and acentric rings on mitotic chromosome spreads. We irradiated A7 and M2 cells with 8 Gy of γ-radiation, at 15 min, 8 hours or 24 hours after the irradiation, mitosis were blocked for 4 hours, and mitotic spreads were prepared. The number of acentric fragments, chromatid breaks, dicentric, and acentric rings were scored in 40 metaphase cells. As shown in Figure 2C, at 8 hours and 24 hours after the irradiation, there were significant higher levels of chromosome aberrations in the filamin-A deficient M2 cells than the filamin-A proficient A7 cells (p<0.01). However, there was no significant difference of the initial level of chromosome breaks shortly (15 min) after irradiation (p>0.05), which suggests a similar level of initial DNA damage in these cells.
Second, when the DSB is not repaired or mis-repaired before the cells enter mitosis, the abnormal chromosome fragments especially these without functional centromeres, often form micronucleus after the separation of the two daughter nuclei. To verify whether the lack of filamin-A would increase micronucleus formations, cells were irradiated and then immediately blocked by cytochalasin B for 24 hours to arrest the cells at cytokinesis. This allows the nuclear division to complete but prevents the cytoplasm separation, thus forming bi-nuclear cells. Then, the rate of micronuclei in bi-nuclear cells was scored. As shown in Figure 2D, lack of filamin-A in M2 cells increased the radiation induced micronucleus formation. These data (Figure 2C and 2D) firmly support the conclusion of Figure 2A–2B that lack of filamin-A impairs DSB repair, although the percentage of cells with micronuclei do not necessary equal to the survival fraction as measured by colony formation.
Knockdown of Filamin-A Reduces RAD51 Focus Formation and Recruitment to Chromatin in Response to Irradiation
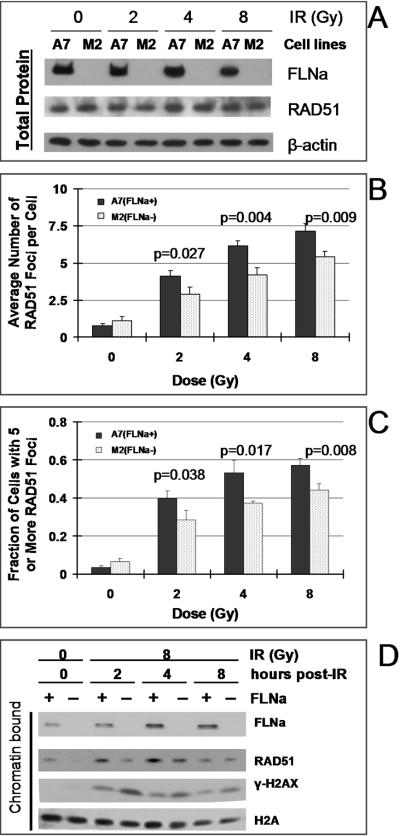
Within 2–6 hours after irradiation, RAD51 redistributes to form nuclear foci (2, 35–41). These DNA damage-induced RAD51 foci are located at single stranded regions where recombination is actively in progress (37). RAD51 focus formation in response to DNA damage is a visual indication for their involvement in HR. Failure or delay in forming RAD51 foci would signal an impaired HR process (1, 42). Because BRCA2 is required for efficient RAD51 focus formation and filamin-A interacts with BRCA2, we predicted that filamin-A deficient cells would show impaired RAD51 focus formations in response to DNA damage. To test this, cells were immunostained with RAD51 antibodies and the RAD51 foci were scored at time points (0 – 8 hours) after the irradiation as described previously (27). Representative images of RAD51 nuclear foci after irradiation are shown in supplement Figure S2. As shown in Figure 3A, lack of filamin-A does not affect the total RAD51 protein level. However, there was a modest but statistically significant reduction of the average number of RAD51 foci per cell (Figure 3B), and the fraction of cells with more than 5 RAD51 nuclear foci (Figure 3C), suggesting a reduced RAD51 recruitment to DNA damage. To confirm this, we measured the levels of chromatin-bound RAD51 by Western blot. As shown in Figure 3D, by four hours after the irradiation, there was an increase of chromatin-bound RAD51 in the A7 (filamin-A proficient) cells, consistent with the RAD51 nuclear focus formation. However, the chromatin-bound RAD51 in M2 (filamin-A deficient cells) was reduced compared with the A7 cells. These data also support a role of filamin-A in the recruitment of RAD51 to DNA damage.
FIGURE 3.
Filamin-A deficiency reduces RAD51 recruitment to DNA damage. Four hours after 2, 4, or 8 Gy of γ-irradiation, the RAD51 nuclear foci and chromatin-bound RAD51 protein were measured. Panel A is a western blot showing that lack of filamin-A in M2 cells does not alter the total RAD51 protein levels either before or after the irradiation. Panel B shows the average numbers of RAD51 nuclear foci per cells. Panel C shows the percentage of cells with five or more RAD51 foci. A7 and M2 cells in exponential phase were grown on cover slips and irradiated with 0, 2, 4 or 8 Gy of gamma radiation. Cells were fixed 4 hours after irradiation and fluorescent immunostaining was performed with anti-RAD51 antibody. RAD51 focus number in individual cells was scored under a fluorescent microscope. At least 250 cells in each experiment were analyzed and data shown are averages of 3 independent experiments. The p-values indicate statistic significance based on t-tests. Panel D shows the chromatin-bound RAD51 proteins at time points after irradiation. Filamin-A deficient cells have less chromatin-bound RAD51 protein between 4–12 hours after irradiation. Histone-2A (H2A) was used as a loading control for chromatin-bound proteins.
Knockdown of Filamin-A Reduces HR-dependent Repair of DSB
After confirming that filamin-A plays a role in RAD51 recruitment to DNA damage, we further tested the hypothesis that lack of filamin-A impairs DSB repair by HR. To do so, we used an in vivo HRR assay system previously established in the HT1080–1885 fibrosarcoma cell line (28). HT1080–1885 cells with filamin-A knockdown were established using the shRNA approach (see Materials and Methods). We isolated 5 control clones (labeled with number 1 to 5 in Figure 4A) that express control shRNA, and 7 clones express shRNA against filamin-A (labeled A to G in Figure 4A). As shown in Figure 4A, the filamin-A shRNA reduced filamin-A expression to varying degrees in individual clones.
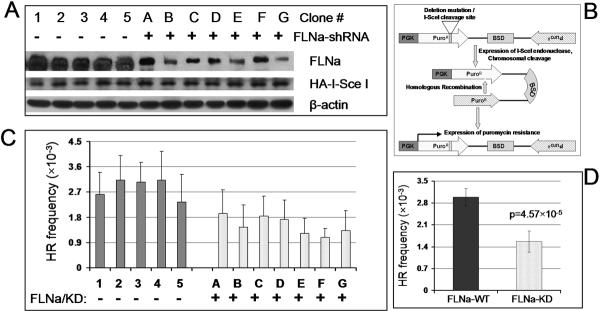
FIGURE 4.
Knockdown of filamin-A inhibits homologous recombinational repair of DSB. The filamin-A expression in HT1080–1885 cells, which host an HR assay substrate, was knocked down by shRNA. Five control clones (labeled as 1–5) and seven knockdown clones (labeled as A-G) were isolated. Break-induced HR was measured in these clones (See Results for a description of the HR assay). Panel A shows the level of filamin-A protein levels in control and knockdown clones. Clones 1–5 are cells expressing a control shRNA, and clones A-G are cells expressing filamin-A specific shRNA. HA-ISceI blot (bottom panel) shows the expression level of the I-SceI enzymes that introduces the site specific DSB in the HT1080–1885 cells. Panel B illustrates the recombination substrate and the HR mechanism resulting in the reconstitution of a functional puromycin resistance gene. Panel C shows the HR frequencies of individual clones. Shown are means and standard deviations of 6–8 experiments for each clone. Panel D summarizes HR frequencies for the filamin-A knockdown clones and the control clones. Shown are the means and standard deviations. The p-value indicates the statistical significance of the difference between means of the control clones and the filamin-A knock down clones (t-test).
HT1080–1885 cells have a single copy of a chromosomal recombination substrate organized as inverted repeats of the puromycin gene (Pur) (28). As shown in Figure 4B, the recipient copy of pur is not functional due to an 80 bp deletion of coding sequence, into which an 18 bp I-SceI endonuclease recognition site has been inserted. The donor copy of pur is not expressed as it lacks a promoter. When a gene conversion type of recombination between the donor and recipient copies occurs, a functional pur gene will be generated and expressed at the recipient copy, conferring puromycin resistance. Therefore, by determining the frequency of puromycin resistance after I-SceI expression, one can measure the HR frequency. After transfection of I-SceI expression vector into the individual clones (shown in Figure 4A), the HR frequency was measured. As shown in Figure 4C, all filamin-A knockdown clones had reduced HR frequencies. The seven knockdown clones had an average of 2-fold HR reduction in comparison with the five control clones (Figure 4D). These data indicate that filamin-A is required for fully efficient DSB repair by HR.
Filamin-A is a homodimer, and each monomer contains 24 tandem repeats. A hinge-2 domain is located between repeats 23 and 24, which enables flexibility of repeat 24. Our previous reported showed that the BRCA2 interacting domain is located between repeats 21 and 24 (14). To confirm that the filamin-A and BRCA2 interaction has direct involvement in the HR regulation, four short filamin-A fragments were constructed (bottom panel of Figure 5A): ABD is the actin binding domain; H2/R24 contains hinge-2 and repeat 24; R23/R24 contains the region between repeats 23 and 24; and R21/R24 contains the region between repeats 21 and 24. These fragments were expressed in the filamin-A wild type HT1080–1885 cells. As shown in Figure 5B, expression of the ABD does not inhibit HR. However, other three fragments inhibited the HR at various degrees. The fragment R21/R24 that contains the BRCA2 interacting domain has the strongest inhibition. It is worthwhile to point out that fragment H2/R24 that contains the filamin-A dimerization domain also inhibited the HR. However, this inhibition is not as strong as the R21/R24 fragment (p=0.025), suggesting that both the interaction between the BRCA2 and the dimerization may be required for efficient HR.
FIGURE 5.
Inhibition of homologous recombination by dominant negative interference. Four EYFP-fused short filamin-A fragments of filamin-A were expressed in HT1080–1885 cells. The same procedure as Figure 4 was used to measure ISceI induced HR, except transfection (instead of electroporation) was used to enhance the ISceI transfection. Panel A illustrates the location of four dominant negative fragments of filamin-A. Panel B shows the HR frequency. Panel 3 illustrates the expression of EYFG-tagged filamin-A fragments.
Filamin-A Truncations Fail to Rescue the Repair Defect in M2 cells
To complement the approach in Figure 5, we expressed three filamin-A truncation mutants in the filamin-A deficient M2 cells. These truncated filamin-A lack the C-terminus dimerization and BRCA2 interacting domain. Because M2 cells do not contain the HR substrate, we measured the γH2AX in response to irradiation to assess the DNA repair capability. As shown in Figure 6, shortly after the irradiation, all cells have similar level of γH2AX. However, at 2 hours and 8 hours after the irradiation, the full-length filamin-A complemented cells remove γH2AX much more efficiently than the M2 cells. The cells expressing the truncated filamin-A all failed to restore the repair ability, suggesting that the C-terminus (repeats 21–24) of filamin-A is required for its function in DNA repair. Altogether, Figures 5 and 6 suggest that the interaction with BRCA2 and self-dimerization of filamin-A are critical for the efficient repair of DSB.
FIGURE 6.
Filamin-A truncations fail to complement the DSB repair defects. Several truncation mutants of filamin-A as illustrated in 6A (bottom panel) was expressed in the M2 filamin-A deficient cells. See text for full description of the truncation mutants. The cell line expressing the full length filamin-A was used as positive control, and cell line carrying an empty vector was used as negative control. Panel B shows the average number of γH2AX foci per cells shortly after irradiation (0.25 hr), and after 2 and 8 hours of repair for M2-Vector (negative control), A7 (M2 cells with full length filamin-A), and several M2 cells expressing truncated filamin-A . Panel C are Western blots used as an alterative method to measure the level of γH2AX at various times after irradiation among the same cell lines. Anti-Actin blot was used as loading control for γH2AX blot. Panel C (bottom panel) also shows the confirmed expression of filamin-A truncation mutants.
Discussion
In this report, we have provided several lines of evidence for a role of the cytoskeleton protein filamin-A in supporting homologous recombinational repair of DNA damage. These measurements include radiation sensitivity, DSB repair kinetics as reflected by γH2AX foci, RAD51 recruitment to DNA damage, and homologous recombination at a DSB site in the cells. We also showed that lack of filamin-A increases the risk of genomic instability after irradiation.
How filamin-A contributes to HR is an issue of discussion. It is known that DSB repair, especially homologous recombinational repair, is a multi-step process that requires not only enzymatic proteins but also a large number of accessory proteins. Some of these proteins, such as RAD51, may act as an essential enzyme in the HR reactions, others such as BRCA2 may serve as auxiliary proteins to promote the assembly of the HR enzymes. It is anticipated that the HR machinery may be associated with the cytoskeletal structures in order to facilitate assembly of the repair complex. It is possible that filamin-A may serve as a nuclear anchor for BRCA2 and HR machinery during HR. In this sense, we suggest that filamin-A is required for an efficient HR repair, but not necessary absolutely required for the HR process. This is reflected in the modest reduction of HR frequency and RAD51 focus formation in filamin-A deficient cells.
Because filamin-A was originally identified as an actin binding protein that likely to be involved in cytoplasm function, and BRCA2 was originally identified as mainly a nuclear protein, it is worthy to point out that a fraction of filamin-A also resides in the nucleus. This conclusion was reached by several independent groups (14, 43–45). Furthermore, the majority of a naturally occurred cleavage fragment of filamin-A resides in the nucleus, and participate in other nuclear functions (43, 44).
The identification of a role of filamin-A in homologous recombinational repair not only explains the sensitivity of filamin-A deficient cells to ionizing radiation, but also explains another previously observed phenotype. We previously reported that the filamin-A deficient cells had a delayed recovery of G2 arrest after DNA damage (25). Because HR plays a major role in DNA repair during late S and G2 phases, it is understandable that there should be a delayed recovery from G2 arrest after DNA damage for filamin-A deficient cells due to a delayed repair of DNA damage in S and G2 phase.
Our findings also have a potential clinical implication for cancer therapy, as our data have suggested that whether or not filamin-A is expressed may influence the cell sensitivity to DNA damage. The next question is whether there actually are alterations of filamin-A expression in cancers. If there indeed is a difference of filamin-A expression, then there would be an opportunity to categorize the cancer into two groups based on filamin-A status and treat each group differently. The filamin-A negative cancer would be sensitive to therapeutic DNA damage and would be an ideal candidate for DNA damage based treatment. We have recently measured the filamin-A expression in a few melanoma tissues, and we found that indeed some melanomas are filamin-A negative (Supplement Figure S3). This also raises the issues of whether filamin-A can be used as a target to sensitize filamin-A positive cancer cells to radiation or other DNA damage-based cancer therapy, and whether the lack of filamin-A expression in cancer can be used as an indicator and prognostic marker for DNA damage-based therapy. As shown in Figure 1, there is a 30%–50% increase in sensitivity of filamin-A deficient cells after 2 Gy of irradiation. This difference has potential significance in radiation therapy to treat cancers with different filamin-A expression status, as a typical course of radiation therapy generally involves multiple doses of 2 Gy irradiation. If each 2 Gy fraction results in 30%–50% sensitivity, a treatment course of 25 fractions of 2 Gy would significantly affect the outcome.
In summary, our study suggests that the cytoskeleton protein filamin-A is required for a fully efficient homologous recombinational repair of DNA double strand breaks. Lack of filamin-A may render the cells more sensitive to DNA damage while further increase the genomic instability. Thus filamin-A may be useful as a new marker for DNA damage-based cancer therapy.
Supplementary Material
Acknowledgements
This research was supported by National Institute of Health grant CA115488. We would like to thank Ms. Jingmei Liu for her technical support and critical reading of the manuscript.
Abbreviations
- FLNa
filamin-A or ABP-280
- DSB
double strand breaks
- HR
homologous recombination
- PBS
phosphate buffered saline
References
- 1.Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–92. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Silver DP, Walpita D, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–28. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 3.Katagiri T, Saito H, Shinohara A, et al. Multiple possible sites of BRCA2 interacting with DNA repair protein RAD51. Genes Chromosomes Cancer. 1998;21:217–22. [PubMed] [Google Scholar]
- 4.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc Natl Acad Sci U S A. 1998;95:13869–74. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–4. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 6.Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- 7.Davies AA, Masson JY, McIlwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 8.Larminat F, Germanier M, Papouli E, Defais M. Deficiency in BRCA2 leads to increase in non-conservative homologous recombination. Oncogene. 2002;21:5188–92. doi: 10.1038/sj.onc.1205659. [DOI] [PubMed] [Google Scholar]
- 9.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 10.Xia F, Taghian DG, DeFrank JS, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci U S A. 2001;98:8644–9. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JJ, Silver D, Cantor S, Livingston DM, Scully R. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999;59:1752s–56s. [PubMed] [Google Scholar]
- 12.Sharan SK, Bradley A. Murine Brca2: sequence, map position, and expression pattern. Genomics. 1997;40:234–41. doi: 10.1006/geno.1996.4573. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Jeffrey PD, Miller J, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Shen Z. Interaction with BRCA2 suggests a role for filamin-1 (hsFLNa) in DNA damage response. J Biol Chem. 2001;276:48318–24. doi: 10.1074/jbc.M102557200. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Yuan Y, Huan J, Shen Z. Inhibition of breast and brain cancer cell growth by BCCIPalpha, an evolutionarily conserved nuclear protein that interacts with BRCA2. Oncogene. 2001;20:336–45. doi: 10.1038/sj.onc.1204098. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Ash JF, Singer SJ. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc Natl Acad Sci U S A. 1975;72:4483–6. doi: 10.1073/pnas.72.11.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K. Filamin, a new high-molecular-weight protein found in smooth muscle and nonmuscle cells. Purification and properties of chicken gizzard filamin. Biochemistry. 1977;16:1857–65. doi: 10.1021/bi00628a015. [DOI] [PubMed] [Google Scholar]
- 18.Gorlin JB, Yamin R, Egan S, et al. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–45. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham CC, Vegners R, Bucki R, et al. Cell permeant polyphosphoinositide-binding peptides that block cell motility and actin assembly. J Biol Chem. 2001;276:43390–9. doi: 10.1074/jbc.M105289200. [DOI] [PubMed] [Google Scholar]
- 21.Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2:888–92. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- 22.van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–8. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 24.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–9. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Meng X, Yuan Y, Maestas A, Shen Z. Recovery from DNA Damage-induced G2 Arrest Requires Actin-binding Protein Filamin-A/Actin-binding Protein 280. J Biol Chem. 2004;279:6098–105. doi: 10.1074/jbc.M306794200. [DOI] [PubMed] [Google Scholar]
- 26.Lu H, Yue J, Meng X, Nickoloff JA, Shen Z. BCCIP regulates homologous recombination by distinct domains and suppresses spontaneous DNA damage. Nucleic Acids Res. 2007;35:7160–70. doi: 10.1093/nar/gkm732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H, Guo X, Meng X, et al. The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous recombinational repair. Mol Cell Biol. 2005;25:1949–57. doi: 10.1128/MCB.25.5.1949-1957.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lio YC, Schild D, Brenneman MA, Redpath JL, Chen DJ. Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phase cells. J Biol Chem. 2004;279:42313–20. doi: 10.1074/jbc.M405212200. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham CC, Gorlin JB, Kwiatkowski DJ, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–7. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 30.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 31.Sedelnikova OA, Rogakou EP, Panyutin IG, Bonner WM. Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat Res. 2002;158:486–92. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Fillingham J, Keogh MC, Krogan NJ, et al. GammaH2AX and its role in DNA double-strand break repair Auger electrons--a nanoprobe for structural, molecular and cellular processes. Biochem Cell Biol. 2006;84:568–77. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 33.Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81:123–9. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi A, Ohnishi T. Does gammaH2AX foci formation depend on the presence of DNA double strand breaks? Cancer Lett. 2005;229:171–9. doi: 10.1016/j.canlet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 35.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A. 1995;92:2298–302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Li M, Lee EY, Maizels N. Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr Biol. 1999;9:975–8. doi: 10.1016/s0960-9822(99)80427-8. [DOI] [PubMed] [Google Scholar]
- 37.Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci U S A. 1999;96:1921–6. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Maizels N. Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO Rep. 2000;1:85–90. doi: 10.1093/embo-reports/kvd002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–82. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Essers Jp, Houtsmuller AB, van Veelen L, et al. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–37. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–5. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 43.Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci U S A. 2003;100:4562–7. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Kreisberg JI, Bedolla RG, Mikhailova M, Devere White RW, Ghosh PM. A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210435. [DOI] [PubMed] [Google Scholar]
- 45.Ozanne DM, Brady ME, Cook S, Gaughan L, Neal DE, Robson CN. Androgen receptor nuclear translocation is facilitated by the f-actin cross-linking protein filamin. Mol Endocrinol. 2000;14:1618–26. doi: 10.1210/mend.14.10.0541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.