Abstract
The Wnt/β-catenin pathway plays an important role in the initiation in most, if not all, colon cancers. Prior work has provided important insights into the regulation of β-catenin stability in the cytoplasm; however, relatively little is known regarding the mechanism by which β-catenin activates gene transcription in the nucleus. Using genetic approaches, studies in human colon cancers and Drosophila have identified CDK8 as a colon cancer oncogene that regulates β-catenin transcriptional activity. These convergent observations provide new insights into the regulation of nuclear β-catenin activity and identify a novel therapeutic target for β-catenin-driven malignancies.
Keywords: CDK8, colon cancer, β-catenin, Mediator complex, genomics
Introduction
β-catenin is the central nuclear effector of the Wg (wingless) and Int (Wnt) signaling pathway and resides in two intracellular pools. In the cytoplasm, a destruction complex containing APC, AXIN and GSK3 phosphorylates β-catenin, which leads to its ubiquitination by β-TrCP and proteasomal degradation. Under physiological conditions, the binding of Wnt ligands to Frizzled (Fzd)-LRP5/6 receptor complexes activates a signaling cascade that inactivates the destruction complex and leads to stabilization of β-catenin. Once stabilized, β-catenin accumulates and translocates to the nucleus, where it participates in the transcriptional activation of target genes, such as MYC and CCND1.
Dysregulation of the Wnt/β-catenin pathway plays a central role in the initiation of colon cancer and has been implicated in other cancers, such as ovarian, liver and skin cancer. In particular, patients afflicted with the colon cancer syndrome Familial Adenomatous Polyposis (FAP) harbor germline loss-of-function mutations in the APC tumor suppressor gene. In addition, APC mutations are also found in 85% of sporadic colorectal cancers. Mice that harbor such APC mutations, such as the APCmin (multiple intestinal neoplasia) mouse, develop intestinal adenomas with high penetrance, demonstrating the key role of APC and its regulation of β-catenin in colon cancer initiation.
β-catenin can also be activated by mutations in the N-terminus of β-catenin that abrogate phosphorylation sites that target the β-catenin protein for degradation and lead to increased β-catenin stability. Although the mechanisms that lead to stabilization of the cytoplasmic pool of β-catenin and its translocation into the nucleus have been intensively studied, our understanding of the mechanism(s) by which β-catenin activates target gene transcription in the nucleus remains incomplete. This review will focus on recent work that provides new insights into the regulation of oncogenic β-catenin transcriptional activity in the nuclear compartment.
The regulation of nuclearβ-catenin activity
In the nucleus, β-catenin interacts with the TCF/LEF transcription factors to drive the transcription of Wnt target genes. Although it is clear that nuclear translocation of β-catenin is necessary for its oncogenic activity, accumulating evidence suggests that other interactions regulate the activity of nuclear β-catenin during cell transformation. For example, Phelps et al. recently showed that loss of APC leads to a differentiation defect that correlates with stabilization of β-catenin but that a second event, sometimes involving KRAS, is required for proliferation and β-catenin nuclear localization (1). Consistent with these findings, Rac1 signaling has been shown to be required for nuclear localization of β-catenin (2). Moreover, recent work has shown that the interaction of β-catenin with specific TCF/LEF family members affects DNA binding specificity and target gene expression (3). In addition, other proteins such as BCL9 (4) and transcriptional activators such as PYGO (5) and the Mediator complex (6) affect the transcriptional activity of β-catenin/TCF/LEF complexes.
In addition to these protein-protein interactions, the activity of nuclear β-catenin is regulated through post-translational modifications. While the role of phosphorylation in β-catenin signaling has been extensively studied in the context of GSK3-dependent regulation of the destruction complex to regulate β-catenin stability in the cytoplasm, converging evidence indicates that β-catenin phosphorylation at other non-GSK3 target residues can enhance β-catenin activity. Specifically, phosphorylation of β-catenin at Ser552 by AKT (7), at Ser191 by JNK2 (2), at Tyr654 by BCR-ABL (8) and SRC (9) correlates with increased accumulation or translocation of nuclear β-catenin and enhanced transcriptional activity.
Beyond phosphorylation, nuclear acetyltransferases, such as the CREB-binding protein (CBP)/p300 and PCAF have been shown to interact and acetylate β-catenin. In the case of CBP/p300, the acetylated form of β-catenin exhibits enhanced affinity for TCF4 and is a more potent transcriptional activator (10). Acetylation of β-catenin by PCAF has been proposed to inhibit ubiquitin-dependent degradation of β-catenin and increase its nuclear accumulation and transcriptional activity (11). Conversely, we recently showed that the NAD+-dependent deacetylase SIRT1 mediates the deacetylation and subsequent inhibition of β-catenin transcriptional activation (12). Together, these observations indicate that the activation of β-catenin activity involves several steps beyond its stabilization in the cytoplasm.
CDK8: Multiple paths to Wnt/β-catenin signaling
To identify other post-translational modifiers of β-catenin that participate in colon cancer, we performed two high-throughput, RNAi-based loss-of-function screens to identify kinases and phosphatases required for both β-catenin transcriptional activity and colon cancer cell proliferation (13). Using a lentivirally delivered short hairpin RNA (shRNA) library targeting more than 95% of human kinases and phosphatases, we identified nine genes that were required for both β-catenin transcriptional activity and colon cancer cell proliferation. Although several of these genes had previously been implicated in the regulation of β-catenin, we found that only one of these genes, the cyclin-dependent kinase (CDK) CDK8, resided in a region of copy number gain in nearly half of 120 colon cancers. Indeed, when we analyzed CDK8 amplifications in a second set of colon cancers, we found that 22% of colon cancers showed amplification or copy number gain of the specific portion of chromosome 13 where CDK8 is located, while an additional 40% showed broad gain of chromosome 13. Expression of CDK8 but not a kinase inactive version of CDK8 transformed immortal murine cells. Taken together, these observations identified CDK8 as a colon cancer oncogene amplified in a substantial subset of colon cancers.
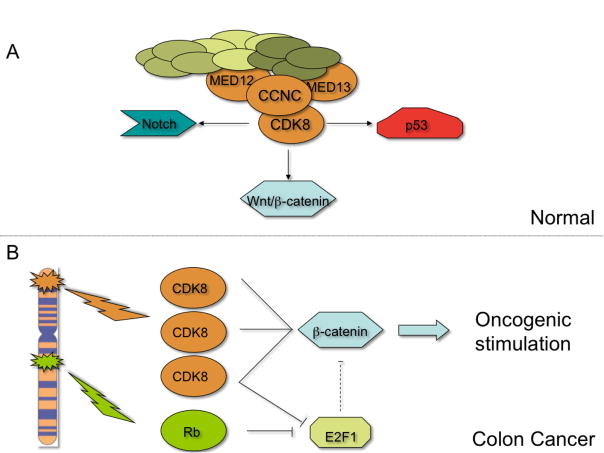
Although most CDKs play key roles in cell cycle regulation, a subset of CDKs, namely CDK7, CDK8 and CDK9 act to regulate gene expression through direct interactions with the transcriptional machinery. CDK8 together with its partner Cyclin C, MED12 and MED13 form the “CDK8 module” of a large complex of proteins called Mediator complex, which plays a key role in regulating both basal and regulated transcription. At least 36 Mediator complex components, which are highly conserved from yeast to humans, have been identified to date (14). Specific components of the Mediator complex bind directly to the activation domains of transcription factors (14) and serve to recruit the Mediator complex to these transcription factors. The CDK8 module, for example, has been found to couple the basal transcriptional machinery to sequence-specific transcription factors such as Notch and p53 (Figure 1A). Moreover, as part of the Mediator complex, CDK8 has been shown both to repress transcription by phosphorylating RNA polymerase II and to activate the transcription of other genes (15). Thus, the mechanism by which the CDK8 module functions is likely to be context-specific. In addition to its functions as part of Mediator, CDK8 has been shown to act as part of a separate complex as a histone kinase implying that Mediator-independent roles for CDK8 activity also exist (16).
Figure 1. Roles for CDK8 Transcriptional Activity in Normal and Colon Cancer Cells.
A. CDK8 participates in regulating several transcriptional programs as part of the Mediator complex. These pathways (Notch, Wnt/β-catenin, and p53) are illustrated. B. In colon cancer, amplification of CDK8 and/or copy number gain of RB lead to stimulation of β-catenin activity either directly or via suppression of E2F1.
Prior work has implicated the CDK8 module components MED12 and MED13 as regulators of β-catenin activity. In C. elegans, let-19 and dpy-22, homologs of the human MED13 and MED12, respectively, are required for Wnt-regulated cell fusion and suppress the transcription of Wnt/β-catenin target genes (17). In contrast, in flies and mammals, MED12 and MED13 activate β-catenin signaling (6, 18). Specifically, the fly MED12 and MED13 homologs, kohtalo and skuld, respectively, activate Wnt/β-catenin target genes through direct interactions with Wnt pathway component Pygopus and recruitment of the Mediator complex (18). The involvement of Mediator subunits in both activation and repression of Wnt-mediated transcription is consistent with the known ability of Mediator to positively and negatively influence regulated transcription (14). The mechanisms through which this occurs, however, remains incompletely understood.
In consonance with observations from model organisms, we found that suppression of CDK8 in colon cancer cells inhibits the expression of a subset of Wnt/β-catenin target genes. These effects are thought to involve the actions of the Mediator complex since suppression of Cyclin C or MED12 induced similar effects. Since Ser-Pro residues important in other CDK consensus sites are found in β-catenin (i.e. Ser191, Ser246 and Ser605), one possibility is that CDK8 directly phosphorylates β-catenin. However, future studies are necessary to identify the CDK8 substrates critical for the regulation of β-catenin activity.
Indeed, genetic studies in flies and mammals suggest that CDK8 might also regulate β-catenin activity indirectly (19). Using Drosophila, Morris et al. screened for genes that modulated E2F1-dependent apoptosis and found that E2F1 represses Wnt/β-catenin activity in both flies and human cancer cell lines. In parallel to these studies, they also identified CDK8 as a strong suppressor of E2F1. Using both Drosophila and human cell line models, they showed that both CDK8 and E2F1 localize to β-catenin target genes such as c-Myc and regulate its transcription. Furthermore, they found CDK8 promotes the phosphorylation of E2F1 and that the ability of CDK8 to repress E2F1 function depends on its kinase activity. These observations implicate CDK8 and E2F1 as regulators of β-catenin activity. Interestingly, colon cancers are unusual in that a substantial fraction show amplification or gain of portions of chromosome 13 where both CDK8 and RB are located (Figure 1B). Consistent with recent work (20), these observations suggest that CDK8 may regulate β-catenin through both Mediator-dependent and independent paths.
Conclusion
Taken together, these recent observations implicate the kinase CDK8 in the regulation of nuclear β-catenin activity. The identification of CDK8 and other Mediator components as essential regulators of β-catenin activity links the basal transcriptional machinery to the TCF/LEF transcription factors. Although ubiquitously expressed, the finding that CDK8 is amplified and acts as an oncogene in colon cancers suggests that this genetic alteration hijacks the physiologic function of the Mediator complex. Since dysregulation of β-catenin activity permits adenoma formation but fails to drive malignant progression in the absence of other cooperating mutations, CDK8 may activate β-catenin and other genes to drive colon cancer progression. Although it is clear that further work is necessary to elucidate the relationship among CDK8, the Mediator complex, RB and E2F1, these studies identify a new level of β-catenin transcriptional regulation and identify new potential targets for cancers that depend on β-catenin activity.
Acknowledgments
This work was supported in part by a grant from the U.S. National Institutes of Health (R33 CA128625).
References
- 1.Phelps RA, Chidester S, Dehghanizadeh S, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–34. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–53. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hecht A, Kemler R. Curbing the nuclear activities of beta-catenin. Control over Wnt target gene expression. EMBO Rep. 2000;1:24–8. doi: 10.1093/embo-reports/kvd012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sustmann C, Flach H, Ebert H, Eastman Q, Grosschedl R. Cell-type-specific function of BCL9 involves a transcriptional activation domain that synergizes with beta-catenin. Mol Cell Biol. 2008;28:3526–37. doi: 10.1128/MCB.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mieszczanek J, de la Roche M, Bienz M. A role of Pygopus as an anti-repressor in facilitating Wnt-dependent transcription. Proc Natl Acad Sci U S A. 2008;105:19324–9. doi: 10.1073/pnas.0806098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/beta-catenin signaling. J Biol Chem. 2006;281:14066–75. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 7.He XC, Yin T, Grindley JC, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat Genet. 2007;39:189–98. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coluccia AM, Vacca A, Dunach M, et al. Bcr-Abl stabilizes beta-catenin in chronic myeloid leukemia through its tyrosine phosphorylation. EMBO J. 2007;26:1456–66. doi: 10.1038/sj.emboj.7601485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piedra J, Miravet S, Castano J, et al. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin Interaction. Mol Cell Biol. 2003;23:2287–97. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy L, Wei Y, Labalette C, et al. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol Cell Biol. 2004;24:3404–14. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge X, Jin Q, Zhang F, Yan T, Zhai Q. PCAF acetylates {beta}-catenin and improves its stability. Mol Biol Cell. 2009;20:419–27. doi: 10.1091/mbc.E08-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firestein R, Blander G, Michan S, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firestein R, Bass AJ, Kim SY, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–51. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89:1439–46. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Gold MO, Rice AP. Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res. 1998;26:3784–8. doi: 10.1093/nar/26.16.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29:650–61. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoda A, Kouike H, Okano H, Sawa H. Components of the transcriptional Mediator complex are required for asymmetric cell division in C. elegans . Development. 2005;132:1885–93. doi: 10.1242/dev.01776. [DOI] [PubMed] [Google Scholar]
- 18.Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci U S A. 2008;105:6644–9. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris EJ, Ji JY, Yang F, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–6. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–51. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]