Abstract
During neuron development, the biosynthetic needs of the axon initially outweigh those of dendrites. However, while a localized role for the early secretory pathway in dendrite development has been observed, such a role in axon growth remains undefined. We therefore studied the localization of Sar1, a small GTPase that controls ER export, during early stages of neuronal development that are characterized by selective and robust axon growth. At these early stages, Sar1 was selectively targeted to the axon where it gradually concentrated within varicosities in which additional proteins that function in the early secretory pathway were detected. Sar1 targeting to the axon followed axon specification and was dependent on localized actin instability. Changes in Sar1 expression levels at these early development stages modulated axon growth. Specifically, reduced expression of Sar1, which was initially only detectable in the axon, correlated with reduced axon growth, while over-expression of Sar1 supported the growth of longer axons. In support of the former finding, expression of dominant negative Sar1 inhibited axon growth. Thus, as observed in lower organisms, mammalian cells utilize temporal and spatial regulation of ERES to address developmental biosynthetic demands. Furthermore, axons, like dendrites, rely on ERES targeting and assembly for growth.
Keywords: Sar1, endoplasmic reticulum, axon, neuron, development, COPII, Golgi
Introduction
Newly synthesized proteins leave the Endoplasmic Reticulum (ER) at the first sorting site of the secretory pathway, peripheral and perinuclear ER Exit Sites (ERES) (1). ERES derived carriers containing cargo proteins converge to a centrally localized perinuclear and perhaps homogenous Golgi complex en route from the early secretory pathway to the trans Golgi network (TGN). The sorting of membrane proteins to their final destination at the apical or basolateral cell surfaces is largely carried out in the TGN. Cargo proteins, such as receptors and ion channels, exported from the TGN are targeted to the plasma membrane (PM) in a polarized manner, defining the identity and function of polarized cells (2). Although a role for the early secretory pathway in regulating cell development and polarization has yet to be defined, extensive expansion of the ER is observed during the developmental differentiation of B cells into antibody secreting plasma cells (3). In lower eukaryotic yeast cells, which have a morphologically unorganized secretory pathway, distinct ERES are differentially utilized to package proteins into distinct vesicles budding from the ER (4). In a higher eukaryote (drosophila), the spatial organization of ERES is utilized for the polarized deposition of membrane proteins at the PM of oocytes (5). In addition, the drosophila trailer hitch ribonucleoprotein complex is recruited to ERES to support local translation of selected mRNAs required for transport out of the ER (6). This spatial activity is required for the establishment of cell polarity. These findings led us to hypothesize that ER exit is not a uniform process, and propose that regulation of selective ERES supports cell development and polarization. In agreement with this hypothesis, we previously characterized a targeted increase in localized dendritic ERES numbers that correlated well with the development of dendrites in primary neuronal cultures (7). These observations were recently corroborated with functional studies using drosophila as a genetic model as well as siRNA studies in primary hippocampal neuron cultures (8). In these studies, there was a selective sensitivity to reduced activity of ERES and the early secretory pathway in the development of dendrites. These findings have lead some to suggest that the secretory pathway is selectively utilized only for dendrite growth (9). We hypothesize, that both spatial and temporal regulation is exerted on ERES to selectively direct their activity in order to support both axonal, and subsequently dendritic outgrowth during development. We therefore analyzed the localization of Sar1 in primary hippocampal neuron development in vitro and the role of Sar1 in axonal growth.
Results
Sar1 is selectively targeted to the developing axon
We analyzed the localization of the cytosolic small GTPase Sar1 during neuronal development. The Sar1 GTPase is a limiting component of the cytosolic coat protein complex II (COPII), the sorting machinery that mediates vesicular export from the ER. Sar1 activation initiates the assembly of organized ER exit sites, recruits the cytosolic COPII coat proteins Sec23/24 and Sec16, induces membrane deformation to control vesicle formation and fission, and together with COPII components mediates cargo export from the ER (10, 11). As such, Sar1 localization provides a highly selective and sensitive marker for ER exit sites, which are the first sorting sites in the secretory pathway. Embryonic rat hippocampal neurons fixed at the indicated times post plating were stained with antibodies to Sar1 (Fig. 1–Fig. 3). In vitro, embryonic hippocampal neurons acquire their characteristic polarized appearance by progressing through defined morphological stages (12). Shortly after plating, the cells will present lamellipodia (stage 1) that will develop within several hours to short immature neurites of somewhat equal length (stage 2). Stage 2 neurons exhibited intense staining of Sar1 within the cell soma and to a lesser extent within the neurites (Fig. 1A). By 24 hours in vitro approximately half (52%; n=90) of the neurons in our cultures had a morphologically identifiable axon—a neurite whose length was at least twice as long as the diameter of the soma and twice the length of it’s brethren (stage 3). At this time point, Sar1 appeared concentrated in one neurite in 30% (6 out of 20) of the stage 2 neurons in 24-hour cultures (Fig. 1B). In addition, Sar1 appeared to be selectively targeted to the axon in 88% (44 out of 50) of stage 3 neurons (Fig. 1C). Indeed, quantitative analysis of the subcellular localization of Sar1 in stage 3, 1 day in vitro (DIV) neurons revealed that Sar1 was unevenly distributed between the axon, soma, and dendrites (F2,33 = 53.39, p < 0.001). Post hoc testing showed that total Sar1 fluorescence in the axon was significantly greater than that in the soma (p < 0.05) or all minor neurites combined (p < 0.001). During the transition from stage 3 to 4 of neuronal development axonal Sar1 redistributed from being concentrated near the soma forming a proximal to distal gradient to being concentrated in varicosities located mostly in distal segments of the axon (compare Fig 1C & Fig. 2A–B to Figs 2C–D & and Supplemental Fig. 1). Importantly, a similar Sar1 distribution was found in more than 85% of the neurons in our rat hippocampal cultures and in developing mouse hippocampal neurons (Fig. 2B). By developmental stage 5 Sar1 appeared to be concentrated in the soma and more evenly disbursed between the dendritic arbor and axon (Fig. 3 and Supplemental Fig. 1, which are strongly representative of Sar1 distribution in mature cultures). Therefore, the apparent selective targeting of Sar1 to the axon was lost upon neuronal maturation.
Figure 1. Sar1 marks the axon during early stages of neuronal development.
Rat hippocampal neurons were fixed at indicated times post plating and labeled with phalloidin (A1, B1, and C1; red in A3, B3, and C3) and antibodies to Sar1 (A2, B2, and C2; green in A3, B3, and C3). Sar1 distribution in stage 2 neurons (6 hrs in vitro) is shown in panel A. By 12 hours in vitro Sar1 is targeted to a single elongated neurite (B). By stage 3 of development, Sar1 is heavily concentrated into the differentially extended neurite (C–24 hours in vitro). Bars equal 10 µm.
Figure 3. Sar1 distribution in mature neurons.
Rat primary hippocampal neurons were fixed at indicated times post plating. Neurons in A (8 DIV) and C (14 DIV) were stained for MAP2 (A1 and C1; red in A3 and C3) and Sar1 (A2 and C2; green in A3 and C3), the one in B (12 DIV) was stained for Tau (B1; red in B3) and Sar1 (B2; green in B3). Note the incoming axons in A and B (arrows) from neighboring neurons that are heavily labeled for Sar1. Also, note that in C Sar1 expression in the dendrites and the apical region of the axon (arrow) is similar. Bars equal 10 µm.
Figure 2. Sar1 distribution in 2 and 3 DIV neurons.
Rat (A & C) and mouse (B) hippocampal neurons were fixed at indicated times post plating. (A–B) Cultures were labeled with phalloidin (A1 and B1; red in A3 and B3) and antibodies to Sar1 (A2 and B2; green in A3 and B3). (C) Cultures were transfected with GFP (C2; green in C4) prior to plating and labeled with antibodies to Sar1 (C1; red in C4) and Map2 (C3; green in C4). Beginning at 3 DIV axonal Sar1 starts to associate with varicosities (arrows in C). C1-4 were made using two fields stitched together. Bars equal 10 µm.
Sar1 facilitates axonal outgrowth
Our data suggests that Sar1 levels may play a role in axon development. However, in a recent study by Ye and colleagues (8), which used mutant analysis in drosophila and the expression of shRNAs in hippocampal cultures to study the role Sar1 plays in neuron development, a decrease in Sar1 levels was found to significantly affect dendrite but not axonal growth. To further explore the hypothesis that Sar1 directly plays a role in axonal elongation we performed two sets of studies. First, we compared the effect over-expressed Sar1-wt (hamster Sar1a) and a trans dominant Sar1 mutant had on early stages of neuronal development (Fig. 4). In these experiments we used an inactive form of Sar1 (Sar1-GDP; Sar1 T39N mutation) that is deficient in GTP binding. We chose Sar1-GDP over the Sar1-GTP (H79G mutation) mutant, which is deficient in GTP hydrolysis (10, 13), because neurons over-expressing Sar1-GTP died by 2 day in vitro (DIV; data not shown). Neurons were transfected prior to plating with either GFP (Fig. 4A1), GFP + Flag tagged Sar1-wt (Sar1-wt-Flag; Fig. 4A2), or GFP + Sar1-GDP-Flag (Fig. 4A3). Similar to endogenous Sar1, exogenously expressed Sar1 was found in axons (Supplemental Fig. 2A). Quantitative analyses of axon length in developing transfected neurons expressing either Sar1-wt-Flag, Sar1-GDP-Flag, or GFP at 2 and 3 DIV found significant differences in axon length between groups (F2,82 = 23.78, p < 0.001 and F2,84 = 14.33, p < 0.001, respectively). Post hoc testing revealed that the axons of Sar1-wt-Flag transfected neurons [269.7 µm (SEM 11); n = 44] were significantly longer than GFP transfected controls [211.06 µm (SEM 20.7); n = 20; p < 0.05], while axons of Sar1-GDP-Flag transfected neurons were significantly shorter than those of controls [134.4 µm (SEM 12.1); n = 21; p < 0.005; Fig. 4B]. By 3 DIV the difference in axon length between the Sar1-wt-Flag transfected [381.87 µm (SEM 18.7); n = 23] and control neurons [342.7 µm (SEM 30.6); n = 43] was no longer significant, while the axons of Sar1-GDP-Flag transfected neurons remained highly significantly shorter than those of controls [156.7 µm (SEM 12.4); n = 21; p < 0.001; Fig. 4C).
Figure 4. Sar1 facilitates axonal elongation.
(A1–A4) Primary rat hippocampal neurons were transfected with GFP (A1), GFP and Flag-tagged hamster Sar1a (Sar1-wt-Flag; A2), GFP and Sar1-GDP-Flag (A3), or constructs that expressed GFP and the Sar1-shRNAs prior to plating and fixed at 2 DIV. (B–E) The effect of Sar1 levels on axonal elongation was determined at 2 and 3 DIV. For these experiments the effect of the different transfection conditions in A1–A4 on axon growth were determined by analyzing GFP expressing cells. As an additional control some of the cultures transfected with the Sar1-shRNAs were also transfected with Sar1-wt-Flag, which appears to be resistant to the Sar1-shRNAs (see Supplemental Fig. 1). Values equal mean ± SEM. An asterisk designates a significant difference between the condition and control GFP expressing neurons. See the main text for statistical significance. A2 is made of two images stitched together. The fields were overlapping, but skewed to one another resulting in a non-overlapping area in the upper left corner. For the purpose of presentation the area, which is outlined in white, was filled in with black. The bar in A4 is also for A1–A2; bars equal 10 µm.
In the second set of experiments, we introduced small hairpin RNAs that specifically and effectively knock down the expression of Sar1 by transfecting neurons with Sar1a-shRNA and Sar1b-shRNA lentiviral vectors (collectively called Sar1-shRNAs) (8). In their study, Ye and colleagues transfected cultured neurons with Sar1-shRNAs at 2 DIV and analyzed neurons at 5 DIV (8). However, we have shown here that substantial amounts of Sar1 have already trafficked to the axon by 2 DIV. Therefore, neurons were transfected prior to plating with a GFP control construct (Fig. 4A1), Sar1-shRNAs, which also express GFP (Fig. 4A4), or Sar1-shRNAs + Sar1-wt-Flag and axonal length was analyzed at 2 and 3 DIV (Fig. 4D–E). At both 2 and 3 DIV a significant difference between groups was found (F2,128 = 26.4, p < 0.001 and F2,187 = 61.18, p < 0.001, respectively). Post hoc testing revealed that at 2 DIV, axonal length of control and Sar1-shRNAs transfected neurons were significantly different [195.8 µm (n = 49; SEM 11.3) and 107.9 µm (n = 47; SEM 6.3), respectively (p < 0.001)]. In addition, the axonal length of control and Sar1-shRNAs transfected neurons were also significantly different at 3 DIV [342.7 µm (n = 43; SEM 30.6) and 135.2 µm (n = 116; SEM 6.3), respectively (p < 0.001)]. In contrast, at 2 and 3 DIV, the axons of neurons transfected with Sar1-shRNAs + Sar1-wt-Flag, which was resistant to the shRNAs (see supplement Fig. 1F), (n = 35 and 31, respectively) were statistically the same as controls [178.1 (SEM 9.2) and 281.9 (SEM 13.9), respectively]. Together, these findings strongly suggest that Sar1 plays a direct role in axon development.
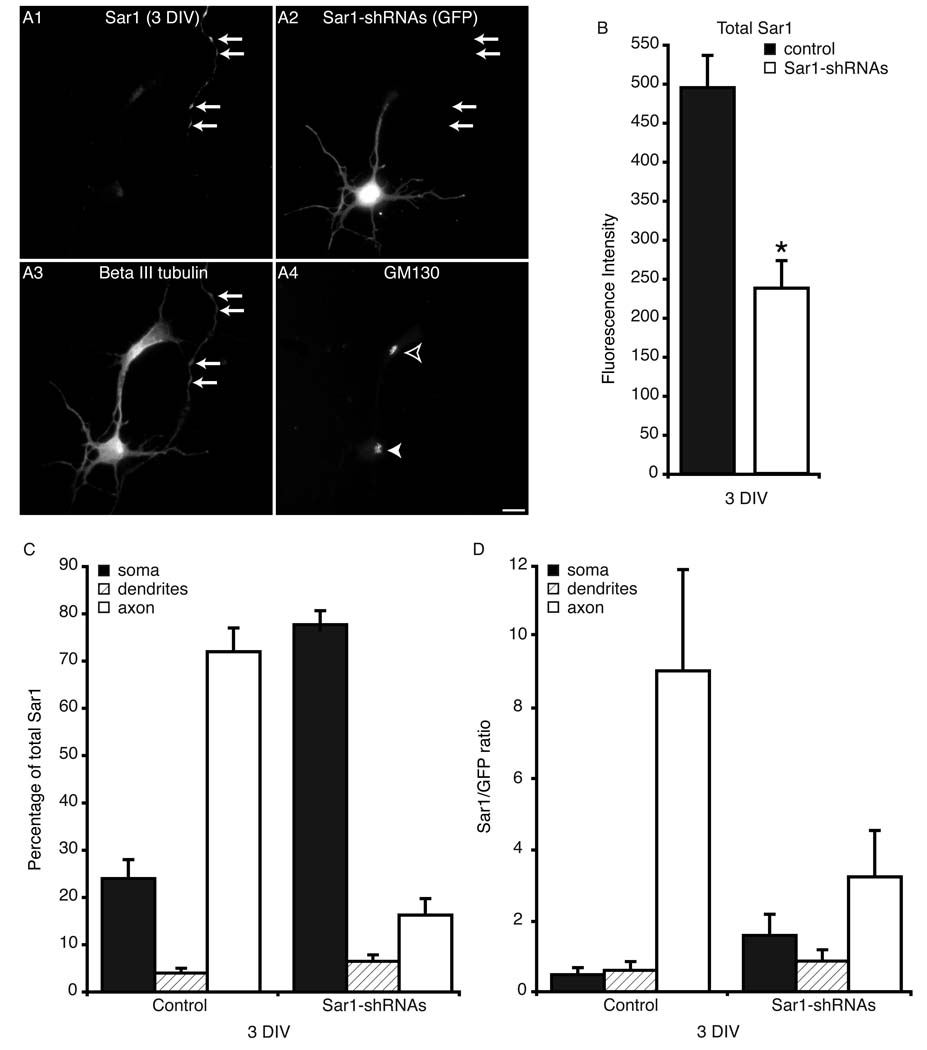
Since Sar1 provides basic cellular function, it is intriguing that depletion of the protein selectively affects the elongation of axons (Fig. 5B) and dendrites (8). Thus, it appears that cells can adapt to partial Sar1 depletion by selectively preserving key cellular functions such as ER to Golgi traffic in the soma, while compromising developmental activities such as local ERES assembly and traffic in the growing processes. We hypothesize that Sar1 depletion will initially lead to a selective reduction of Sar1 targeting and local assembly of ERES in the axon. Such seemingly local depletion would lead to the observed inhibition of axon growth, while maintaining normal traffic in the soma. To test this hypothesis, a quantitative analysis of the subcellular localization of Sar1 at 3 DIV was performed. We chose to perform our analyses at 3 DIV because this was the first time point at which total neuronal Sar1 expression was significantly reduced (52%) in neurons transfected with Sar1-shRNAs compared to controls (n = 10/group; t9 = 2.38, p < 0.05; Fig. 5). For these experiments cultures were transfected with a GFP control construct or Sar1-shRNAs. At 3 DIV total axonal Sar1 fluorescence was significantly greater than that in the soma (n = 14, t13 = 2.46, p < 0.05) or all minor neurites combined (n = 14, t13 = 2.69, p < 0.05) in control cultures. In addition, somatic Sar1 fluorescence was significantly greater than dendritic (n = 14, t13 = 4.045, p = 0.001). In contrast, in cultures transfected with Sar1-shRNAs the soma contained significantly more Sar1 than either the axon (n = 15, t14 = 5.77, p < 0.001) or all dendrites combined (n = 15, t14 = 6.654, p < 0.001). In addition, there was no statistical difference in axonal Sar1 fluorescence compared to that in all dendrites combined. To control for volume, we measured the level of GFP fluorescence in the soma, axon, and dendrites and then compared the Sar1/GFP ratio of each compartment. In controls, the Sar1/GFP ratio of the axon was significantly greater than that of both the soma (n = 14, t13 = 2.35, p < 0.05) and all dendrites combined (n = 14, t13 = 2.34, p < 0.05; Fig. 5D). In contrast, there were no significant differences in this ratio between dendrites and axons in neurons transfected with the Sar1-shRNAs. Importantly, no statistical difference in total GFP expression between control and Sar1-shRNAs transfected cultures was detected. In these experiments we noted that although there was a significant reduction in Sar1 expression by 3 DIV in neurons transfected with the Sar1-shRNAs, there were no clear morphological changes in the Golgi complex of these cells as visualized with a GM130 antibody (Fig. 5A4). Normal Golgi morphology composed of compacted Golgi cisternae is indicative of functional ER to Golgi traffic in the soma. In contrast, the Golgi complex was clearly dispersed by 5 DIV in neurons transfected with the Sar1-shRNAs (Supplemental Fig. 2B). Thus, in support of our hypothesis ERES assembly and function is selectively regulated during development. Normally, at the very early stages of development Sar1 is preferentially targeted to the axon. However, under conditions of Sar1 depletion the secretory functions in the cell body are preserved at the expense of axonal growth. The observed relative redistribution of ERES to the soma under conditions of Sar1 depletion may be required to preserve cell survival and thus highlight a cellular strategy utilized to address a limited supply of Sar1 and compromised activity of COPII.
Figure 5. Quantitative analysis of Sar1 levels in 3 DIV neurons.
(A) Primary rat hippocampal neurons were transfected with Sar1-shRNAs prior to plating and labeled with antibodies to Sar1, beta III tubulin, and GM130 at 3 DIV. (B–C) Sar1 levels in neurons transfected with GFP (control) or Sar1-shRNAs were quantified by immunofluorescence microscopy. Values equal mean ± SEM. The asterisk in B designates a significant difference between the two conditions. (B–D) See main text for statistical analysis of the data. The arrows in A1 point to Sar1 staining in an axon coming from a neuron that is outside the field of view. The arrows in A2 & A3 point to the same region as those in A1. The filled arrowhead in A4 points to the Golgi of the neuron in the field that has been transfected with the Sar1-shRNAs constructs, while the open arrowhead points to that of an untransfected neighboring cell. The bar equals 10 µm.
Sar1 distribution is altered by cytochalasin D treatment
Pharmacological destabilization of the actin cytoskeleton with cytochalasin D (CytD) at early stages of neuron development leads to the formation of neurons with multiple elongating axons (14). In this system, if Sar1 were required for rapid process elongation at early stages of differentiation, one would predict that Sar1 would be evenly distributed between the processes. Thus, we analyzed the distribution of endogenous Sar1 in CytD treated cultures. When neurons were treated with CytD 4 hours after plating, multiple elongated processes developed (Fig. 6B1) as previously reported (14). Under these conditions, endogenous Sar1 appeared to be distributed equally between all developing neurites of all the neurons in CytD treated cultures (Fig. 6B2), suggesting that selective Sar1 distribution may be regulated by cytoskeletal dynamics. In control experiments (Fig. 6A1–2), neuronal morphology and Sar1 distribution were normal. Therefore, interference with the cytoskeletal rearrangement that is required for the specification of a single axon changed Sar1 distribution. These findings support the hypothesis that Sar1 is required for rapid process elongation at early stages of neuronal differentiation.
Figure 6. Redistribution of endogenous Sar1 after cytochalasin D induced actin destabilization.
(A–B) Rat hippocampal neurons were treated with DMSO only or CytD for 24 hours starting at 4 hours post plating. At 28 hours post plating cultures were fixed and labeled with phalloidin (A1 & B1) and antibodies to Sar1 (A2 & B2). (C–F) Neurons (1 DIV) were rinsed with PBS, permeabilized, and washed as described in Methods. Cells were then incubated in the presence of buffer (C), 5 µg Sar1-GTP (D), 5 µg Sar1-GTP and rat liver cytosol (E), or 5 µg Sar1-GDP and rat liver cytosol (F). At the end of the incubations, the distribution of Sar1 (C–D) and Sec13 (E–F) was determined using IF microscopy. Open arrowhead in D points to ERES in a minor neurite. The bar in F is also for C–E; bars equal 10 µm.
Axonal ERES can be reconstituted in vitro
We next reconstituted the formation of axonal ERES in vitro. Hippocampal neurons (1 DIV) were permeabilized, washed, and incubated in vitro as described in Methods. When permeabilized neurons were incubated in the absence of COPII coat machinery, residual punctate Sar1 staining was observed, particularly in the axon (Fig. 6C). Permeabilized neurons incubated with Sar1-GTP (Sar1 H79G) to stabilize Sar1-membrane binding and ERES assembly, exhibited localized membrane proliferation and constriction throughout the developing axon (Fig. 6D), which is typical of ERES assembly (7, 10, 15, 16). Importantly, Sar1 tubular domains were not observed (data not shown) when incubations were performed in the presence of an inactive Sar1 mutant (Sar1-GDP). When permeabilized neurons were incubated with Sar1-GTP and rat liver cytosol (RLC; a source of COPII components Sec23/24 and Sec13/31), stabilized ERES recruited and nucleated COPII as analyzed by the assembly of the COPII outer layer Sec13 component (Fig. 6E). Importantly, Sec13 was not recruited when permeabilized neurons were incubated with Sar1-GDP and RLC (Fig. 6F). Together, these data establish that ERES formation can occur in the axons of developing neurons.
Key components of the secretory pathway localize to axons
If ERES targeting to developing axons is required for local elevated biosynthetic activity, additional elements of the secretory pathway should also be targeted to the developing axon. For these studies we analyzed the subcellular distribution of several key components of the secretory pathway. The folding chaperone calnexin that participates in protein folding is localized in the axon (Fig. 7I), which is in agreement with previous studies (17). In addition, an over-expressed mEmerald-KDEL fusion protein that is retained in the ER is highly localized to the elongating axon (Fig. 8D). The guanine nucleotide exchange factor for Sar1, Sec12 which activates Sar1 to initiate ERES assembly on the ER (Fig. 7C) and Yip1a (Fig. 8A), an evolutionary conserved membrane protein involved in ER to Golgi transport that was implicated in the formation and the subsequent fusion of COPII vesicles (18), also localized in the developing axon. Similarly, exogenous over-expressed Yip1a was found in the axon (GFP or Myc tagged; Fig. 8B & 8C, respectively), Thus components of protein folding and export machinery from the ER are localized in the developing axon.
Figure 7. Distribution of the secretory pathway during neuronal development.
Rat primary hippocampal neurons were fixed at 1 (A–J) and 4 (K) DIV. Cultures were immunostained for proteins of the secretory pathway as indicated in the figure. In A–J the longest neurite is oriented such that it extends away from the soma in the direction the arrow is pointing in A. (K) 4 DIV neurons were labeled with phalloidin (K1) and antibodies to Sec23 (K2). Bars equal 10 µm.
Figure 8. Exogenously expressed ERES markers are targeted to axons.
(A–C) Localization of endogenous and over-expressed Yip1a, a membrane ERES marker, in the developing axon. (A) Untransfected early stage 3 neurons were stained using phalloidin (A1; red in A3) and an antibody directed against Yip1a (A2; green in A3). (B) Neuronal cultures transfected with a GFP-Yip1a (B2; green in B3) construct on 2 DIV, were fixed and stained with phalloidin (B1; red in B3) at 3 DIV. (C) Neuron cultures were transfected at 2 DIV with Myc-Yip1, fixed at 3 DIV, and stained for over-expressed Myc-Yip1 (C1; red in C3) and Sec23 (C2; green in C3) or Sar1 (c2a; green in c3a). The inset image c3b mag. is a blowup of the designated region in C3 (c3b). (D) Shows a 2 DIV rat hippocampal neuron, which was transfected prior to plating, expressing the ER marker mEmerald-KDEL. Bars equal 10 µm.
The presence and activity of Golgi outposts in dendrites is well documented (19). Golgi elements may also function during specific times of development to support local biosynthetic transport in the growing axon. To test this hypothesis, we analyzed the distribution of Golgi markers within the axon. Although Golgi markers were not expected to appear highly concentrated because of the lack of organized Golgi structures in axons (data not shown), pre Golgi (Giantin; Fig. 7D), Golgi (GM130; Fig. 7G), and trans Golgi network markers (clathrin and AP1; Fig. 7E & F, respectively) were detected in the axon. Therefore, assembly of axonal ERES was followed by mobilization of secretory pathway machinery proteins. We noted however that the distribution of Golgi markers was not uniform. Specifically, while all axons surveyed expressed Giantin, which participates in fusion events delivering incoming pre Golgi intermediates to the Golgi, axonal GM130 immunoreactivity was only found in 17% of analyzed neurons. Future studies need to address the possibility that axonal Golgi vesicles may present non-uniform composition, exhibit differential processing properties, and form de novo in the axon.
The pathogenic axonal protein α-synuclein functions together with Rab1 to regulate ER to Golgi transport (20). In agreement with the above findings, alpha-GDI, which regulates Rab protein function, was found in the axon (Fig. 7H). In contrast to the above findings, as previously reported (21), the transferrin receptor was not found in the developing axon (Fig. 7J).
Sec23 distribution becomes polarized during high biosynthetic demand in the axon
Sec23 is a subunit of the COPII coat that activates the GTPase activity of Sar1 (Sar1-GAP) and forms part of the inner layer pre-budding complex of the coat that selects cargo proteins for ER exit. Endogenous Sec23 subunits were also located in the axon (Fig. 7B and Fig. 8C). However, the selective axonal distribution of Sar1 was much more pronounced (for example, compare the distribution of Sar1 in Fig. 7A to the staining pattern of Sec23 in Fig. 7B). This difference in overall distribution continued into later stages of development. For example, at 4 DIV the expression of Sec23 is mostly somatic, while the expression of Sar1 is still highly axonal (compare Fig. 7K to Supplemental Fig. 1A2). Importantly, both Sar1 and Sec23 colocalized with over-expressed Myc tagged Yip1a in axonal varicosities (Fig. 8C).
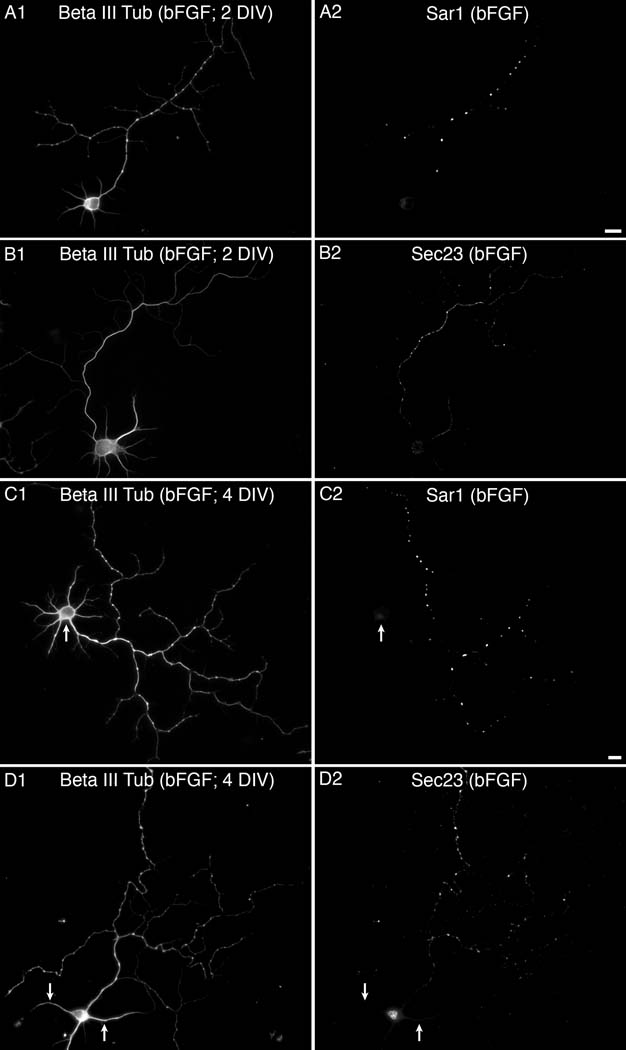
Importantly, the location of Sar1 and Sec23 reports on the activity of ERES. Using reconstruction from serial thin sections and high-resolution quantitative EM microscopy previous studies demonstrated that ERES present regular morphology. Several ER cisternae are grouped together in a cup-like structure from which a number of COPII coated bud profiles protrude toward the center of the domain. Budded vesicles fuse to form vesicular tubular clusters (the ER-Golgi intermediate compartment). ERES typically vary in the number of buds within a unit with an average of 4.4 buds per site and a range of 2–20 buds per site in non-polarized cells. COPII-coated bud numbers vary with the position of ERES in the cell with ERES positioned at the Golgi region showing increased number of ER membranes and budding profiles (22). As the size of a single bud or COPII vesicle falls below the optical resolution of light microscopy, analysis of COPII markers by immunofluorescence reports on the complexity of the assembled site (i.e. number of grouped ER cisternae and bud profiles). In response to increased biosynthetic cargo load ERES can undergo growth events (defined as nucleation or fusion events) (23–25). Cargo load also regulates the kinetics of COPII-membrane interactions and vesicle formation activities (26). Axonal ERES may show reduced number of associated ER membranes, buds, and overall complexity in particular given the lack of morphologically defined Golgi elements in the axon (not shown). The sites may become more elaborate when general (or specific) biosynthetic demand will increase. Sar1 is a limiting component of the COPII vesicle formation reaction, and thus is better represented at COPII assembly sites. However, we hypothesized that when the axon requires an increase in biosynthetic cargo the distribution of COPII subunits is polarized to the axon, mirroring the local distribution of Sar1. To test this hypothesis, we analyzed Sar1 and Sec23 distribution in basic FGF (bFGF) treated cultures. bFGF has been previously shown to enhance axonal outgrowth (27–29). Although bFGF does not alter the elongation rate at the tip of axons, the axons of neurons exposed to bFGF in vitro are significantly more branched than untreated controls, and thus the total length of the axon is increased. For our studies, we added bFGF (10 ng/ml) to the culture media for 48 hours prior to analysis. Specifically, bFGF was either added at 2 hrs post plating or at 2 DIV and cultures were fixed at 2 and 4 DIV, respectively (Fig. 9). Cultures exposed to bFGF had enhanced axons as previously described. In both experiments Sar1 preferentially targeted the axons of bFGF-exposed neurons where it was mostly associated with varicosities (Fig. 9A). In contrast, the preferential association of Sar1 with varicosities was not usually observed until 3–5 DIV in untreated cultures (compare Fig. 9A with Fig. 2). An analysis of the distribution of Sec23 found that in the bFGF treated cultures that were fixed at 2 DIV Sec23 appeared to be preferentially targeted to the axon. However, Sec23 did not appear to associate as strongly with varicosities as Sar1 (compare 9A to 9B). In the bFGF treated cultures fixed at 4 DIV the distribution of Sec23 appeared to more closely resemble that of Sar1. Thus, in bFGF treated neurons the distribution of Sec23 appears to shift from being mostly somatic with some expression in both the axon and dendrites to being highly expressed in the axon. Therefore the local and temporal increase in biosynthetic demand targeted both Sar1 and the COPII coat to the growing axon.
Figure 9. An increase in demand leads to enhanced recruitment of Sec23 to the growing axon.
bFGF (10 ng/ml) was added to the culture media of rat hippocampal neurons at 2 hours post plating (A–B) or 2 DIV (C–D). Cultures were subsequently fixed at 2 and 4 DIV, respectively, and the distribution of Sar1 (A & C) and Sec23 (B & D) was assessed by immunofluorescence. An antibody against Beta III tubulin was used to visualize neuronal morphology (A1–D1). The arrows point to the same region within corresponding images. The bar in A is also for B and the one in C is also for D. Bars equal 10 µm.
Discussion
Using primary hippocampal cultures, we examined the role of Sar1 in the early stages of axonal outgrowth. Sar1 was found to selectively target the specified axon at the very early stages of rapid axonal elongation (Fig. 1). Sar1 activity could be reconstituted in defined in vitro assays (Fig. 6C–F). A reduction in Sar1 activity resulted in significantly shorter axons (Fig. 4). Furthermore, the selective axonal targeting of Sar1 was found to require cytoskeletal rearrangements that participate in initial neuronal polarization (Fig. 6A–B). Taken together, the findings presented here strongly suggest that Sar1 is required to support enhanced axonal elongation. We propose that similar differential regulation of Sar1-regulated ERES assembly, highlighted here because of the distinctive morphology of the neuron, exists in other cell types to support local biosynthetic demand during periods of development.
Sar1 activity is required for axonal elongation
Findings presented here demonstrate that Sar1 activity is required for axon growth. Specifically, Sar1 is targeted in a robust manner to the newly specified axon in accordance with a preferential utilization of the protein during the initial phase of axon growth (Fig. 1–Fig. 3 and Fig. 5). In addition, the expression of trans dominant negative Sar1 mutants leads to axon growth inhibition (Fig. 4). Furthermore, a reduction in Sar1 expression dramatically decreases axonal length (Fig. 4). Collectively, these findings strongly support the hypothesis that Sar1 activity is required for axonal elongation.
Our finding that a reduction in Sar1 protein levels leads to a significant decrease in axonal length differs from findings described in a previous study that found reductions in Sar1 expression in primary hippocampal neurons dramatically decreased the total length of dendrites, but not axons (8). Importantly, in both studies the identical Sar1-shRNA vectors were used to reduce Sar1 expression. These seemingly contrasting findings most likely represent the difference in timing at which Sar1 expression was reduced. Specifically, Ye and colleagues transfected cultures at 2 DIV, and did not detect a significant decrease in Sar1 levels until 5 DIV. In contrast, in the studies performed here neurons were transfected prior to plating in an attempt to interfere with the robust mobolization of sar1 to the axon at the very early stages of development (Fig. 1, Fig. 2, and Fig. 5). We hypothesize that the axonal Sar1 already present at 2 DIV was sufficient to maintain normal axonal rates of elongation in the previous study. Importantly, our Sar1-shRNA findings are supported by results obtained with trans dominant Sar1 proteins (Fig. 4). Specifically, we show that when the activity of Sar1 is inhibited using a Sar1-GDP mutant axonal growth is significantly reduced. Therefore, collectively the current and previous studies establish a role of Sar1 in both dendrite and axon growth.
Two isoforms of Sar1, Sar1a and Sar1b are expressed at similar levels in hypocampal neurons and the expression level of both proteins did not change significantly over the course of development (Supplemental Fig. 1E). In our study we depleted both isoforms for analysis. The two isoforms are highly homologous yet a differential role in neuronal development may be envisioned. However, null mutations in the human Sar1b specifically lead to the development of chylomicron retention or Anderson disease to suggest that overall the activity of the two Sar1 proteins in the neuron is at least partially redundant (30).
Localized Sar1 assembly and axon growth
Although it is unknown to what extent Sar1 activities within the soma support axonal growth, the robust localization of Sar1 in the growing axon suggests that locally assembled Sar1 supports axon growth. In support of this suggestion, in the shRNA experiments performed here a significant reduction in Sar1 axonal expression, but not somatic expression was associated with a significant reduction in axonal length after 3 days post Sar1-shRNA transfection. In addition, although there was a significant reduction in total neuronal Sar1 fluorescence at this time point, there were no clear morphological differences in the Golgi complex of neurons transfected with Sar1-shRNA vectors, suggesting that somatic ER export was still functioning near normal levels (Fig. 5). Importantly, the affects of reduced Sar1 levels on the Golgi complex structure were observed at later time points (Supplemental Fig. 2B). These results suggest that Sar1 activities are preferentially and locally preserved to protect cells from Sar1 depletion. These observations also support a role for the observed local axonal Sar1 assembly in axon outgrowth.
What role(s) may axonal Sar1 and ERES play in axon outgrowth? The activity of Sar1 has been extensively studied and its only known role is in vesicular transport. We have previously demonstrated that Sar1 initiates ERES morphogenesis prior to coat assembly (15, 16). Thus, the buildup of Sar1 within the axon observed here (Fig. 1–Fig. 2) provides the earliest indication for ERES assembly, which are also marked with COPII Sec23 subunit (Fig. 7–Fig. 9). Previous studies have demonstrated that the neuronal ER is a continuous structure that can be divided into two major subdomains: the smooth (SER) and rough ER domains. The SER, which extends into the axon and dendrites, contains both chaperons that function in protein folding (Fig. 7–Fig. 8) and lipid synthesizing enzymes such as HMG co-A reductase (17). Although lipid traffic may operate in a non-vesicular manner (31), axonal ER may be required to support both lipid and protein transport. Thus, newly synthesized proteins utilized for axon elongation may be processed through axonal ERES and/or cargo loaded ERES may be mobilized into the growing axon to support its growth. Axonal ERES may also be used to facilitate axon guidance by regulating the delivery of membrane proteins such as the EphA2 receptor to the PM (32).
Previous studies have suggested a preferential delivery and membrane deposition of membrane cargo (CD8α) directly at the axonal growth cone (33). In contrast, our studies show that Sar1 is localized throughout the growing axon, and these results are in agreement with previous studies demonstrating that protein delivery to the PM (34) as well as selective lipid deposition (35) can take place through the entire length of the axon. During early stages of development Sar1 presented uniform distribution throughout the future axon (Fig. 1), whereas localized appearance at axonal varicosities was observed at later developmental stages (Fig. 2, Fig. 3, and Fig. 9). It may very well be that temporal control governs the site of protein deposition in the growing axon. Future experiments should explore the site of cargo delivery in the axon to determine whether the site of membrane delivery is also cargo specific.
Sar1 supports outgrowth, not specification of the axon
One of the early events that mark a neurite for axon selection is the local destabilization of the actin network (12). The Rho family of GTPases are involved in the regulation of neuronal polarity probably by acting as regulators of the axonal actin cytoskeleton. Together the regulated effects imposed on microtubule dynamics and actin stability by Rac, CDC42, N-wasp, and CRMP-2 (to name a few) during the initial stages of development may enable the microtubule network to extend well into the neurite selected to become the axon. These microtubule tracks may be required for the delivery of organelles that support axonal outgrowth. The specific targeting of Sar1 to a single neurite is disrupted in cytD treated cells (Fig. 6). Under these conditions the perturbation of actin rearrangements lead to the formation of multiple axons. These results and the effects of Sar1 on axon growth collectively suggest that Sar1 is required for axon outgrowth rather than axon specification. An attractive hypothesis derived from our results suggests that the recruitment and activation of Sar1 may operate to couple ERES to microtubule arrays that extend into the selected neurite where actin is destabilized. In support of this theory, our previous studies have demonstrated that Sar1 recruits conventional kinesin to ERES (15). These results are supported by recent findings that KIF5B is required for the assembly of ERES and for COPII mediated export from the ER (36). Conventional kinesins including KIF5B play a key role in axon development and function (37, 38).
COPII activities in development
Recently, a genetic screen aimed at defining activities that affect dendrite arborization (Dar) in drosophila identified Sar1 as a key protein in dendrite development (8). Although loss of drosophila Sar1 function is lethal, these studies were possible because embryonic (and thus some neuronal) development progressed sufficiently to reveal selective defects. Similarly, mutations in Rab1 or Sec23 lead to inhibition of dendrite development, suggesting that a dedicated role exists for the early secretory pathway in dendrite growth. Importantly, progression of embryonic development in organisms deficient in COPII functions is also observed in zebrafish depleted for Sec23a and b proteins (39) and in C. elegans harboring loss of function mutation in the single C. elegans Sec23 allele (40). While eventually lethal, the main notable defect in these developing embryos is the inability to secrete collagen (41). A similar defect in collagen secretion is observed in humans caring mutation in Sec23a (42). These findings raise the question: What is the mechanism by which progression in embryogenesis occurs in the presence of disrupted COPII trafficking. Overall the results may suggest that additional COPII independent secretion pathways operate to support embryogenesis. Alternatively, robust maternal contribution of COPII proteins may be sufficient for maintaining key cellular secretion functions leading to the progression through earlier stages of embryogenesis. Studies in C. elegans support the former possibility. Here the somewhat limited phenotypes observed when Sec23 is mutated were indeed attributed to maternal contribution. Preventing maternal contribution was not possible so far as functional Sec23 is required for oogenesis (40). Perhaps in a similar manner Sar1-null drosophila embryos could not be generated due to the sterility of the generated flies.
Localized secretory pathways and axon development
The local axonal delivery and translation of mRNAs encoding receptors that regulate axon functions including the κ-opioid receptor (43), or the EphA2 receptor has been demonstrated (44). While up to a third of the human genome product is sorted at ERES, it is yet to be defined whether only locally translated cargoes are selectively mobilized through axonal ERES. Importantly, such receptors require a functional secretory pathway for delivery to the axonal plasma membrane. In most mammalian cells, such cargo proteins are transported from ERES to an organized Golgi, yet stacked Golgi cisternae have not been visualized in maturing axons. However, our analysis found that selective Golgi proteins are present in axons albeit in low abundance (Fig. 7), and thus unorganized Golgi outposts may support local biosynthetic traffic. In agreement with our study, a recent study has also demonstrated that growing axons with protein synthetic activity contain ER and Golgi components needed for classical protein synthesis and secretion. In support of the developmentally-regulated role for ERES during axon growth described here, the study demonstrated that the capacity of local secretion in axons increased by injury (45). The role of Golgi organization is not well defined and recent studies have shown that unstacking the Golgi complex leads to increase in vesicle formation (46). These results suggest that a disassembled or unorganized Golgi may be required during periods of high biosynthetic demands to promote effective traffic. A non-canonical Golgi independent pathway may also operate from local ERES in the axon. Recent studies have shown that during defined stages of developmental epithelial remodeling in drosophila integrins are trafficked in a non-canonical Golgi independent manner (47). However, in agreement with the presence of some Golgi proteins in the axon (Fig. 7), a recent study has demonstrated that the axonal delivery of the GABA transporter-1 is dependent not only on COPII activity (Sec24 subunit) but also on ARF-GAP1 which regulates sequential ARF1-dependent traffic to, within, and from the Golgi complex (48).
An additional Golgi-independent role for axonal ERES may also be in the sorting of proteins required for the assembly of compartments dedicated to calcium storage. In skeletal muscle the Golgi-independent assembly of sarcoplasmic reticulum is dependent on Sar1 mediated sorting at ERES (49). Future studies are required to define sorting and secretion pathways that mediate axonal traffic following exit from the ER.
In conclusion, our results combined with recent studies demonstrating the selective function of the secretory pathway in dendrite development strongly support our hypothesis that spatial and temporal regulation of the secretory pathway is required to support cell polarization and development.
Methods
Materials
Antibodies to Sar1, Sec23, Sec12, Sec13 and α-GDI were kindly provided by Dr. W.E. Balch (The Scripps Research Institute, La Jolla, CA). Expression vectors for Sar1-wt-Flag and Sar1-GDP-Flag were made from an expression vector for Sar1-GTP-Flag, which was kindly provided by Dr. S.I. Bannykh (Yale University, New Haven, CT) The antibody against Flag was purchased from Fisher Scientific (Pittsburgh, PA). Recombinant bFGF was purchased from R&D Systems (Minneapolis, MN). Antibody to Giantin was kindly provided by Dr. A. Linstedt, (Carnegie Mellon University, Pittsburgh PA). Antibodies to calnexin, clathrin, TfR, and Adaptin γ was from BD Biosciences (San Jose, CA). The Myc antibody (9E10) was from Upstate Biotechnology (Lake Placid, N.Y.). Antibodies to MAP2 and Tau were provided by Dr. Shelley Halpain (The University of California at San Diego, CA). Rhodamine phalloidin was purchased from Invitrogen (Carlsbad, CA) and the antibody to GM130 was purchased from BD Bioscience Pharmigen (San Diego, CA). Antibody to Yip1a and expression vector encoding Myc-tagged Yip1a were kindly provided by Dr. W. Hong (Institute of Molecular and Cell Biology, Singapore). The expression vector encoding Yip1a-GFP was kindly provided by Dr. M. Murata (University of Tokyo, Tokyo Japan). The expression vector for mEmerald-KDEL was kindly provided by Dr. M. Davidson (NHMFL at FSU, Tallahassee, FL). Species specific secondary antibodies conjugated to Alexa Fluor fluorophores were purchased from Invitrogen. Dissociated cultures were mounted using Fluormount-G purchased from Electron Microscopy Sciences (Hatfield, PA).
Sar1-shRNAs
There are two Sar1 genes in rodents: Sar1a and Sar1b. In this study, Sar1a and Sar1b pLentiLox3.7 shRNA vectors (Sar1a-shRNA and Sar1b-shRNA, respectively), which were kindly provided by Dr. B. Ye (University of Michigan, Ann Arbor, MI), were used to knock down Sar1 gene expression in neuron cultures. The sequences of the sense and antisense oligos used by Dr. Ye to generate Sar1a-shRNA and Sar1b-shRNA (collectively called Sar1-shRNAs) were:
Sar1a-shRNA oligos Sense: TGAACCACTCTTCTTCACATGTTCAAGAGACATGTGAAGAAGAGTGGTTCTTTTTTGGAAC
Antisense: TCGAGTTCCAAAAAAGAACCACTCTTCTTCACATGTCTCTTGAACATGTGAAGAAGAGTGGTTCA
Sar1b-shRNA oligos Sense:TGAACTACCTTCCTGCTATCATTCAAGAGATGATAGCAGGAAGGTAGTTCTTTTTTGGAAC
Antisense:TCGAGTTCCAAAAAAGAACTACCTTCCTGCTATCATCTCTTGAATGATAGCAGGAAGGTAGTTCA
The effectiveness of Sar1a-shRNA and Sar1b-shRNA in knocking down exogenously over-expressed HA epitope tagged Sar1a and Sar1b was previously tested by Western blot in Cos-7 cells (8). In addition, these shRNAs were shown to reduce endogenous Sar1 expression in neurons using fluorescence microscopy (8). Furthermore, by Western blot (Supplemental Fig. 1F) and quantitative fluorescence microscopy (Fig. 5B) we show that endogenous Sar1 expression is also reduced in HEK293 and neuronal cells transfected with Sar1-shRNAs.
To confirm the resistance of hamster Sar1a to knockdown by Sar1-shRNAs, HEK293 cells were co-transfected with the Sar1-shRNAs and Flag-tagged hamster Sar1a (Sar1-wt-Flag). Hamster Sar1a is a species ortholog of the rat Sar1b gene that has one nucleotide difference (AATTACCTTCCTGCTATCA) from the rat sequence targeted by Sar1b-shRNA (AACTACCTTCCTGCTATCA). The nucleotide difference appears to provide sufficient resistance to Sar1b-shRNA, and is also resistant to Sar1a-shRNA, which differs substantially in the targeted nucleotide sequence. Specifically, in neurons co-transfected with Sar1-wt-Flag and Sar1-shRNAs, Sar1-wt-Flag was efficiently expressed and targeted to the developing axon (Supplemental Fig. 2A). In addition, Sar1-wt-Flag expression was not reduced in HEK293 cells co-transfected with Sar1-wt-Flag + Sar1-shRNAs compared to those transfected with Sar1-wt-Flag alone (Supplemental Fig. 1F). Furthermore, in co-transfection experiments Sar1-wt-Flag rescued the negative effect Sar1-shRNAs have on axonal growth (Fig. 4).
Dissociated Neuron Cultures
Neurons were cultured at low density from embryonic day 18 (E18) Sprague Dawley rats or E15 B6129SF2/J mouse hippocampi as previously described (7, 50). Briefly, hippocampal neurons were plated onto poly-L-lysine-coated glass coverslips that are inverted over a monolayer of glial cells after 2 hr incubation. Paraffin dots attached to the coverslips were used to keep the neurons separated from the cells making up the glial feeder. Cells were plated at a density of 2700 cells per cm2 to achieve low-density cultures, which was required for visualization of ER export sites and to accurately measure individual axonal lengths. Most neurons, >90%, developed the characteristic mature morphology of spiny neurons between 16–21 DIV. Neurons were either transfected with lipofectamine 2000 (Invitrogen) at the indicated times post plating or were electroporated prior to plating using a Nucleofector II device (Amaxa Inc. Gaithersburg, MD) using the manufacturer’s rat neuron kit (VPG-1003). For the experiment that used bFGF to simulate axonal branching and growth, bFGF (10 ng/ml) was added to the culture medium at 2 hrs post plating or at 2 DIV and cultures were fixed and analyzed at 2 and 4 DIV, respectively.
Microscopy
Images were collected on a custom built two-headed (Up/Down) microscope. This system consists of a BX51WI Olympus microscope (up) base equipped with an Olympus DSU spinning disk confocal that sits on top of an inverted Olympus IX81 multispectral, prismless, TIRF microscope (down). The system is equipped with two Hamamatsu C4742-98 CCD cameras. The scopes share a high precision Prior motorized XY stage and are controlled through the same software (SlideBook 4.2; Intelligent Imaging Innovations, Inc; Denver, CO).
To quantify axonal length 5 sequential slices taken 0.2 µm apart through the midplane of the axon were collected with fixed illumination using a 40× 1.3 plan fluorite objective or a 60× 1.4 NA plan apochromat objective. Projection images were produced in SlideBook and exported to the program ImageJ. Using the semiautomatic neurite tracing plugin NeuronJ in ImageJ (51) we measured axonal length based on threshold segmentation. Using these experimental parameters, the axons from a minimum of 20 neurons from at least three different experiments per condition were measured.
To quantify Sar1 and GFP fluorescence image stacks consisting of sequential sections taken 0.2 µm apart and covering the entire neuron height were taken of each neuron using a 40× 1.3 plan fluorite objective. The Sar1 channel was deconvolved using the Agard/Sadat inverse matrix algorithm. When differences in illumination were necessary to prevent detector saturation intensity values were transformed using a linear relation to compare values. This is possible because of the highly linear nature of our imaging system (52). The volume of an axon can be particularly difficult to accurately measure in vitro because it is usually in contact with the coverglass, which can cause it to flatten out (become very thin in spots). Therefore, after performing background subtraction we created projection images of each neuron using the sum pixels setting. This resulted in a two-dimensional image made up of pixels that contained the intensity information of all voxels residing within the neuron that existed above the pixel. Using threshold segmentation methodologies previously described (52), we masked the axon, dendrites, and soma in order to measure the total fluorescence within each compartment. To correlate Sar1 fluorescence intensity between the axon, dendrites, and soma on the basis of volume we determined the Sar1/GFP fluorescence ratio of each compartment.
All experiments were repeated ≥ 3 times. E18 pups from a time pregnant female were used to obtain neurons for each replicate. Neurons were plated on 18 mm diameter coverslips in 60 mm tissue culture dishes as described above (5 coverslips/60 mm dish). Experimental conditions were performed on a per dish bases. For each replicate, neurons on ≥ 2 coverslips per experimental condition were imaged, with similar numbers of neurons being analyzed per coverslip and per replicate. In the text, the “n” value is the total number of neurons analyzed. When the data shown was for qualitative purposes only (e.g. Fig. 7) the images shown were chosen because they represented the protein staining pattern visualized across all experiments.
In order to completely display some neurons in their entirety, it was sometimes necessary to stitch overlapping fields together to get the entire axon in the image. In such cases, exposure times were kept constant and fields were stitched together without modifying background levels.
Statistical Analyses
Unless stated otherwise, one-way analysis of variance with post-hoc comparison via Tukey's Honestly Significant Difference was used to evaluate between group differences. In all cases, prior to analysis diagnostic statistics were used to confirm that the data were normally distributed.
ER Export Assay and Digitonin Permeabilization
Cultures were rinsed with phosphate buffered saline (PBS) followed by permeabilization with 25 µg/ml digitonin in KHM buffer (110mM KOAc, 20mM Hepes 7.4, 2mM MgOAc) at RT for 5 minutes then washed 3 times with KHM. Cells were prepared and incubated with the indicated components as described in the text. At the end of the incubations, the cells were rinsed, fixed, and prepared for imaging as described (7).
Supplementary Material
Rat hippocampal neurons were fixed at indicated times post plating. (A) Cultures were triple-labeled with phalloidin (A1; red in A4) and antibodies to Sar1 (A2; green in A4) and Map2 (A3; blue in A4). (B) 14 DIV cultures were labeled with antibodies to MAP2 (B1; red in B3) and Sar1 (B2; green in B3). (C) 21 DIV neurons were fixed and stained for MAP2 (C1; red in C3) and Sar1 (C2; green in C3). (D) A part of an axon in a 14 DIV stained for Tau (D1; red in D3) and Sar1 (D2; green in D3). When present, arrows point to the same place in corresponding images. (E) Analysis of Sar1 expression in the rat hippocampus, cortex (ctx), and cerebellum (cb). Samples were collected at the indicated date of development, normalized according to total protein (100 µg/lane) and analyzed by western blot with Sar1 antibodies. The antibody detected a doublet: Sar1b (top band) and Sar1a (lower band) proteins. Note that similar level of expression for Sar1a and Sar1b was detected in hippocampus, cortex, and cerebellum. In addition, no change in Sar1 expression levels was detected throughout the indicated development period. (F) HEK293 cells were transfected with equal amounts of DNA as follows: (1) Control (EBFP only); (2) Sar1-shRNAs+ EBFP (3&5) Sar1-wt-Flag +EBFP; or (4&6) Sar1-shRNAs + Sar1-wt-Flag, and analyzed at 3 days post transfection. A quantitative analysis (n=2) that compared the ratio of total Sar1 or Flag to β-actin between experimental conditions found total endogenous Sar1 expression to be reduced 80.6% in cultures transfected with Sar1-shRNAs compared to controls (compare lane 1 to lane 2). Total endogenous Sar1 was reduced 44.5% in cultures co-tranfected with Sar1- shRNAs + Sar1-wt-Flag verses those transfected with Sar1-wt-Flag alone (compare lanes 3 and 4). In contrast, Sar1-wt-Flag was not reduced in the former versus the later transfected cultures (increased 25.3%). A1–4 were made using two fields stitched together. Bars equal 10 µm.
(A) Primary rat hippocampal neurons were transfected with Sar1-wt-Flag and Sar1-shRNAs constructs prior to plating and fixed at 3 DIV. Sar1-wt-Flag (detected by anti-Flag antibodies) was targeted to the developing axon and reversed the axonal defect associated with reduced Sar1 expression (see Fig. 4 and main text). The arrows point to the corresponding place in each panel. (B) Primary rat hippocampal neurons were transfected with Sar1-shRNAs-Sar1-AB expressing constructs, which also express GFP, prior to plating and fixed at 5 DIV. The morphology of the Golgi complex in Sar1 depleted neurons expressing GFP (B2; green in B4) was compared to that in untransfected neurons using antibodies against GM130 (B1; red in B4) and MAP2 (B3; blue in B4). The filled arrowhead in B1 points to the Golgi of the neuron in the field that has been transfected with the Sar1-shRNAs constructs, while the open arrowhead points to that of an untransfected neighboring cell. The bar equals 10 µm.
Acknowledgements
Supported by NIH grants DK062318 (MA) and MH064372 (KNF), and a NARSAD Young Investigator Award (KNF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health. We are extremely grateful to Mrs. Rachel Aliotta and Mr. David Klinkenberg for their technical assistance.
References
- 1.Bannykh SI, Balch WE. Membrane dynamics at the endoplasmic reticulum-Golgi interface. J Cell Biol. 1997;138(1):1–4. doi: 10.1083/jcb.138.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostov K, Su T, ter Beest M. Polarized epithelial membrane traffic: conservation and plasticity. Nat Cell Biol. 2003;5(4):287–293. doi: 10.1038/ncb0403-287. [DOI] [PubMed] [Google Scholar]
- 3.Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y. Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110(5):1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morsomme P, Prescianotto-Baschong C, Riezman H. The ER v-SNAREs are required for GPI-anchored protein sorting from other secretory proteins upon exit from the ER. J Cell Biol. 2003;162(3):403–412. doi: 10.1083/jcb.200212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herpers B, Rabouille C. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-golgi units involved in gurken transport in Drosophila oocytes. Mol Biol Cell. 2004;15(12):5306–5317. doi: 10.1091/mbc.E04-05-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wihelm J, Buszczak M, Sayles S. Efficient Protein Trafficking Requires Trailer Hitch, a Component of a Ribonucleoprotein COmplex Localized to the ER in Drosophila. Developmental Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Aridor M, Guzik AK, Bielli A, Fish KN. Endoplasmic reticulum export site formation and function in dendrites. J Neurosci. 2004;24(15):3770–3776. doi: 10.1523/JNEUROSCI.4775-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130(4):717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehlers MD. Secrets of the secretory pathway in dendrite growth. Neuron. 2007;55(5):686–689. doi: 10.1016/j.neuron.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Bielli A, Haney CJ, Gabreski G, Watkins SC, Bannykh SI, Aridor M. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol. 2005;171(6):919–924. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iinuma T, Shiga A, Nakamoto K, O'Brien MB, Aridor M, Arimitsu N, Tagaya M, Tani K. Mammalian Sec16/p250 Plays a Role in Membrane Traffic from the Endoplasmic Reticulum. J Biol Chem. 2007;282(24):17632–17639. doi: 10.1074/jbc.M611237200. [DOI] [PubMed] [Google Scholar]
- 12.Fukata Y, Kimura T, Kaibuchi K. Axon specification in hippocampal neurons. Neurosci Res. 2002;43(4):305–315. doi: 10.1016/s0168-0102(02)00062-7. [DOI] [PubMed] [Google Scholar]
- 13.Aridor M, S.I B, Rowe T, Balch WE. Sequential Coupling Between COPII and COPI Vesicle Coats in Endoplasmic Reticulum to Golgi Transport. J Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradke F, Dotti CG. The role of local actin instability in axon formation. Science. 1999;283(5409):1931–1934. doi: 10.1126/science.283.5409.1931. [DOI] [PubMed] [Google Scholar]
- 15.Aridor M, Fish KN, Bannykh S, Weissman J, Roberts TH, Lippincott-Schwartz J, Balch WE. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol. 2001;152(1):213–229. doi: 10.1083/jcb.152.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumental-Perry A, Haney CJ, Weixel KM, Watkins SC, Weisz OA, Aridor M. Phosphatidylinositol 4-phosphate formation at ER exit sites regulates ER export. Dev Cell. 2006;11(5):671–682. doi: 10.1016/j.devcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Krijnse-Locker J, Parton RG, Fuller SD, Griffiths G, Dotti CG. The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol Biol Cell. 1995;6(10):1315–1332. doi: 10.1091/mbc.6.10.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CZ, Collins RN. Insights into biological functions across species: examining the role of Rab proteins in YIP1 family function. Biochem Soc Trans. 2005;33(Pt 4):614–618. doi: 10.1042/BST0330614. [DOI] [PubMed] [Google Scholar]
- 19.Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23(15):6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science. 2006;313(5785):324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mundigl O, Matteoli M, Daniell L, Thomas-Reetz A, Metcalf A, Jahn R, De Camilli P. Synaptic vesicle proteins and early endosomes in cultured hippocampal neurons: differential effects of Brefeldin A in axon and dendrites. J Cell Biol. 1993;122(6):1207–1221. doi: 10.1083/jcb.122.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135(1):19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens DJ. De novo formation, fusion and fission of mammalian COPII-coated endoplasmic reticulum exit sites. EMBO Rep. 2003;4(2):210–217. doi: 10.1038/sj.embor.embor736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aridor M, Bannykh SI, Rowe T, Balch WE. Cargo can modulate COPII vesicle formation from the endoplasmic reticulum. J Biol Chem. 1999;274:4389–4399. doi: 10.1074/jbc.274.7.4389. [DOI] [PubMed] [Google Scholar]
- 25.Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. Embo J. 2008;27(15):2043–2054. doi: 10.1038/emboj.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forster R, Weiss M, Zimmermann T, Reynaud EG, Verissimo F, Stephens DJ, Pepperkok R. Secretory cargo regulates the turnover of COPII subunits at single ER exit sites. Curr Biol. 2006;16(2):173–179. doi: 10.1016/j.cub.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 27.Katsuki H, Itsukaichi Y, Matsuki N. Distinct signaling pathways involved in multiple effects of basic fibroblast growth factor on cultured rat hippocampal neurons. Brain research. 2000;885(2):240–250. doi: 10.1016/s0006-8993(00)02953-x. [DOI] [PubMed] [Google Scholar]
- 28.Szebenyi G, Dent EW, Callaway JL, Seys C, Lueth H, Kalil K. Fibroblast growth factor-2 promotes axon branching of cortical neurons by influencing morphology and behavior of the primary growth cone. J Neurosci. 2001;21(11):3932–3941. doi: 10.1523/JNEUROSCI.21-11-03932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walicke P, Cowan WM, Ueno N, Baird A, Guillemin R. Fibroblast growth factor promotes survival of dissociated hippocampal neurons and enhances neurite extension. Proc Natl Acad Sci U S A. 1986;83(9):3012–3016. doi: 10.1073/pnas.83.9.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charcosset M, Sassolas A, Peretti N, Roy CC, Deslandres C, Sinnett D, Levy E, Lachaux A. Anderson or chylomicron retention disease: molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Molecular genetics and metabolism. 2008;93(1):74–84. doi: 10.1016/j.ymgme.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 31.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426(6968):803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 32.Martin KC. Local protein synthesis during axon guidance and synaptic plasticity. Curr Opin Neurobiol. 2004;14(3):305–310. doi: 10.1016/j.conb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Craig AM, Wyborski RJ, Banker G. Preferential addition of newly synthesized membrane protein at axonal growth cones. Nature. 1995;375(6532):592–594. doi: 10.1038/375592a0. [DOI] [PubMed] [Google Scholar]
- 34.Griffin JW, Price DL, Drachman DB, Morris J. Incorporation of axonally transported glycoproteins into axolemma during nerve regeneration. J Cell Biol. 1981;88(1):205–214. doi: 10.1083/jcb.88.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toews AD, Armstrong R, Ray R, Gould RM, Morell P. Deposition and transfer of axonally transported phospholipids in rat sciatic nerve. J Neurosci. 1988;8(2):593–601. doi: 10.1523/JNEUROSCI.08-02-00593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta V, Palmer KJ, Spence P, Hudson A, Stephens DJ. Kinesin-1 (uKHC/KIF5B) is required for bidirectional motility of ER exit sites and efficient ER-to-Golgi transport. Traffic. 2008;9(11):1850–1866. doi: 10.1111/j.1600-0854.2008.00811.x. [DOI] [PubMed] [Google Scholar]
- 37.Cai Q, Pan PY, Sheng ZH. Syntabulin-kinesin-1 family member 5B-mediated axonal transport contributes to activity-dependent presynaptic assembly. J Neurosci. 2007;27(27):7284–7296. doi: 10.1523/JNEUROSCI.0731-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman A, Kamal A, Roberts EA, Goldstein LS. Defective kinesin heavy chain behavior in mouse kinesin light chain mutants. J Cell Biol. 1999;146(6):1277–1288. doi: 10.1083/jcb.146.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nature genetics. 2006;38(10):1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- 40.Roberts B, Clucas C, Johnstone IL. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol Biol Cell. 2003;14(11):4414–4426. doi: 10.1091/mbc.E03-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci. 2008;121(Pt 18):3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- 42.Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, Orci L, Eyaid W. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nature genetics. 2006;38(10):1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 43.Bi J, Tsai NP, Lin YP, Loh HH, Wei LN. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc Natl Acad Sci U S A. 2006;103(52):19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110(2):223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- 45.Merianda TT, Lin AC, Lam JS, Vuppalanchi D, Willis DE, Karin N, Holt CE, Twiss JL. A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol Cell Neurosci. 2009;40(2):128–142. doi: 10.1016/j.mcn.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Wei JH, Bisel B, Tang D, Seemann J. Golgi Cisternal Unstacking Stimulates COPI Vesicle Budding and Protein Transport. PLoS ONE. 2008;3(2):e1647. doi: 10.1371/journal.pone.0001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotman H, Karhinen L, Rabouille C. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev Cell. 2008;14(2):171–182. doi: 10.1016/j.devcel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Reiterer V, Maier S, Sitte HH, Kriz A, Ruegg MA, Hauri HP, Freissmuth M, Farhan H. Sec24- and ARFGAP1-dependent trafficking of GABA transporter-1 is a prerequisite for correct axonal targeting. J Neurosci. 2008;28(47):12453–12464. doi: 10.1523/JNEUROSCI.3451-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nori A, Bortoloso E, Frasson F, Valle G, Volpe P. Vesicle budding from endoplasmic reticulum is involved in calsequestrin routing to sarcoplasmic reticulum of skeletal muscles. Biochem J. 2004;379(Pt 2):505–512. doi: 10.1042/BJ20031875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacLaurin SA, Krucker T, Fish KN. Hippocampal dendritic arbor growth in vitro: regulation by Reelin-Disabled-1 signaling. Brain research. 2007;1172:1–9. doi: 10.1016/j.brainres.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58(2):167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 52.Fish KN, Sweet RA, Deo AJ, Lewis DA. An automated segmentation methodology for quantifying immunoreactive puncta number and fluorescence intensity in tissue sections. Brain research. 2008;1240:62–72. doi: 10.1016/j.brainres.2008.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rat hippocampal neurons were fixed at indicated times post plating. (A) Cultures were triple-labeled with phalloidin (A1; red in A4) and antibodies to Sar1 (A2; green in A4) and Map2 (A3; blue in A4). (B) 14 DIV cultures were labeled with antibodies to MAP2 (B1; red in B3) and Sar1 (B2; green in B3). (C) 21 DIV neurons were fixed and stained for MAP2 (C1; red in C3) and Sar1 (C2; green in C3). (D) A part of an axon in a 14 DIV stained for Tau (D1; red in D3) and Sar1 (D2; green in D3). When present, arrows point to the same place in corresponding images. (E) Analysis of Sar1 expression in the rat hippocampus, cortex (ctx), and cerebellum (cb). Samples were collected at the indicated date of development, normalized according to total protein (100 µg/lane) and analyzed by western blot with Sar1 antibodies. The antibody detected a doublet: Sar1b (top band) and Sar1a (lower band) proteins. Note that similar level of expression for Sar1a and Sar1b was detected in hippocampus, cortex, and cerebellum. In addition, no change in Sar1 expression levels was detected throughout the indicated development period. (F) HEK293 cells were transfected with equal amounts of DNA as follows: (1) Control (EBFP only); (2) Sar1-shRNAs+ EBFP (3&5) Sar1-wt-Flag +EBFP; or (4&6) Sar1-shRNAs + Sar1-wt-Flag, and analyzed at 3 days post transfection. A quantitative analysis (n=2) that compared the ratio of total Sar1 or Flag to β-actin between experimental conditions found total endogenous Sar1 expression to be reduced 80.6% in cultures transfected with Sar1-shRNAs compared to controls (compare lane 1 to lane 2). Total endogenous Sar1 was reduced 44.5% in cultures co-tranfected with Sar1- shRNAs + Sar1-wt-Flag verses those transfected with Sar1-wt-Flag alone (compare lanes 3 and 4). In contrast, Sar1-wt-Flag was not reduced in the former versus the later transfected cultures (increased 25.3%). A1–4 were made using two fields stitched together. Bars equal 10 µm.
(A) Primary rat hippocampal neurons were transfected with Sar1-wt-Flag and Sar1-shRNAs constructs prior to plating and fixed at 3 DIV. Sar1-wt-Flag (detected by anti-Flag antibodies) was targeted to the developing axon and reversed the axonal defect associated with reduced Sar1 expression (see Fig. 4 and main text). The arrows point to the corresponding place in each panel. (B) Primary rat hippocampal neurons were transfected with Sar1-shRNAs-Sar1-AB expressing constructs, which also express GFP, prior to plating and fixed at 5 DIV. The morphology of the Golgi complex in Sar1 depleted neurons expressing GFP (B2; green in B4) was compared to that in untransfected neurons using antibodies against GM130 (B1; red in B4) and MAP2 (B3; blue in B4). The filled arrowhead in B1 points to the Golgi of the neuron in the field that has been transfected with the Sar1-shRNAs constructs, while the open arrowhead points to that of an untransfected neighboring cell. The bar equals 10 µm.