Abstract
Bacillus thuringiensis subsp. israelensis, which is used worldwide to control Aedes aegypti larvae, produces Cry11Aa and other toxins during sporulation. In this study, pull-down assays were performed using biotinylated Cry11Aa toxin and solubilized brush border membrane vesicles prepared from midguts of Aedes larvae. Three of the eluted proteins were identified as aminopeptidease N (APN), one of which was a 140 kDa protein, named AaeAPN1 (AAEL 012778 in VectorBase). This protein localizes to the apical side of posterior midgut epithelial cells of larva. The full-length AaeAPN1 was cloned and expressed in E. coli and in Sf21 cells. AaeAPN1 protein expressed in Sf21 cells was enzymatically active, had a GPI-anchor but did not bind Cry11Aa. A truncated AaeAPN1, however, binds Cry11Aa with high affinity, and also Cry11Ba but with lower affinity. BBMV but not Sf21 expressed AaeAPN1 can be detected by wheat germ agglutinin suggesting the native but Sf21 cell expressed APN1 contains N-acetylglucosamine moieties.
Keywords: Bacillus thuringiensis, Cry11Aa toxin, aminopeptidase, receptor, binding affinity, midgut, proteomics
Introduction
Aedes mosquitoes are not only a nuisance but more importantly also transmit viral diseases, including dengue, yellow fever and chikungunya, all of which have seen recent reemergence. Increasingly these Aedes mosquitoes are controlled through use of formulations of B. thuringiensis subsp. israelensis that target the larval instar.
During the sporulation phase, this bacterium produces insecticidal inclusions that contain insecticidal proteins (Cry4Aa, Cry4Ba, Cry10Aa and Cry11Aa) and cytolytic proteins (Cyt1Aa, Cyt2Ba and Cyt1Ca) (Berry et al., 2002). Among them, Cry11Aa is the most active toxin against Ae. aegypti (Chilcott and Ellar, 1988). Upon ingestion by susceptible insect larvae, Cry11Aa inclusions are first solubilized in the alkaline environment of larval midgut, and then the soluble protoxins are processed into 34 and 32 kDa fragments by gut proteases (Dai and Gill, 1993; Yamagiwa et al., 2002). The activated fragments then bind specific receptors in the microvilli of midgut epithelial cells, leading to membrane insertion and pore formation (Bravo et al., 2005; Bravo et al., 2007). These latter processes are thought to lyse midgut cells ultimately killing larval mosquitoes (Soberon et al., 2009).
Toxin selectivity is determined both by specific domains in these toxins and the presence of toxin binding receptor proteins in the midgut of the target insect. There has been far greater understanding of these receptor proteins in lepidopteran insects, with three major proteins identified – cadherins, aminopeptidases (APNs), and alkaline phosphatases (ALPs) (for reviews see (Bravo et al., 2005; Pigott and Ellar, 2007)). Recent evidence suggests mosquitocidal toxins also bind these same set of proteins in the mosquito midgut with identification of cadherin binding Cry4Ba, Cry11Ba and Cry11Aa (Hua et al., 2008; Likitvivatanavong et al., 2009)(Chen et al., submitted), APNs binding Cry11Ba (Abdullah et al., 2006; Zhang et al., 2008), and ALPs binding Cry11Aa (Fernandez et al., 2006).
Since APNs were identified as Cry toxin-binding proteins (Gill et al., 1995; Knight et al., 1995; Sangadala et al., 1994), a number of APNs from lepidopteran insects have been reported to bind Cry1 toxins. Recently, APNs as toxin receptors have been identified from dipteran insects. For example, APNs from Anopheles quadrimaculatus and An. gambiae bind Cry11Ba (Abdullah et al., 2006; Zhang et al., 2008). In this report, we show APN proteins bind also Cry11Aa toxin. These APN proteins are classified into at least five families by phylogenetic analyses (Pigott and Ellar, 2007; Zhang et al., 2008). To date, more than 20 APNs have been shown to bind Cry toxins (Abdullah et al., 2006; Pigott and Ellar, 2007; Zhang et al., 2008; Zhang et al., 2009). Cry toxin interaction with APNs generally is thought to involve glycosylated moieties. For example, N-acetyl galactosamine (GalNAc) is an important determinant of Cry1Ac interaction with 120 kDa APN from M. sexta (Burton et al., 1999; Masson et al., 1995), Heliothis virescens (Gill et al., 1995) and Lymantria dispar (Jenkins et al., 2000). But some APNs are believed to bind toxins in a glycan-independent manner. It was first reported that putative Cry1Aa toxin binding sites in Bombyx mori APN were localized between 135-Ile and 198-Pro by ligand blot (Yaoi et al., 1997; Yaoi et al., 1999) and more recently was confirmed under nondenaturing conditions by ELISA (Atsumi et al., 2005). In case of the 106 kDa APN (AgAPN2) from An. gambiae, partial AgAPN2 expressed in E. coli binds Cry11Ba toxin on dot blot and microtiter plate binding and can inhibit Cry11Ba toxicity against An. gambiae larvae (Zhang et al., 2008).
The biological relevance of interaction between Cry toxin and APN needs additional investigation (Pigott and Ellar, 2007). To date, only a few APNs have been shown to mediate in vivo toxin activity. First, transgenic Drosophila expressing 120 kDa APN from M. sexta developed 100% susceptibility to high concentrations of Cry1Ac toxin (Gill and Ellar, 2002). Second, silencing of midgut APN in Spodoptera litura by dsRNA reduces its expression resulting in lower Cry1C toxicity (Rajagopal et al., 2002). Similarly, Sf21 cells expressing HaAPN1 from Helicoverpa armigera show increased toxin sensitivity, and silencing of HaAPN1 in H. armigera by dsRNA results in decreased larval susceptibility to Cry1Ac toxins (Sivakumar et al., 2007). Finally, the strongest evidence that APN proteins play an important role in Cry toxicity comes from recent evidence that toxin-resistant H. armigera have a mutation in the APN gene (Zhang et al., 2009). Therefore, there is increasing evidence that shows APN is a functional receptor, to which cadherin-induced toxin oligomers can bind according to the sequential toxin binding model (Bravo et al., 2004).
In this study an Ae. aegypti APN, named AaeAPN1, was isolated and identified as a Cry11Aa-binding protein using a biotinylated Cry11Aa toxin pull-down assay. BBMV and cell-expressed AaeAPN1 were also characterized by lectin blot and anti-cross reacting determinants (CRD) antibody detection. AaeAPN1 expressed in E. coli binds Cry11Aa toxin with high affinity.
Materials and methods
Purification, activation and biotinylation of Cry11Aa toxin
Inclusions for Cry11Aa and Cry11Ba were isolated from recombinant strains producing these toxins individually (Chang et al., 1993; Delecluse et al., 1988). B. thuringiensis strains were grown in nutrient broth sporulation medium containing 12.5 μg/ml erythromycin at 30°C (Lereclus et al., 1995). Following cell autolysis spores and inclusions were harvested, washed three times with 1 M NaCl, 10 mM EDTA, pH 8.0 and centrifuged. The resulting pellet was resuspended in 30 ml of the same buffer and purified by NaBr gradients as previously described (Cowles et al., 1995).
Purified inclusions were solubilized in 50 mM Na2CO3, pH 10.5, and activated by trypsin (1:20, w/w). The activated Cry11Aa toxin was purified by ion exchange chromatography (Mono Q, FPLC), biotinylated (GE Healthcare) and then purified using a Sephadex G25 column.
Preparation of membranes from Aedes midguts and Sf21 cells
Brush border membrane vesicles (BBMV) were prepared from fourth instar larval midguts as described previously (Nielsen-Leroux and Charles, 1992). Sf21 cells infected with recombinant baculovirus were harvested and washed with 1×PBS containing 1 mM PMSF and 1× protease inhibitor cocktail (Roche). The cell suspension was sonicated and then centrifuged at 700g for 10 min. The supernatant was centrifuged at 16,000g for 1 h at 4°C. The membrane pellet was stored in −80 °C until needed.
Pull-down experiments and mass spectrometry
Cry11Aa biotinylated toxin (100 μg) were added to 500 μg Aedes midgut BBMV or cell membrane proteins and incubated at 4°C overnight with gentle rocking on a rotating platform. The sample was centrifuged at 16,000 g for 1 h and the pellet was resuspended in 1 ml solubilization buffer (20 mM Tris-HCl, pH 7.5, 1mM EDTA, pH 8.0, 1mM MgSO4, 0.01% NaN3, 10% glycerol, 2% CHAPS, 1 mM PMSF and 1× protease inhibitor cocktail), incubated at 4°C for at least 1 h and then centrifuged at 4°C for 1 h. The supernatant was added to 50 μl of immobilized streptavidin (Pierce) gel slurry and incubated overnight at 4°C. The sample was centrifuged at 1,250 × g for 60 sec, the supernatant discarded, and the pellets were washed with 250 μl of acetate-5M NaCl wash buffers (39:1) three times by gently inverting the columns 5–7 times. After washing, the pellets were mixed with 35 μl 1x protein loading buffer, samples separated by SDS-PAGE, and then visualized by Coomassie-blue or silver staining. Protein bands were excised from the gel, digested by trypsin, and analyzed by nano-ultra performance liquid chromatography/tandem mass spectrometry (nano-UPLC/MS/MS).
For pull-down assays using agarose bound wheat germ agglutinin (WGA), solubilized BBMV was incubated with WGA and then washed three times. Bound proteins were eluted by 500 mM N-acetylglucosamine (GlcNac) (pH 3.0), separated using SDS-Page and the gel stained by Coomassie blue. Bands were excised for mass spectrometry.
Assembly, cloning and analysis of AeaAPN1 gene
Five pairs of primers listed in Table 1 were designed based on sequences of two transcripts, AAEL012778-RA and AAEL012778-RB that encoded one of the proteins identified by mass spectrometry. These primer pairs were used for PCR amplification of full-length or partial AaeAPN1 cDNA. Total RNA was extracted from fourth instar larval midguts using Trizol reagent (Invitrogen). For 3′ amplification, first strand cDNA was synthesized from total RNA with an oligo(dT) primer. Using primers APN1-3R and a gene-specific primer, APN1-3F, a 0.5 kb PCR product was obtained from the 3′ end of the transcript. Because 5′-RACE failed, a gene-specific primer, APN1-5F was designed based on the genome sequence to be between a predicted transcript start site and a translation start condon. Using this primer together with primer APN1-5R from a putative APN ORF gave a 0.3 kb in PCR amplification. This PCR product contained in frame stop condons before the predicted start condon. Based on sequences of the 3′ and 5′ end amplified products, two partial overlapping fragments of APN1 ORF were amplified by using gene-specific primers, APN1-1F/1R and APN1-2F/2R, respectively. These two products were then were assembled into full-length AaeAPN1 ORF using a unique restriction site, NdeI, present in the two PCR products. All PCR products were cloned into the TA cloning vector, pCR2.1 TOPO (Invitrogen) and fully sequenced (Institute of Integrative Genome Biology (IIGB), University of California Riverside).
Table 1.
Primers used in this study
| Primer | Primer sequences(5′–3′) |
|---|---|
| APN1-5F | 5′-AGTTAAAAGGATTTCAAAGCC-3′ |
| APN1-5R | 5′-TCGCTTCTACCACGTTCAAA-3′ |
| APN1-3F | 5′-CGCTTTGATTGAATTCCTGA-3′ |
| APN1-3R | 5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′ |
| APN1-1F | 5′-GGATCCATGATCGTTCTCAAGTGGTCGGTG-3′ |
| APN1-1R | 5′-CATATGGTCGTAGTACCCGATGCCAAG-3′ |
| APN1-2F | 5′-CATATGCCACAAATGAAACAATTTGCCATTCCCG-3′ |
| APN1-2R | 5′-TCGATACTAGTTAAACAACGCTACTGC-3′ |
| APN1-4F | 5′-GGATCCCGAACCTACGAAACCGGAGATA-3′ |
| APN1-4R | 5′-GTCGACAAATATTGGTACTCAGCAACATCG-3′ |
Sequence alignments and other sequence analysis were performed using NCBI blast programs and Lasergene (DNAstar). Signal peptide and glycosylphosphatidylinositol (GPI) anchor were predicted by SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) and DGPI (http://129.194.185.165/dgpi/), respectively. The OGPET v1.0 (http://ogpet.utep.edu/OGPET/) and NetNGlyc v1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) programs were used to determine potential O- and N-glycosylation sites, respectively.
Expression of AaeAPN1 in E. coli and Sf21 cells
A partial gene fragment, AaeAPN1t, within the 3′ variable region was obtained by PCR using primers APN1-4F/4R. The fragment was cloned into the pCR2.1 vector, sequenced and then subcloned into pQE30 (Qiagen), an E.coli expression vector. The construct was transformed into E.coli M15(pREP4) strain, and protein expression induced by addition of 1 mM isopropyl β-D-thiogalactoside (IPTG). The recombinant protein was purified by Ni-NTA resin (Qiagen) using urea and resolved in SDS-PAGE. Gels were stained and destained, the 29 kDa protein band excised, washed three times and then used for antibody development.
The full-length AaeAPN1 ORF was cloned into the baculovirus expression vector, pFastBac1 and donor plasmids generated were then transformed into competent E. coli DH10B cells. White recombinant colonies were selected and presence of the AaeAPN1 ORF confirmed by PCR. Sf21 cells were grown and maintained in TNM-FH medium (Invitrogen) with penicillin and streptomycin at 27°C. Then recombinant bacmid DNA was transfected into Sf21 cells by Cellfectin reagent (Invitrogen). Recombinant AaeAPN1 baculovirus were amplified and P3 virus stocks were used for protein expression.
Immunolocalization of AaeAPN1 in larval midgut of Ae. aegypti
Late fourth instar larvae and isolated guts of Ae. aegypti were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. After fixation, tissues were processed, 8μm thick sections cut and analyzed as described (Kang’ethe et al., 2007). Sections were incubated with protein A-purified rabbit polyclonal anti-AaeAPN1 antibody diluted 1:100 in 1% bovine serum albumin (BSA)/PBST overnight at 40°C. After washing with PBST containing 2% goat serum (GS) and 0.1% BSA, sections were incubated in the dark at room temperature for 40 min with secondary antibodies – Cy3-conjugated goat-anti-rabbit IgG for AaeAPN1 (1:1,000 dilution) and Alexa-488 for actin F (1:100 dilution) – both diluted in 0.1%BSA/2%GS/PBST. Tissues were again washed and mounted in Shur/Mount media (Electron Microscopy Sciences). Images were obtained using a Zeiss Axioplan laser-scanning confocal microscope (LSM 510) located in IIGB. All images were imported in Adobe Photoshop for assembly and annotation.
Dot blot binding assays
Inclusion bodies from E. coli expressing H. virescens cadherin fragment, PP9, and the partial AaeAPN1protein, AaeAPN1t, were solubilized in 50 mM Na2CO3, pH 10.5, 37°C for 1 h. Forty μg of solubilized proteins were dotted onto a PVDF filter directly. Membranes were incubated with 3% BSA and then with 20 nM Cry11Aa toxins for 2 h. Unbound toxins were removed by washing the membrane four times with washing buffer (0.1% Tween 20, in PBS) for 15 min. Membranes were then incubated with rabbit anti-Cry11Aa antibody (1:5000 dilution) followed by a secondary goat anti-rabbit antibody conjugated to horseradish peroxidase (GE Healthcare) (1: 5000 dilution). Finally, membranes were exposed to luminol (ECL™, GE Healthcare).
GPI-anchor and glycosylation detection for Sf21cell-expressed and BBMV AaeAPN1
Cell membrane fractions were separated in SDS-PAGE and electrotransferred to PVDF membranes. For detection of GPI anchor, after mild base pretreatment with CRD buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, pH 8.0, 0.05% Tween 20) plus 3%BSA for 2 hr, the blot was washed with this buffer for 15 min three times. Then the blot was treated with 0.6 units of PI-PLC (Prozyme) in 20 mM Tris-HCl, pH 7.4, 1% Triton-100, 1mM DTT, 3% BSA and washed with the CRD buffer. The presence of a GPI anchor was detected with an anti-CRD antiserum (1:200) (Prozyme). For lectin blot, membranes were blocked with 1×TBST, 5% blocking reagent for 1 h, rinsed with 1×TBST and incubated with biotinylated lectin (0.1μg/ml) at 4°C overnight. The blot was washed with 1×TBST for 15 min four times and then incubated with strepavidin-horseradish peroxidase (HRP) conjugate (1:10,000). After washing for 15 min four times, protein bands were visualized by luminol (GE Healthcare).
Aminopeptidase assay
Enzymatic activity was measured using alanine, leucine or valine p-nitroanilides as substrates. Membrane fraction of Sf21 cells infected with recombinant or vector baculovirus were extracted and incubated in 1 ml solubilization buffer (20 mM Tris-HCl, pH 7.5, 1mM EDTA, pH 8.0, 1mM MgSO4, 0.01% NaN3, 10% glycerol, 2% CHAPS, 1 mM PMSF and 1× protease inhibitor cocktail), for 1 h at 4°C. The mixture was then centrifuged at 4°C at 16,000 g for 1 h and the soluble protein concentration measured using bicinchoninic acid protein assay reagent kit (Pierce). The same procedure was used to solubilize Aedes larval midgut BBMV. All samples were diluted to a final concentration of 500 μg/ml. Then 100 μl of the resultant samples were mixed with 900 μl 22.2 mM Tricine buffer, pH 8.0, 0.44% (v/v) methanol containing 0.28 mM of each substrate. The same concentration of BSA in solubilization buffer was used as a background control. Increase in the absorbance at 405 nm for 5 min was recorded in a microplate reader (Molecular Devices). Increases in absorbance values over the background control were recorded. Enzymatic activity was calculated using a millimolar extinction coefficient of 10.8 nM−1 cm−1 for p-nitroanilide at 405 nm.
Competitive ELISA
A truncated AaeAPN1t dose-dependent binding curve was obtained using a modified ELISA (Perez et al., 2005). In brief, 96-well plates were coated with 0.4 μg Cry11Aa per well and then treated with blocking buffer (PBS, 0.1% Tween 20, 0.5% gelatin) for 1 h at 37°C, washed and then 0–1000 nM AaeAPN1t protein solutions were transferred to the coated plates. After washing, to remove unbound AaeAPN1t protein, anti-AaeAPN1 antibody (1:2000) was added and incubated overnight at 4°C. Subsequently after additional washes, goat-anti-rabbit antibody coupled to alkaline phosphatase (1:2000) was added to wells and incubated for a further 2 h at 37°C. After further washing alkaline phosphatase activity was revealed with freshly prepared substrate (3 mM nitrophenyl phosphate) and absorbance read at 405 nm with a microplate reader (Molecular Devices). The AaeAPN1t concentration that showed a linear range of binding was used for competitive ELISA below.
Kinetics of disassociation of Cry11Aa binding to AaeAPN1t was measured by competitive ELISA (Bravo et al., 2004; Dong et al., 2003; Perez et al., 2005). AaeAPN1t, 80 nM, was equilibrated with increasing concentrations of Cry11Aa toxin (0.2 nM to 1000 nM) in 100 μl PBST for 1 h at room temperature. The mixtures were then transferred to plate wells previously coated with Cry11Aa toxin. The detection procedure was then continued as described above. Data were analyzed using Origin (Origin Lab, Northampton, MA). The concentration corresponding to half maximal absorbance was considered the dissociation constant (Dong et al., 2003; Perez et al., 2005).
For Cry11Ba competition with Cry11Aa binding to AaeAPN1, 80nM AaeAPN1t was incubated with increasing concentrations of Cry11Ba toxin (0.2 nM to 1000 nM) in 100 μl PBST for 1 h at room temperature. The mixtures were then transferred to plate wells previously coated with Cry11Aa toxin and analyzed as described above.
Results
Identification of AaeAPN1 as a Cry11Aa-binding protein
A pull-down assay with biotinylated Cry11Aa toxin was performed using solubilized Ae. aegypti larval midgut BBMV and the proteins eluted were separated by SDS-PAGE gel. Four protein bands with the molecular weight of 140, 95, 45 and 32 kDa, respectively, were detected (Fig. 1). Since the 32 kDa protein is a fragment of Cry11Aa toxin, the other three protein bands were individually exercised, destained, digested with trypsin, and the resulting tryptic peptides were analyzed by nano-UPLC/MS/MS in a data-dependent acquisition mode. All MS/MS spectra were processed and searched against the NCBInr protein database. Proteins identified with a significant score (p<0.01) were considered positive hits. The eluted 140 kDa protein was identified as a zinc metalloprotease with accession number AAEL 012778-PA in VectorBase. The 95 kDa band identified two proteins with accession numbers AAEL008155 and AAEL012774. The 45 kDa protein was identified as actin (Table 2). In this manuscript, we will focus on the 140 kDa zinc metalloprotease, named AaeAPN1. The m/z of the seven identified peptides listed in Fig. 2 ranged from 524.7 to 911.9 and covered 10.6% of the total amino acid sequence of the AaeAPN1 protein.
Figure 1. Biotinylated Cry11Aa toxins pulls-down aminopeptidases from Ae aegypti BBMV.

Eluted proteins in the pull-down using biotinylated Cry11Aa toxins were separated in SDS-PAGE and four protein bands with the molecular weight of 140, 95, 45 and 32 kDa were detected by Coomassie blue staining. Proteins corresponding to the top three bands were excised from the gel and analyzed by mass spectrometry.
Table 2.
Mass identification of eluted proteins from Aedes Aegypti BBMV by biotinylated Cry11Aa toxin pull-down assays
| NCBI no. | VectorBase accession no | Annotation | Predicted Mr, kDa | Additional features |
|---|---|---|---|---|
| gi_108870803 | AAEL012778 | zinc metalloprotease (AaeAPN1) | 112 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_108875835 | AAEL008155 | zinc metalloprotease (AaeAPN2) | 104 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_108870802 | AAEL012774 | zinc metalloprotease (AaeAPN3) | 102 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_108879764 | AAEL004616 | Actin | 42 | Ubiquitous protein involved in the formation of filaments |
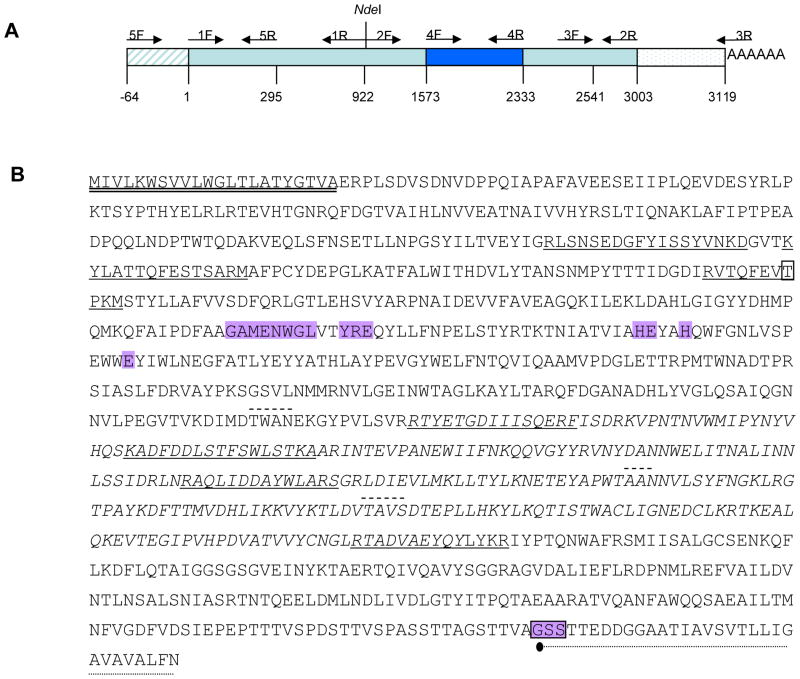
Figure 2. Cloning of AaeAPN1 gene and structure features of the full-length AaeAPN1 protein.
A. Schematic for isolation of full-length AaeAPN1 cDNA, 3′- and 5′-end amplification products, two partial overlapping fragments, and truncated AaeAPN1 (bp 1573 – 2333). Primer positions are indicated by arrows. B. Seven peptides identified by mass spectrometry are underlined. The putative N-terminal signal peptide is double-underlined. The GPI-anchor signal peptide is dot-underlined, the possible GPI anchor site is in shadowed box and • indicated the probable cleavage site of the anchor moiety. The predicted O-glycosylated residue is in box and the putative N-glycosylation sites are dash-underlined. The consensus gluzincin aminopeptidase motif is in shadow. The amino acid sequence of APN1t is in italics.
Isolation of full-length AaeAPN1 and its analysis
Since AaeAPN1 was identified as a putative Cry11Aa toxin receptor, isolation of the full-length AaeAPN1 transcript was carried out. Because this gene is predicted to have two transcripts, AAEL012778-RA and AAEL012778-RB, we performed 3′- and 5′-end amplifications. The 3′-product confirmed AAEL012778-RA as the transcript for the 140 kDa protein we isolated. Since 5′-RACE was not successful an alternative amplification of the 5′ end was performed. We used a forward gene-specific primer designed to a sequence between the predicted transcript start site and putative translation start condon together with a gene-specific reverse primer from the putative AaeAPN1 ORF. A 0.3 kb PCR product was obtained and the sequence of this PCR product confirmed the 5′ end annotation for this gene. Because the full-length ORF could not be obtained by PCR using primers designed to the known 5′ and 3′ sequences, two partial overlapping fragments with a unique restriction site, NdeI, were amplified and finally assembled as a full-length ORF gene of AaeAPN1 (Fig. 2A).
The 3003 bp ORF encodes a full-length AaeAPN1 protein of 112kDa with a consensus zinc binding/catalytic site (gluzincin aminopeptidase motif), GAMENWGX3-YRE-X23-HE- X2-H-X18-E (where X is any amino acid) (Hooper, 1994). The N-terminal sequences contain a putative 22 amino acid signal peptide predicted by SignalP. The C-terminus has a predicted GPI-anchored site with 970Gly a predicted ω-site seven amino acids after a hydrophobic tail. Additionally, there are three consensus Asn-Xaa-Ser/Thr sequences, indicating possible N-glycosylation sites in the protein. Also, 248Thr was predicted as a potential O-glycosylation residue (Fig. 2B).
The AAEL012778 gene contains five exons, giving rise to AAEL012778-RA. The predicted AAEL012778-RB transcript arises from a lack of splicing the intron between exons 4 and 5 lead to a premature stop codon, and is likely due to incorrect annotation. Orthologues proteins in An. gambiae and C. pipiens appear to be AGAP004809-PA and CPIJ001048, respectively. The AGAP004809 gene also has five exons in An. gambiae.
Prokaryotic and eukaryotic expression of AaeAPN1
A partial C-terminal variable region consisting of amino acids 525–778 of the full-length AaeAPN1, named AaeAPN1t, was expressed in E. coli as a 6×His-tagged protein, purified and then used as an antigen for antibody development. Using these antibodies we detected in Aedes BBMV immunoreactive bands of 140, 65, 45 and 32 kDa (Fig. 3A, lane 1). The 140 kDa protein likely represents a fully glycosylated form while the lower molecular bands are likely degradation products. In Sf12 cells infected with recombinant baculovirus containing full-length APN1 the anti-APN1 antibody detected a 110 kDa band (Fig. 3A, lane 2). This size protein was not detected in cells infected with vector alone (Fig. 3A, lane 3), showing the relative specificity of the APN1 antibody. Because we anticipated a protein band of higher molecular mass, we also excised the 110 kDa band for analysis using nano-UPLC/MS/MS. Data from this analysis confirmed that the 110 kDa expressed protein is indeed AAEL012778 (data not shown). Differences in molecular mass of the Sf21 expressed APN1 and that in BBMV are likely due to differences in posttranslational modifications. It is not unusual to observe large differences in the molecular sizes of a differentially glycoslyated APN, as observed with receptor A of Heliothis virescens that is expressed as 130 and 170 kDa proteins in BBMV (Luo et al., 1997; Oltean et al., 1999).
Figure 3. Characterization of Sf21 cells-expressed AaeAPN1.
A. Proteins from BBMV (lane 1), Sf21 cell-expressed (lane 2), control Sf21 cells containing vector alone (lane 3) and E. coli-expressing truncated AaeAPN1 were separated by SDS-PAGE and electrotransferred to PVDF membrane. Proteins were detected using anti-AaeAPN1 antibody. B. After the proteins of Sf21 cells transfected with recombinant baculovirus containing the AaeAPN1 cDNA (lane 5) or vector alone (lane 6) were transferred to PVDF membranes, the blot was treated with 0.6 units of PI-PLC. The presence of GPI anchor was detected by anti-CRD antibody (1:200).
Substrate utilization by AaeAPN1 is summarized in Table 3. When expressed in Sf21 cells it had leucine and alanine aminopeptidase activity but did not utilize valine as a substrate. Membrane fractions from Sf21 cells infected with vector alone had residual aminopeptidase activity that utilizes all three substrates at low levels. Not surprisingly enzymatic levels in APN1 Sf21 cells were substantially lower than those observed in midgut BBMV where expression of a number of aminopeptidases is known (Table 2 and 4). Clearly, leucine, alanine and valine aminopeptidase activity in BBMV represents the total activities of all APNs, and not only the activity of AaeAPN1. Collectively these results indicate Sf21cells-expressed AaeAPN1 was catalytically active. Similarly, APN proteins from several lepidopteran insects expressed in different insect cell lines were also shown to have APN activity (Banks et al., 2003; Luo et al., 1999; Rajagopal et al., 2003; Simpson and Newcomb, 2000; Sivakumar et al., 2007).
Table 3.
Aminopeptidase activity of Aedes BBMV and Sf21 cell-expressed AaeAPN1
| Alanine p- nitroanilide | Leucine p- nitroanilide | Valine p- nitroanilide | |
|---|---|---|---|
| BBMV | 119 ± 5.5c | 1620 ± 9.2c | 168.0 ± 4.6b |
| AaeAPN1 | 36.7 ± 1.5b | 75.3 ± 3.2b | 13.8 ± 2.4a |
| Sf21 | 12.7 ± 1.2a | 12.2 ± 0.9a | 12.2 ± 0.9a |
Activities are expressed as nmol p-nitroanilide formed/min/mg protein. Values followed by different letters within each column show significant differences at P<0.05.
Table 4.
Mass identification of eluated proteins from Aedes Aegypti BBMV by wheat germ agglutinin (WGA) agarose pull-down assays
| NCBI no. | VectorBase accession no | Annotation | Predicted Mr, kDa | Additional features |
|---|---|---|---|---|
| gi_157133543 | AAEL012778 | zinc metalloprotease (AaeAPN1) | 112 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_157118050 | AAEL008155 | zinc metalloprotease (AaeAPN2) | 104 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_157133541 | AAEL012774 | zinc metalloprotease (AaeAPN3) | 102 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_157126493 | AAEL010532 | Alpha-amylase | 69 | Alpha-amylase catalytic domain |
| gi_157133539 | AAEL012776 | zinc metalloprotease | 112 | GPI-anchor, with a consensus gluzincin aminopeptidase motif |
| gi_47679567 | AAEL008600 | carboxypeptidase A | 58 | Belongs to the carboxypeptidase A family |
| gi_47679581 | AAEL001840 | carboxypeptidase B | 48 | Belongs to the carboxypeptidase B family |
| gi_157135663 | AAEL003298 | alkaline phosphatase | 58 | Exists as a dimer, each monomer binding two zinc atoms and one magnesium atom, which are essential for enzymatic activity |
To determine if AaeAPN1 expressed in Sf21 cells contains a GPI-anchor, an anti-CRD antibody that detects GPI-anchors was used. After phosphatidylinositol-specific phospholipase C (PI-PLC) digestion AaeAPN1 was recognized by the antibody (Fig. 3B, lane 5), which suggests AaeAPN1 expressed in Sf21 cells is GPI-anchored as predicted. However, Sf21 cells-expressed AaeAPN1 was not detected by biotinylated concanavalin A (Con A), soybean agglutinin (SBA), WGA (data not shown), or by ligand blot or toxin pull-down assays using Cry11Aa (data not shown). Therefore, it was not surprising that Sf21 cells expressing AaeAPN1 were not susceptible to Cry11Aa toxin (data not shown).
Expression of AaeAPN1 in larval midgut of Aedes aegypti
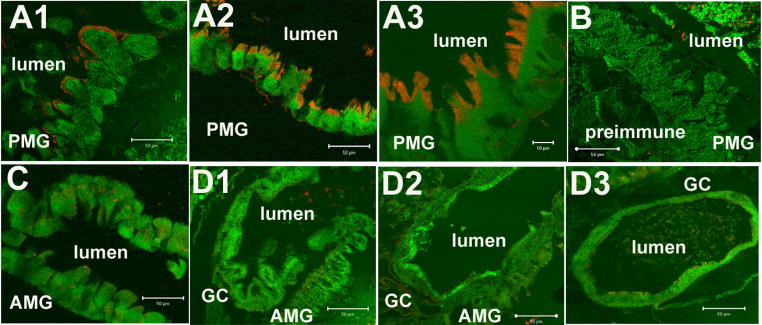
Rabbit-anti-AaeAPN1 polyclonal antibody purified by protein A was used to analyze the expression and distribution of AaeAPN1 in fourth instar larval tissue sections. As shown in Fig. 4A there is intense immunofluorescence (Cy3, red) in the apical side of posterior midgut (PMG) epithelial cells in larval (A1) and gut sections (A2, A3). No AaeAPN1 expression was observed in anterior midgut (Fig. 4C) and gastric caeca cells (D1-D3). Also, no immunofluorescence was found in control when tissues were incubated with preimmune antiserum (1:100 dilution) (Fig. 4B). The antibody is relatively specific since it detects only a single major band in Sf21 cells expressing AaeAPN1 (Fig. 3, lane 2).
Figure 4. AaeAPN1 protein is immunolocalized to the apical membrane of posterior midgut epithelial cells of early fourth instar Ae. aegypti larvae.
Intense immunofluorescence signal (Cy3, red) was observed in apical side of posterior midgut (PMG) epithelial cells. Panels A1, A2 and A3, PMG at different magnifications; A3 – x100, A1, A2 – x40. No immunofluorescence was found in control PMG using preimmune antibody (panel B –x40). No immunoreactivity was observed in anterior midgut (panel C – AMG, x40) and gastric caeca cells (GC - D1, D2 and D3, x40). Panels A2, A3 and C – are gut sections, while other images are from whole fourth instar larvae sections.
Glycosylation status of BBMV AaeAPN1
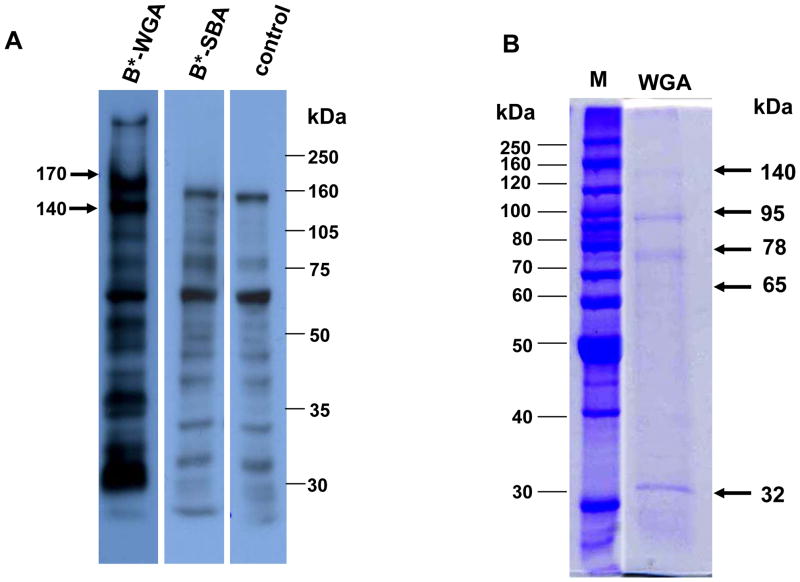
Lectin blots showed 170 and 140 kDa proteins were detected by biotinylated WGA but not by biotinylated SBA (Fig. 5A). Therefore, a pull-down assay using WGA agarose was performed and proteins eluted with acetylglucosamine were resolved by SDS-PAGE. Fig. 5B shows that five protein bands of 140, 95, 78, 65 and 30 kDa were obtained. The top two bands showed a similar molecular size to proteins eluted from the biotinylated Cry11Aa pull-downs. Consequently all five protein bands were excised and analyzed by mass spectrometry. As expected, the 140 and 95 kDa bands were confirmed to be the same proteins as those pulled down using biotinylated Cry11Aa (Table 4). We are uncertain why the 170 kDa protein was not pulled down using WGA agarose since it was observed in lectin blots. The anti-APN1 antibodies do not detect this size band (Fig. 3A) and we do not know whether it is an APN. The 78 and 65 kDa proteins were identified as amylase-like protein and zinc metalloprotease, respectively, but the 32 kDa band was identified to have three proteins: carboxypeptidases A and B and an alkaline phosphatase (ALP) (Table 4). However, the observed sizes of 78 kDa and 32 kDa in the gel were smaller than the predicted molecular sizes of the identified proteins. Among them, the 65 kDa zinc metalloprotease was recently reported as a Cry4Ba toxin-binding protein by proteomic identification and displayed a mass of around 70 kDa in 2-D gel (Bayyareddy et al., 2009). The size discrepancy observed here and in the published report could be caused by proteolytic degradation during sample preparation and/or storage of dissected midguts.
Figure 5. Detection of BBMV protein glycosylation and analysis of pull-down of BBMV proteins using wheat germ agglutinin (WGA).
A. Following separation of BBMV proteins bys SDS-PAGE, nitrocellulose membranes were incubated with biotinylated WGA (lane 1), biotinylated soybean agglutinin (SBA) (lane 2) or no lectin (lane 3). Bound biotinylated lectins were detected with strepavidin-horseradish peroxidase conjugate (1:10,000 dilution) and visualized by luminol. Arrow indicates expected molecular size of AaeAPN1. B. Analysis of WGA agarose pull-down and detected by Coomassie-blue staining (lane 5). Arrows indicate proteins that were analyzed by mass spectrometry. Molecular markers are in lane 4.
Determination of binding affinity between truncated AaeAPN1fragment and Cry11Aa toxin
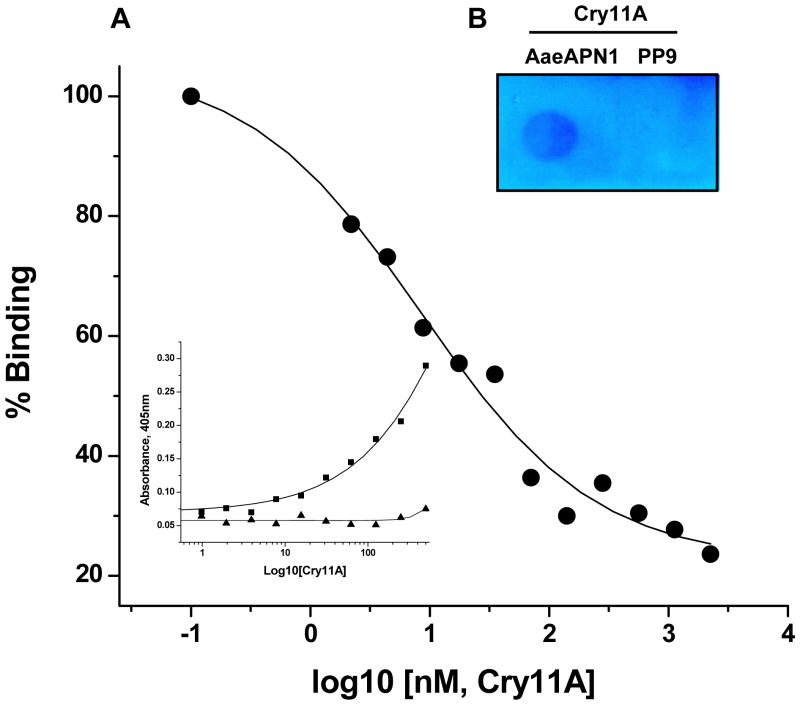
The AaeAPN1t fragment was found to bind Cry11Aa toxin in an immunoassay (Fig 6A) and in a dot blot assay (Fig 6B), but not in a toxin overlay assay using SDS-PAGE separated proteins (data not shown). To measure Cry11Aa affinity to AaeAPN1t, competitive ELISAs were performed. As shown in Figure 6A (insert), the AaeAPN1t protein binds Cry11Aa toxin in a dose-dependent manner. Moreover, a H. virescens cadherin fragment, PP9, that binds the Cry1A toxins, did not bind Cry11Aa even at high concentration levels. AaeAPN1t, 80 nM, that binds Cry11Aa in the linear range was then used for subsequent competitive ELISAs. In competitive ELISA increasing Cry11Aa concentrations (0.1–1000nM) displaced AaeAPN1 from binding immobilized Cry11Aa (Figure 6A). The apparent dissociation constant (Kd) for AaeAPN1t binding to Cry11Aa toxin was 8.5 nM.
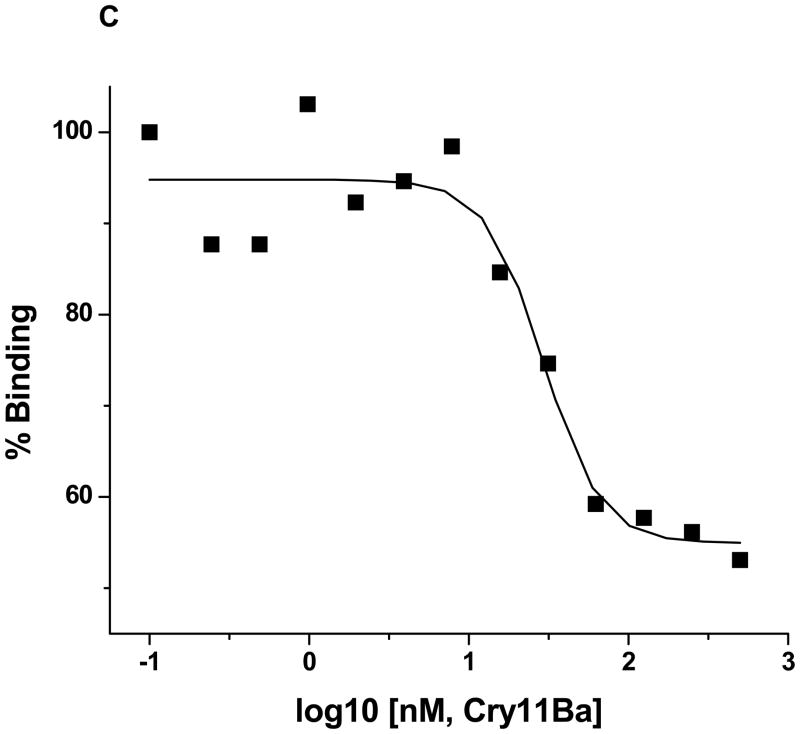
Figure 6. Cry11Aa and Cry11Ba toxins bind truncated AaeAPN1 fragment with high affinity.
A. The insert shows truncated AaeAPN1t fragment shows dose-dependent binding (solid squares) to immobilized Cry11Aa (0.4μg), but a cadherin fragment, PP9, from the Heliothis virescens cadherin (Xie et al., 2005) (solid triangles) does not bind Cry11Aa. The binding affinity, 8.5 nM, of Cry11Aa toxin to truncated AaeAPN1 fragment was determined in a competition assay using 80 nM AaeAPN1 with increasing concentrations of Cry11Aa toxin (0.1–1000 nM). Maximal binding was normalized to the maximal absorbance obtained in the absence of AaeAPN1. B. Cry11Aa binds to truncated AaeAPN1 expressed in E. coli but not to the PP9 cadherin fragment. Forty μg of solubilized AaeAPN1t and PP9 were dotted onto a PVDF filter. Cry11Aa binds AaeAPN1t but not the PP9 cadherin fragment from H. virescens. C. Mosquitocidal Cry11Ba can partially compete with the Cry11Aa binding site on the Aedes AaeAPN1. Binding assays were performed as described in Materials and Methods.
B. thuringiensis subsp. jegathesan produces a highly mosquitocidal toxin, Cry11Ba (Delecluse et al., 1995; Kawalek et al., 1995). This toxin can compete with Cry11Aa binding but at a higher concentration (Fig. 6C), suggesting it has a lower affinity to AaeAPN1 than does Cry11Aa.
Discussion
Availability of the complete Ae. aegypti genome enabled us to utilize a proteomic approach to identify Cry11Aa binding proteins. Thus biotinylated Cry11Aa toxin was used as bait in pull-down experiments performed with BBMV prepared from the midgut of larval Aedes. With the exception of actin all pulled down proteins were identified as APNs, already characterized as functional receptors in lepidopteran insects. One of the proteins identified, AaeAPN1, was then cloned and expressed for further analysis. Both Sf21 cell-expressed AaeAPN1 and that in BBMV were characterized for enzymatic activity, and toxin and lectin binding. Also, Cry11Aa toxin bound this APN with high affinity. Characterization of toxin binding to AaeAPN2 is under investigation.
A similar pull-down assay using biotinylated Cry11Aa was done by Fernadez et al., (2006), however, BBMV proteins were cleaved with PI-PLC before use. In this case a 65 kDa protein and trace amounts of a 62 kDa protein were eluted by Cry11Aa affinity chromotogragphy. No APN proteins were observed. Here biotinylated Cry11Aa was incubated with BBMV directly then solubilized and the toxin-protein complexes isolated using streptavidin agarose. Thus differences in the methods used probably account for the separate identification of APN and ALP proteins. In addition, use of BBMV here allowed for interaction of Cry11Aa with proteins and complexes found in membranes. Thus actin was also identified as a Cry11Aa interacting protein (Fig. 1). Binding of actin to Cry toxins has been observed in a number of different insects through proteomic analysis (Bayyareddy et al., 2009; Krishnamoorthy et al., 2007; McNall and Adang, 2003). It is possible that after toxin inserts into cell membranes, it may interact with actin, which could disrupt cytoskeletal links causing subsequent cell death (Montell, 2006).
Both cadherin and ALP proteins are expressed in the apical side of distal and proximal caeca and apical regions of posterior midgut of larvae Ae. aegypti ((Fernandez et al., 2006); Chen, J., Aimanova, K. and Gill S. S., unpublished). Of note these are the same tissues which bind Cry toxins from subsp. israelensis (Ravoahangimalala et al., 1993) and show subsequent toxin pathogenicity (Charles and de Barjac, 1983; Zalunin et al., 2002). However, AaeAPN1 was localized in the apical side of posterior midgut epithelial cells, but not in anterior midgut and gastric caeca cells (Fig. 4). In contrast AaeAPN2 was expressed not in posterior midgut and anterior midgut cells but in gastric caeca cells (Chen, J., Aimanova, K. and Gill S. S., unpublished). Interestingly, distribution of these two APNs collectively is consistent with those of cadherin and ALP proteins.
These distribution patterns of Cry11Aa toxin binding proteins suggest the sequential model of Cry toxin action observed in lepidopterans (Bravo et al., 2004; Soberon et al., 2009) also is likely observed in dipteran insects. It is probable that mosquitocidal Cry toxins initially bind cadherin as monomeric toxins. Subsequently upon oligomerization the toxin bind to a second GPI-anchored receptor as APN or ALP that allows for enrichment in lipid rafts (Bravo et al., 2004; Zhuang et al., 2002) and subsequent membrane insertion and pore formation (Bravo et al., 2007).
There are, however, some important differences in Cry toxin action in mosquitoes. For example, Cry11Ba, a toxin related to Cry11Aa, binds an APN with high affinity in two anopheline mosquitoes (Abdullah et al., 2006; Zhang et al., 2009). In this study, Cry11Aa toxin also binds AaeAPN1 with high affinity (Fig. 6). These observations suggest Cry11 toxins could bind APNs directly without prior binding to cadherin and oligomerization. If these initial observations are further validated in mosquitoes, this evidence would suggest there is additional complexity to the mechanism of action of lepidopteran-specific toxins.
In this study, attempts to test if AaeAPN1 is involved in toxin action were unsuccessful. In large part this was because soluble Cry11Aa has very low toxicity making it difficult to have a significant excess of the anti-AaeAPN1 antibody or the AaeAPN1t peptide. Furthermore, a decrease in Cry11Aa toxicity was also observed in control experiments when pre-immune serum or inclusion bodies of a partial Heliothis virescens cadherin fragment, PP9 (1:100 mass ratio) (Xie et al., 2005) were used, indicating attenuation of Cry11Aa toxicity was not specific to anti-AaeAPN1 or the AaeAPN1t peptide. In contrast Cry11Ba is significantly more toxic and such experiments are possible as shown for A. gambiae AgAPN2 (Zhang et al., 2008). Since RNAi techniques for Ae. aegypti larva are now under investigation by other investigators these methods will likely provide more reliable data.
AaeAPN1 expressed in E. coli binds Cry11Aa by dot blot and competitive ELISA (Fig. 6A), which indicates AaeAPN1-Cry11Aa interaction is glycan-independent and one of the Cry11Aa binding regions is localized to amino acids 525–778 of APN1. This region is in the C-terminal of AaeAPN1 and it differs from that observed in B. mori APN (Nakanishi et al., 2002; Yaoi et al., 1999), which is localized in the N-terminal region. Surprisingly, Sf21 cells-expressed AaeAPN1 failed to bind to Cry11Aa either in denatured conditions or under native conditions. The reason for this discrepancy is unclear and requires further investigation, but potentially improper glycosylation could mask the glycan-independent toxin binding site. Sf21 cells-expressed AaeAPN1 is a membrane bound GPI-anchored protein (Fig. 3B) and has APN enzymatic activity (Table 3). But its molecular weight is lower than that observed in BBMV (Fig. 3A), suggesting either a lack of or different glycosylation (data not shown). It indicates that posttranslational modification in Sf21 cells might be different from that in epithelial cells of Aedes midguts.
Sf21 cells-expressing AaeAPN1 were not susceptible to monomeric Cry11Aa toxin (data not shown). This finding suggests as indicated above that Cry11Aa binds sequentially to cadherin and to a GPI-anchored APN or ALP, which would mean that oligomeric toxins could potentially be toxic to these Sf21 cells. So far, testing of APN function using cell expression has been relatively unsuccessful because obtaining proper glycosylation seems to be the main obstacle (Banks et al., 2003; Garner et al., 1999; Luo et al., 1997; Pigott and Ellar, 2007). An exception is demonstration of Cry1Ac toxicity to Sf21 cells expressing Helicoverpa armigera APN1 (Sivakumar et al., 2007). In vivo assays, as the use of transgenic techniques (Gill and Ellar, 2002) and silencing of target gene by corresponding dsRNA (Rajagopal et al., 2002; Sivakumar et al., 2007), have instead been more reliable.
Mosquitoes exposed to only Cry11Aa develop resistance after selection for 16–20 generations with higher levels of resistance observed after further selection. Resistance development was slower when this toxin was used in combination with the Cry4A and Cry4B toxins (Georghiou and Wirth, 1997). The mosquitoes also developed resistance when only the Cry4A and Cry4B toxins were used (Georghiou and Wirth, 1997). Some Cry1A resistance observed in lepidopteran larvae is associated with mutations or deletions in APN (Herrero et al., 2005; Zhang et al., 2009), so it is possible that changes in APN genes can also lead to Cry11Aa resistance observed in Culex.
Acknowledgments
We appreciate the gift of baculovirus expression vectors provided by Professor J. A. Traugh, Department of Biochemistry, University of California Riverside. Research was funded in part through grants from the National Institutes of Health, 1R01 AI066014 and the University of California Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdullah MA, Valaitis AP, Dean DH. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Mizuno E, Hara H, Nakanishi K, Kitami M, Miura N, Tabunoki H, Watanabe A, Sato R. Location of the Bombyx mori aminopeptidase N type 1 binding site on Bacillus thuringiensis Cry1Aa toxin. Appl Environ Microbiol. 2005;71:3966–3977. doi: 10.1128/AEM.71.7.3966-3977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DJ, Hua G, Adang MJ. Cloning of a Heliothis virescens 110 kDa aminopeptidase N and expression in Drosophila S2 cells. Insect Biochem Mol Biol. 2003;33:499–508. doi: 10.1016/s0965-1748(03)00022-5. [DOI] [PubMed] [Google Scholar]
- Bayyareddy K, Andacht TM, Abdullah MA, Adang MJ. Proteomic identification of Bacillus thuringiensis subsp. israelensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera, Culicidae) larvae. Insect Biochem Mol Biol. 2009;39:279–286. doi: 10.1016/j.ibmb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zaritsky A, Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2002;68:5082–5095. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gill SS, Soberon M. Bacillus thuringiensis: Mechanisms and Use. In: Gilbert LI, Kostas I, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 6. Elsevier; 2005. pp. 175–205. [Google Scholar]
- Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Burton SL, Ellar DJ, Li J, Derbyshire DJ. N-acetylgalactosamine on the putative insect receptor aminopeptidase N is recognised by a site on the domain III lectin-like fold of a Bacillus thuringiensis insecticidal toxin. J Mol Biol. 1999;287:1011–1022. doi: 10.1006/jmbi.1999.2649. [DOI] [PubMed] [Google Scholar]
- Chang C, Yu YM, Dai SM, Law SK, Gill SS. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, de Barjac H. Action of crystals of Bacillus thuringiensis var. israelensis on the midgut of Aedes aegypti L. larvae, studied by electron microscopy. Ann Microbiol (Paris) 1983;134A:197–218. [PubMed] [Google Scholar]
- Chilcott CN, Ellar DJ. Comparative toxicity of Bacillus thuringiensis var. israelensis crystal proteins in vivo and in vitro. J Gen Microbiol. 1988;134:2551–2558. doi: 10.1099/00221287-134-9-2551. [DOI] [PubMed] [Google Scholar]
- Cowles EA, Yunovitz H, Charles JF, Gill SS. Comparison of toxin overlay and solid-phase binding assays to identify diverse CryIA(c) toxin-binding proteins in Heliothis virescens midgut. Appl Environ Microbiol. 1995;61:2738–2744. doi: 10.1128/aem.61.7.2738-2744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai SM, Gill SS. In vitro and in vivo proteolysis of the Bacillus thuringiensis subsp. israelensis CryIVD protein by Culex quinquefasciatus larval midgut proteases. Insect Biochem Mol Biol. 1993;23:273–283. doi: 10.1016/0965-1748(93)90008-g. [DOI] [PubMed] [Google Scholar]
- Delecluse A, Bourgouin C, Klier A, Rapoport G. Specificity of action on mosquito larvae of Bacillus thuringiensis israelensis toxins encoded by two different genes. Mol Gen Genet. 1988;214:42–47. doi: 10.1007/BF00340177. [DOI] [PubMed] [Google Scholar]
- Delecluse A, Rosso ML, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. 1995;61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Chen S, Bartsch U, Schachner M. Generation of affinity matured scFv antibodies against mouse neural cell adhesion molecule L1 by phage display. Biochem Biophys Res Commun. 2003;301:60–70. doi: 10.1016/s0006-291x(02)02933-9. [DOI] [PubMed] [Google Scholar]
- Fernandez LE, Aimanova KG, Gill SS, Bravo A, Soberon M. A GPI- anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegypti larvae. Biochem J. 2006;394:77–84. doi: 10.1042/BJ20051517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner KJ, Hiremath S, Lehtoma K, Valaitis AP. Cloning and complete sequence characterization of two gypsy moth aminopeptidase-N cDNAs, including the receptor for Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol. 1999;29:527–535. doi: 10.1016/s0965-1748(99)00027-2. [DOI] [PubMed] [Google Scholar]
- Georghiou GP, Wirth MC. Influence of exposure to single versus multiple toxins of Bacillus thuringiensis subsp. israelensis on development of resistance in the mosquito Culex quinquefasciatus (Diptera: Culicidae) Appl Environ Microbiol. 1997;63:1095–1101. doi: 10.1128/aem.63.3.1095-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M, Ellar D. Transgenic Drosophila reveals a functional in vivo receptor for the Bacillus thuringiensis toxin Cry1Ac1. Insect Mol Biol. 2002;11:619–625. doi: 10.1046/j.1365-2583.2002.00373.x. [DOI] [PubMed] [Google Scholar]
- Gill SS, Cowles EA, Francis V. Identification, isolation, and cloning of a Bacillus thuringiensis CryIAc toxin-binding protein from the midgut of the lepidopteran insect Heliothis virescens. J Biol Chem. 1995;270:27277–27282. doi: 10.1074/jbc.270.45.27277. [DOI] [PubMed] [Google Scholar]
- Herrero S, Gechev T, Bakker PL, Moar WJ, de Maagd RA. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four Aminopeptidase N genes. BMC Genomics. 2005;6:96. doi: 10.1186/1471-2164-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Hua G, Zhang R, Abdullah MA, Adang MJ. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochemistry. 2008;47:5101–5110. doi: 10.1021/bi7023578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JL, Lee MK, Valaitis AP, Curtiss A, Dean DH. Bivalent sequential binding model of a Bacillus thuringiensis toxin to gypsy moth aminopeptidase N receptor. J Biol Chem. 2000;275:14423–14431. doi: 10.1074/jbc.275.19.14423. [DOI] [PubMed] [Google Scholar]
- Kang’ethe W, Aimanova KG, Pullikuth AK, Gill SS. NHE8 mediates amiloride-sensitive Na+/H+ exchange across mosquito Malpighian tubules and catalyzes Na+ and K+ transport in reconstituted proteoliposomes. Am J Physiol Renal Physiol. 2007;292:F1501–1512. doi: 10.1152/ajprenal.00487.2005. [DOI] [PubMed] [Google Scholar]
- Kawalek MD, Benjamin S, Lee HL, Gill SS. Isolation and Identification of novel toxins from a new mosquitocidal isolate from Malaysia, Bacillus thuringiensis subsp. jegathesan. Appl Environ Microbiol. 1995;61:2965–2969. doi: 10.1128/aem.61.8.2965-2969.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PJ, Knowles BH, Ellar DJ. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy M, Jurat-Fuentes JL, McNall RJ, Andacht T, Adang MJ. Identification of novel Cry1Ac binding proteins in midgut membranes from Heliothis virescens using proteomic analyses. Insect Biochem Mol Biol. 2007;37:189–201. doi: 10.1016/j.ibmb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Lereclus D, Agaisse H, Gominet M, Chaufaux J. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis spo0A mutant. Biotechnology (N Y) 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- Likitvivatanavong S, Aimanova K, Gill SS. Loop residues of the receptor binding domain of Bacillus thuringiensis Cry11Ba toxin are important for mosquitocidal activity. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, McLachlin JR, Brown MR, Adang MJ. Expression of a glycosylphosphatidylinositol-linked Manduca sexta aminopeptidase N in insect cells. Protein Expr Purif. 1999;17:113–122. doi: 10.1006/prep.1999.1122. [DOI] [PubMed] [Google Scholar]
- Luo K, Sangadala S, Masson L, Mazza A, Brousseau R, Adang MJ. The Heliothis virescens 170 kDa aminopeptidase functions as “receptor A” by mediating specific Bacillus thuringiensis Cry1A delta-endotoxin binding and pore formation. Insect Biochem Mol Biol. 1997;27:735–743. doi: 10.1016/s0965-1748(97)00052-0. [DOI] [PubMed] [Google Scholar]
- Masson L, Lu YJ, Mazza A, Brousseau R, Adang MJ. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J Biol Chem. 1995;270:20309–20315. doi: 10.1074/jbc.270.35.20309. [DOI] [PubMed] [Google Scholar]
- McNall RJ, Adang MJ. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem Mol Biol. 2003;33:999–1010. doi: 10.1016/s0965-1748(03)00114-0. [DOI] [PubMed] [Google Scholar]
- Montell DJ. A kinase gets caspases into shape. Cell. 2006;126:450–452. doi: 10.1016/j.cell.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Yaoi K, Nagino Y, Hara H, Kitami M, Atsumi S, Miura N, Sato R. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella -- their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 2002;519:215–220. doi: 10.1016/s0014-5793(02)02708-4. [DOI] [PubMed] [Google Scholar]
- Nielsen-Leroux C, Charles JF. Binding of Bacillus sphaericus binary toxin to a specific receptor on midgut brush-border membranes from mosquito larvae. Eur J Biochem. 1992;210:585–590. doi: 10.1111/j.1432-1033.1992.tb17458.x. [DOI] [PubMed] [Google Scholar]
- Oltean DI, Pullikuth AK, Lee HK, Gill SS. Partial purification and characterization of Bacillus thuringiensis Cry1A toxin receptor A from Heliothis virescens and cloning of the corresponding cDNA. Appl Environ Microbiol. 1999;65:4760–4766. doi: 10.1128/aem.65.11.4760-4766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C, Fernandez LE, Sun J, Folch JL, Gill SS, Soberon M, Bravo A. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci U S A. 2005;102:18303–18308. doi: 10.1073/pnas.0505494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255–281. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R, Agrawal N, Selvapandiyan A, Sivakumar S, Ahmad S, Bhatnagar RK. Recombinantly expressed isoenzymic aminopeptidases from Helicoverpa armigera (American cotton bollworm) midgut display differential interaction with closely related Bacillus thuringiensis insecticidal proteins. Biochem J. 2003;370:971–978. doi: 10.1042/BJ20021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. Silencing of midgut aminopeptidase N of Spodoptera litura by dsRNA establishes its role as BT toxin receptor. J Biol Chem. 2002;107:1. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- Ravoahangimalala O, Charles JF, Schoeller-Raccaud J. Immunological localization of Bacillus thuringiensis serovar israelensis toxins in midgut cells of intoxicated Anopheles gambiae larvae (Diptera: Culicidae) Res Microbiol. 1993;144:271–278. doi: 10.1016/0923-2508(93)90011-p. [DOI] [PubMed] [Google Scholar]
- Sangadala S, Walters FS, English LH, Adang MJ. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)- K+ efflux in vitro. J Biol Chem. 1994;269:10088–10092. [PubMed] [Google Scholar]
- Simpson RM, Newcomb RD. Binding of Bacillus thuringiensis delta-endotoxins Cry1Ac and Cry1Ba to a 120-kDa aminopeptidase-N of Epiphyas postvittana purified from both brush border membrane vesicles and baculovirus-infected Sf9 cells. Insect Biochem Mol Biol. 2000;30:1069–1078. doi: 10.1016/s0965-1748(00)00082-5. [DOI] [PubMed] [Google Scholar]
- Sivakumar S, Rajagopal R, Venkatesh GR, Srivastava A, Bhatnagar RK. Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J Biol Chem. 2007;282:7312–7319. doi: 10.1074/jbc.M607442200. [DOI] [PubMed] [Google Scholar]
- Soberon M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66:1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Zhuang M, Ross LS, Gomez I, Oltean DI, Bravo A, Soberon M, Gill SS. Single amino acid mutations in the cadherin receptor from Heliothis virescens affect its toxin binding ability to Cry1A toxins. J Biol Chem. 2005;280:8416–8425. doi: 10.1074/jbc.M408403200. [DOI] [PubMed] [Google Scholar]
- Yamagiwa M, Ogawa R, Yasuda K, Natsuyama H, Sen K, Sakai H. Active form of dipteran-specific insecticidal protein cryllA produced by Bacillus thuringiensis subsp. israelensis. Biosci Biotechnol Biochem. 2002;66:516–522. doi: 10.1271/bbb.66.516. [DOI] [PubMed] [Google Scholar]
- Yaoi K, Kadotani T, Kuwana H, Shinkawa A, Takahashi T, Iwahana H, Sato R. Aminopeptidase N from Bombyx mori as a candidate for the receptor of Bacillus thuringiensis Cry1Aa toxin. Eur J Biochem. 1997;246:652–657. doi: 10.1111/j.1432-1033.1997.t01-1-00652.x. [DOI] [PubMed] [Google Scholar]
- Yaoi K, Nakanishi K, Kadotani T, Imamura M, Koizumi N, Iwahana H, Sato R. Bacillus thuringiensis Cry1Aa toxin-binding region of Bombyx mori aminopeptidase N. FEBS Lett. 1999;463:221–224. doi: 10.1016/s0014-5793(99)01626-9. [DOI] [PubMed] [Google Scholar]
- Zalunin IA, Chaika S, Dronina MA, Revina LP. Cytopathological effect of Bacillus thuringiensis israelensis endotoxins on the intestines of Aedes aegypti mosquito larvae. Parazitologiia. 2002;36:337–344. [PubMed] [Google Scholar]
- Zhang R, Hua G, Andacht TM, Adang MJ. A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochemistry. 2008;47:11263–11272. doi: 10.1021/bi801181g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cheng H, Gao Y, Wang G, Liang G, Wu K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol. 2009 doi: 10.1016/j.ibmb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Oltean DI, Gomez I, Pullikuth AK, Soberon M, Bravo A, Gill SS. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J Biol Chem. 2002;277:13863–13872. doi: 10.1074/jbc.M110057200. [DOI] [PubMed] [Google Scholar]