Abstract
The androgen receptor (AR), which mediates the signals of androgens, plays a crucial role in prostate related diseases. Although widely used, currently marketed anti-androgenic drugs have significant side effects. Several studies have revealed that non-steroidal anti-inflammatory drugs, like flufenamic acid, block AR transcriptional activity. Herein we describe the development of small molecule analogs of flufenamic acid that antagonize AR. This novel class of AR inhibitors binds to the hormone binding site, blocks AR transcription activity, and acts on AR target genes.
Keywords: Androgen receptor, transcription factor, anti-androgen, flufenamic acid
INTRODUCTION
The Androgen Receptor (AR) is crucial for development and maintenance of the sexual characteristics, bone density, and muscle. AR is also an important mediator for diseases like prostate benign hyperplasia (BPH) and prostate cancer (1). AR belongs to the steroid receptor subclass of the nuclear receptor family (NRs), intracellular transcription factors that respond directly to their ligands (2–3). In the absence of androgens like dihydrotestosterone (DHT), AR resides in the cytoplasm, bound to chaperone heat shock proteins (HSP). Upon ligand binding, inactive AR dissociates from heat shock proteins, undergoes a series of conformational changes, and translocates to the nucleus (4). In the nucleus, ligand-activated AR binds to specific androgen response elements (ARE), recruits coregulatory proteins, and starts the regulation of a distinct set of genes (Figure 1) (5–8). Complex interactions with either coactivators (CoA) or corepressors (CoR) fine tune AR mediated gene expression (9–10). Like other NRs, ligand-bound AR exhibits extra binding sites on its surface such as the activation function 2 (AF2) that recruits coregulators such as the steroid receptor coactivator (SRC) family (11–14). We have previously identified another allosterically regulated binding site, BF3, on the surface of AR that can affect AF2 function (15).
Figure 1.
Alternative modes of binding to the Androgen Receptor (AR). In the absence of endogenous ligand, dihydrotestosterone (DHT), inactive AR resides in the cytoplasm bound to chaperone heat shock proteins (HSP). Upon ligand binding, inactive AR dissociates from HSP, undergoes a series of conformational changes, becomes active and translocates to the nucleus. DHT (a.) or competitive agonists (b.) bind to the hormone binding site and allow AR in an activated conformation to recruit coactivators (CoA), which in turn exert gene regulation by recruiting the transcription machinery. DHT competitive antagonists or anti-androgens (c.) bind to the hormone binding site but lock the AR in an inactive conformation where CoA cannot bind and gene transcription is blocked. CoA competitive antagonists (d.) bind to the surface of ligand-bound AR preventing the recruitment of CoA and transcription machinery without affecting androgen binding.
Anti-androgens like hydroxyflutamide or bicalutamide are used successfully against androgen dependent prostate cancer but exhibit strong side effects including gynecomastia, impotence, osteoporosis, and cardiovascular diseases. Additionally, tumors treated with anti-androgens become resistant to that therapy within several years of treatment (16–18). For these reasons current pharmacological strategies targeting AR are focused on the development of SARMs (selective Androgen Receptor Modulators) that interact with the ligand binding pocket of AR and regulate AR mediated gene transcription in a tissue selective manner (19–20). Since AR plays an important oncogenic role in hormone refractory prostate cancer, targeting AR signaling in the setting of tumor resistance remains particularly crucial (21). It has been reported that tumor resistance can be due to AR mutations occurring in the ligand binding domain (LBD) that produce a receptor hypersensitive to its natural ligand, other endogenous hormones, and synthetic anti-androgens (22–23). One can envision that drugs acting through mechanisms complementary to ligand antagonism might prove useful in combination with SARMS.
We identified flufenamic acid (1) (FLF) and other novel AR inhibitors using high throughput screening and showed that this non-steroidal anti-inflammatory drug (NSAID) is able to block AR transcription activity (15). Other studies have shown that FLF inhibits cell growth (24), the expression of the AR in LNCaP (25) and aldo-keto reductases (AKR) which can affect androgen signaling at the level of hormone metabolism (26–27). Herein, we describe the synthesis and evaluation of FLF analogs.
RESULTS AND DISCUSSION
Biochemical Structure-activity relationships of Flufenamic Acid Analogs
Based upon our identification of flufenamic acid (FLF) (1) as an inhibitor of AR transcriptional activity (15), we carried out a structure activity relationship (SAR) study by synthesizing structural analogs of FLF (Figure 2A). To address the importance of the carboxylic acid group we prepared the corresponding methyl ester (2) and amide (3). To evaluate the role of the amine function, we replaced the secondary nitrogen with oxygen (3) or sulfur (4) atoms. Additionally, we elongated the spacer function Y between the two aromatic rings by introducing an additional methylene (5 and 6) or a carbonyl group (7). Finally, the effects of structural rigidification were explored using both commercially available and synthesized tricyclic analogs of FLF (9–13). All FLF derivatives were tested to determine their ability to bind to the AR and independently their ability to disrupt binding of transcriptional cofactors.
Figure 2.
Preliminary structure-activity relationships of Flufenamic acid (1) derivatives. A) Synthesized analogs of FLF. B) Heat maps reflecting the binding affinities of FLF analogs 1-13 for the AR-LBD (IC50 values available in table 3 SI): 1) DHT binding site: a Scintillation Proximity Assay (SPA) measuring [3H]-DHT displacement from AR-LBD (28). 2) CoA binding: A Fluorescence Polarization assay (FP) measuring fluorescently labeled SRC2-3 peptide displacement from DHT-bound AR-LBD (15). 3) Solubility limits were quantified by UV absorbance under conditions reflecting binding assays conditions. Values are means of two independent experiments in triplicates. The general error limits are ± 5%.
These FLF derivatives were tested for their ability to directly displace the endogenous radioactive ligand [3H]-DHT from AR-LBD using a Scintillation Proximity assay (SPA) (Figure 2B, IC50 values available in table 3 SI) (28). The synthesized FLF analogs were tested for their ability to compete with a fluorescently labeled coregulatory peptide, mimicking the coregulatory protein SRC2-3, for binding to DHT-bound AR-LBD (15). These two biochemical assays distinguish the ability of the small molecules to competitively inhibit ligand binding, thus allosterically regulating coregulatory protein recruitment, or directly inhibiting the recruitment of coregulatory proteins (Figure 1).
The carboxylic acid and amine moieties of flufenamic acid were required for binding to the AR as demonstrated by the finding that the derivatives 2–5 were inactive in both assays (Figure 2B, table 3 SI). The introduction of a longer spacer in between the two aromatic rings yielded inactive compounds as well (6–8). More rigid structures like the tricyclic analogs 9–13 did not show any particular improvement in binding to the AR-LBD. In addition, we confirmed the findings of Bisson et al. (29) that phenothiazines are able to inhibit AR since phenothiazine 11 showed a weak affinity for the hormone binding site.
With these basic elements of SAR set, a focused library was prepared to give better understanding of the role of substituents on the B-ring of FLF analogs (Figure 3, IC50 values available in table 3 SI).
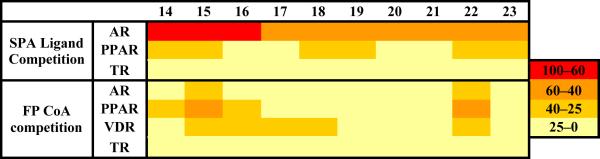
Figure 3.
Structure-activity relationships of the distal B phenyl ring of Flufenamic acid derivatives. A) Heat maps reflecting the binding affinities of FLF analogs 14-75 for the AR-LBD (IC50 values available in table 3 SI): 1) DHT binding site: a Scintillation Proximity Assay (SPA) measuring [3H]-DHT displacement from AR-LBD (28). 2) CoA binding: A Fluorescence Polarization assay (FP) measuring fluorescently labeled SRC2-3 peptide displacement from DHT-bound AR-LBD (15). 3) Solubility limits were quantified by UV absorbance under conditions reflecting binding assays conditions. Values are means of two independent experiments in triplicates. The general error limits are ± 5%. B) Correlation plots of log IC50 obtained in DHT and CoA competition binding assays against hydrophobic character of each para R substituents represented by π values (30).
A focused library of FLF derivatives was produced by Ullmann coupling between the o-chlorobenzoic acid and locally synthesized or commercially available anilines (Scheme 1 SI). This library was tested in biochemical assays described above (Figure 3A). Two subsets of compounds emerged from this study. One set was able to displace the coregulator peptide without affecting DHT binding. Compounds 36, 37, 38, and 39 had a slightly improved ability to displace peptide from the AF2 pocket relative to FLF. The second set of FLF derivatives inhibited both interactions between coactivator peptide and the AR and between AR and DHT. Although the canonical model states that compounds that displace the hormone will allosterically block binding of the coregulatory peptide, we observed a very poor correlation between the two assays (r2 = 0,187, Figure 6 SI).
Hydrogen bonding interactions are often important contributors to the binding between small molecules and proteins. Flufenamic acid has a weakly basic diphenylamine functionality and an acidic aromatic carboxylic group and both of these functionalities are essential for binding to the AR. The ability of flufenamic acid to generate an intermolecular hydrogen bond with AR could be influenced by different electron donating or withdrawing groups on the aromatic rings. Comparing FLF analogs with different B-ring substituents in para position to the nitrogen reveals similar binding affinities for electron-donating substituted derivatives (compounds 50, 62, and 64) and electron-withdrawing substituted derivatives (compounds 37, 47, 48, and 75). Additional substitution on the B-ring in presence of a m-CF3 group, with electron-withdrawing groups (p-NO2 24) or with electron-donating groups (p-OPh 26, p-OPh(m-OMe) 33, p-OPh(p-OMe) 44) also had little effect on potency. Correlation plots of the electronic effect of introduced substituents (characterized by σ values (30)) and both competition assays showed no evidence of a Hammett relationship (r2 < 0.5, figure 7 SI). Interestingly, analog 24 that incorporates a 4-nitro-3-trifluoromethylaniline moiety, a structural feature found in the anti-androgen hydroxyflutamide, did not show any activity either assays.
However, the hydrophobic character of the substituents (π values) was found to influence the activity of the FLF derivatives (Figure 3B). Plotting the π value of each para substituent against log IC50 gave significant correlations for both the SPA (r2 = 0.88, p < 0.0001, n = 15) and FP (r2 = 0.81, p = 0.0003, n = 10) assays. We observed a stronger relationship for the hormone displacement assay when comparing the slopes of the two linear plots (|slopeSPA| = |−0.69 ± 0.07| > |slopeFP| = |−0.33 ± 0.05|). Compounds with long alkyl chains (O-n-Hexyl 17, n-Hexyl 18) had stronger affinities for the AR than compounds with shorter hydrophobic groups like methyl (65), iso-propyl (51), tert-butyl (42). A similar trend was seen in comparing pentyl ester 22 to methyl ester 75. Additionally, phenyl substituted derivatives such as 15 (thiophenyl), 20 (benzyl), or 23 (phenoxy) gave potency in the low micromolar range in contrast to phenylsulfonyl compound (58). Finally, the pyridinyl compound 27 showed lower activity than the phenoxy analog 23. In a subsequent small directed library, introduction of electro-withdrawing (NO2 14, CF3 16, F 21) or electro-donating (Me 19) groups on the third aromatic ring had little effect on the potency of the compounds.
There were relatively strong positional effects for the substituents on the B-ring: para-substituted derivatives had generally better potencies than meta or ortho substituted compounds. For instance, the para-benzyl analog 20 is more potent than both the meta-benzyl analog 40 and the ortho-benzyl analog 41. Indeed, the most active compounds are all para-substituted FLF derivatives (14–23). No active compounds was observed among the di-substituted FLF analogs such as 3,5-Me 55, 3,5-OMe 59, 3,5-OPh 31, (o-Me, m-Cl) 32, (2-NH2, 3-Cl) 68.
Since the most potent analogs had additional aliphatic or aromatic substituents, their increased hydrophobic character may reduce their solubility in aqueous media. Thus the solubility limit of each FLF analog was determined in a buffer containing 5% DMSO (pH = 7.4) reflecting the assay conditions. Overall, the derivatives showed solubility limits significantly higher than their observed potencies. Therefore, solubility is not believed to interfere with the assays.
To investigate the specificity of these FLF analogs towards the androgen receptor we selected the ten most potent compounds 14–23 and determined their activities with respect to other nuclear receptors. Ligand displacement was monitored for the AR, peroxisome proliferator-activated (PPARγ) and thyroid (TR) receptors (28) and co-activator displacement for the AR, PPARγ, vitamin D (VDR) and TR receptors (15, 31–32) (Figure 4, % inhibition values available in figure 8 SI).
Figure 4.
Summary of selectivity of FLF acid analogs for AR. Each ligand was tested at a single concentration of 10 μM against various nuclear receptors to determine effects on ligand binding (SPA assay) (28) or coactivator binding (FP assay) (31–32). Results are shown as % inhibition relative to controls (values available in figure 8 SI). Values are means of two independent experiments in triplicates. The general error limits are ± 5%.
This set of compounds had no effect whatsoever on the TR. However some of the analogs showed some activity at 10 μM against both the VDR and PPARγ receptors. Overall, we could conclude that compounds 14 and 16 exhibit a reasonable but not strong specificity for the AR.
Transcriptional Structure-Activity Relationships of Flufenamic Acid Analogs
The most potent FLF derivatives 14–23 were tested to determine their effects upon AR mediated gene transcription. The inhibition of DHT induced transcription in the presence of FLF analogs was determined in MDA-kb2, a cell line stably expressing an AR responsive luciferase reporter gene (33) driven by a MMTV promoter (Table 1 and 3 SI). The cells were treated with compounds in the presence DHT and transcriptional AR mediated signal was measured by luminescence after 20 hours using BrightGLo® (Promega). Two concentrations (0.2 nM and 8 nM) of DHT were chosen for the assay representing the previously determined EC50 and EC90 of DHT under identical conditions (data not shown). Additionally, we evaluated the cell permeability and membrane retention of FLF analogs using a parallel artificial membrane permeation assay (PAMPA) (34–35). This assay was carried out with a 0.5% DMSO content at pH 7.4, reflecting the conditions of the cell-based assays.
Table 1.
Summary of transcriptional activities and cytotoxicities of FLF analogs in MDA-kb2 (33) and cellular permeabilities and membrane retentions determined by a PAMPA assay (34–35).
| # | R | Transcriptional Inhibition (IC50) in MDA-kb2a | Cytotoxicity (IC50) in MDA-kb2 (μM)b | Permeability (10‒6 cm s‒1)c [Retention in membrane (%)]d | |
|---|---|---|---|---|---|
| 0.2 nM DHT(μM) | 8 nM DHT (μM) | ||||
| 14 | p-O(p-NO2)Ph | 1.9 ± 0.9 [100] | 38.6 ± 4.7 [100] | Non toxic | 191 ± 18 [87] |
| 15 | p-SPh | 6.5 ± 5.7 [75] | 14.4 ± 5.5 [30] | Non toxic | 180 ± 16 [93] |
| 16 | p-O(p-CF3)Ph | 3.7 ± 1.8 [100] | 53.0 ± 5.1 [100] | Non toxic | 118 ± 29 [92] |
| 17 | p-nOHex | 20.5 ± 18.7 [75] | >100 | Non toxic | 55 ± 10 [68] |
| 18 | p-nHex | 7.2 ± 3.9 [75] | 77.2 ± 17.7 [50] | Non toxic | 160 ± 116 [88] |
| 19 | p-O(p-Me)Ph | 0.4 ± 0.2 [50] | >100 | Non toxic | 106 ± 8 [87] |
| 20 | p-Bn | 2.2 ± 1.0 [75] | >100 | Non toxic | 98 ± 31 [89] |
| 21 | p-O(p-F)Ph | 3.5 ± 2.4 [25] | >100 | Non toxic | 113 ± 12 [88] |
| 22 | p-CO2C5H11 | 0.7 ± 0.3 [100] | 83.8 ± 20.7 [100] | Non toxic | 294 ± 40 [91] |
| 23 | p-OPh | 0.9 ± 0.3 [50] | >100 | 181.4 ± 99.4 | 153 ± 29 [84] |
| Hydroxyflutamide | 0.06 ± 0.03 [100] | 7.2 ± 5.5 [100] | Non toxic | 183 ± 29 [75] | |
| Bicalutamide | 0.5 ± 0.2 [100] | 11.1 ± 4.8 [100] | Non toxic | 628 ±22 [88] | |
Transcription assay was performed in MDA-kb2 (33), stably transformed cell line expressing a MMTV-Luc reporter gene. The assay was conducted in presence of either 0.2 nM or 8 nM of DHT corresponding to the EC50 and EC90 values determined for DHT. % inhibition is reported in brackets.
Cytotoxicity was measured with Cell-Titer Glo®.
In the presence of 0.2 nM DHT, all FLF analogs (except 17) inhibited AR transcription with IC50 below 10 μM. The most potent full inhibitor 22 had an IC50 of 700 nM. Three of these compounds (19, 21, and 23) exhibited partial antagonism. When tested in presence of 8 nM DHT (EC90 of DHT in this assay), we observed increased IC50 values for all compounds. This indicates that direct competition for DHT is a significant component of the mechanism of action for the tested FLF analogs. A similar IC50 shift was observed for control anti-androgens, hydroxyflutamide and bicalutamide. In parallel, we determined that none of the FLF derivatives directly inhibited the luciferase enzyme obtained from lysed MDA-kb2 cells grown in the presence of DHT thus ensuring unambiguous analysis (Table 2 SI). Additionally, the cytotoxicities of these compounds were determined under identical conditions. The LD50 values observed were in general higher than 150 μM. Other mammalian cell lines (HepG2, Raji, Hek293, BJ) were used to determine general and specific toxicity (Tables 2 and 3 SI). None of the compounds exhibited any significant cytotoxicity independently of any cell line. The permeabilities of the FLF analogs tested were within a range indicating reasonable cell access by passive permeation (360 > Pe > 40*10−6 cm s−1) although they did have relatively elevated membrane retention, congruent with their hydrophobicity. Theses values are comparable with marketed anti-androgens.
Specific Regulation of FKBP51 gene transcription by Flufenamic Acid Analogs
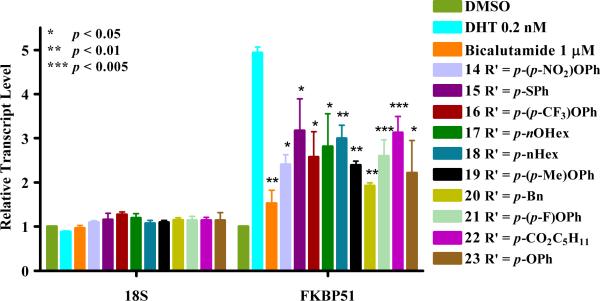
FKBP51 is a FK506-binding protein with isopropyl peptidyl isomerase activity that associates with heat shock proteins (HSP90, HSP70, and HSP40) and plays a role in the proper folding, binding and intracellular trafficking of steroid hormone receptors (5, 36). This AR target gene is upregulated in the presence of androgens in various cell lines including CWR22 (5), LNCaP (6), PC3, and DU145 (37). In control experiments with MDA-kb2 cells FKBP51 was induced by 4–5 fold in presence of 0.2 nM DHT. The quantification of the mRNA transcription levels of FKBP51, 18S and GAPDH in MDA-kb2 cells treated with DHT and FLF analogs was carried out using rt-PCR after an exposure time of 20 hours (no toxicity was detected at that time point, see Table 1). The results are presented in Figure 5.
Figure 5.
The effects of FLF analogs on 18S and FKBP51 transcription in MDA-kb2 cells. Cells were exposed to drugs (10 μM) for 20 hours. 18S and FKBP51 transcription was monitored by qRT-PCR. The levels for the tested genes are normalized to GAPDH transcript level and to DMSO control condition. The ΔΔCt method was used to measure the fold change in expression of genes. Standard deviations were calculated from 3 biological independent experiments performed in triplicates.
Bicalutamide was chosen as a control drug since OHF acts as a mixed agonist in MDA-kb2 (33). No dramatic effect was observed on house-keeping gene expression (GAPDH and 18S), confirming that FLF analogs were not general transcription inhibitors. All compounds were able to decrease transcription levels of FKBP51 in the presence of 0.2 nM DHT after 20 hours exposure when tested at a concentration of 10 μM. The most active compound was compound 20, exhibiting a similar efficacy to bicalutamide in presence of 0.2 nM of DHT.
Based on the initial discovery that Flufenamic acid could bind to the androgen receptor and modulate its function, we synthesized a focused library of flufenamic acid analogs and tested their ability to block binding of endogenous ligand and coactivator. Like FLF, a small set of these compounds (36, 37, 38, and 39) were found to preferentially displace the coactivator rather than competing with the endogenous ligand DHT. However, unlike FLF, these compounds did not inhibit AR mediated transcription or exhibit general toxicity (Table 3 SI). Overall, none of these FLF derivatives exhibits significantly tighter affinity than FLF for the coactivator binding site.
Remarkably, a larger set of the FLF analogs competitively inhibited ligand binding, rather than competing directly for the co-activator binding site or binding to the BF3 site. Many of these compounds were potent inhibitors of DHT mediated transcription. The novel anti-androgens we identified have a totally different structure than the currently marketed anti-androgenic drugs. Indeed, compound 24, structurally similar to hydroxyflutamide, was not active in either biochemical or cell-based assays. However these FLF derivatives (14–23) have several common structural features that are crucial for interacting with the AR including: a free carboxylic acid, a secondary amine linking the two aryl rings, and a single hydrophobic group in para position on the B-ring. Based on correlation plots (Figure 3B) of our SAR study on the FLF analogs, hydrophobicity is a leading component for the preferential binding of these compounds to the hormone binding site rather than to the coactivator binding site.
In general, these analogs, which are able to displace the hormone in low micromolar range, were also able to displace the coactivator peptide. This may suggest that for this particular class of compounds long alkyl chain or articulated phenyl groups in this para position are well sited and bulky enough to prevent the formation of the AF2 or other external sites where coregulators are being recruited.
Except for compound 17, we noted a good correlation between biochemical inhibition and inhibition of transcription among this set of compounds. Indeed, we measured for this particular analog a higher IC50 (20.5 ± 18.7 μM) in the transcription assay in comparison with an IC50 value of 4.5 μM in the DHT competition assay. Compound 17 showed the lowest cell permeability of this series (55×10−6 cm/s), which could explain its reduced efficiency to perform in cells than in biochemical conditions. Additionally, analogs 19, 21, and 23 were not able to fully inhibit AR transcription signal in cells whereas they did compete completely for ligand binding under biochemical conditions. Those para-phenoxy derivatives showed partial antagonism whereas compounds 14 and 16 showed full antagonism. We hypothesize that electron- withdrawing groups introduced on this third aromatic ring lead to full antagonists.
The competitive mode of action of those compounds was also confirmed in cells since a loss of activity was observed when hormone concentration was increased. Finally, the FLF analogs act on AR target genes and are not general transcription inhibitors.
In conclusion, FLF derivatives exhibit a range of biological activities with respect to the androgen receptor. A subgroup of compounds, which were DHT antagonists, were active in cells and inhibited transcription of an AR target gene. Overall, this series contains novel non-toxic antagonists of AR with potencies comparable to currently marketed anti-androgens. For these reasons, the continued examination of this compound series is warranted.
METHODS
General Considerations
Synthetic procedures and copies of NMR spectra for each material described are collected in the Supporting Information. Hydroxyflutamide was purchased from LKT Laboratories, Inc. Bicalutamide was purchased from Toronto Research Chemicals, Inc. Radiolabeled dihydrotestosterone ([3H]-DHT) ([1,2,4,5,6,7-3H(N)]-(5α-androstan-17β-ol-3-one), 110 Ci/mmol, was obtained from PerkinElmer.
All cell lines are purchased from ATCC. All media were supplemented with 10% FBS, 2 mM L-Glutamine and other indicated reagents. Rajii cells were maintained at 37°C in RPMI 1640/HEPES media (Gibco BRL) supplemented with sodium pyruvate (1 mM) and HEPES (10mM). HepG2, Hek293, and BJ cells were maintained at 37°C in D-MEM media (Gibco BRL) supplemented with NEAA (0.1mM) and sodium pyruvate (1 mM). MDA-kb2 cells were maintained at 37°C, without CO2 in L-15 media (Gibco BRL) supplemented with geniticin (500 μg mL−1), penicillin (100 U mL−1), and streptomycin (100 μg mL−1).
Fluorescence polarization and luminescence experiments were performed with an EnVision (PerkinElmer). Radiation experiments were performed with a TopCount Microplate Scintillation and Luminescence Counter (Packard Instrument Company).
All data were analyzed using GraphPad Prism 4.03 (GraphPad Software, San Diego, CA) and IC50 values were obtained by fitting data to equation (Sigmoidal dose-response (variable slope) or “four parameter logistic equation”): y = Bottom + (Top- Bottom)/(1+10^((LogEC50 − x)*Hillslope)); x is the logarithm of concentration; y is the response. Values are means of two independent experiments in triplicates. The general error limits are ± 5%.
Biochemical Assays
Protein Expression and Purification
cAR-LBD (His6; residues 663-919) was expressed in E. coli and purified to homogeneity in the presence of dihydrotestosterone (DHT, 10 μM) using a modified version of published protocols (28). The protein was stored with twice the amount of DHT (20 μM) to ensure fully liganded AR-LBD. Protein concentrations were measured by Bradford and BCA protein assays. Usually 6-8 mg of protein per liter of cell culture were obtained. The protein was dialyzed overnight against buffer (50 mM HEPES pH 7.2, 150 mM Li2SO4, 10% glycerol, 0.2 mM TCEP, 20 μM DHT) and stored at −80 °C in buffer.
Radiolabeled Ligand Competition Binding Assays
The assays were performed following a previously published procedure (28).
NR / Co-activator peptide Competition FP Assays. AR-LBD / SRC2-3 Competition Assay
The small molecules were serially diluted from 10000-2.44 μM in DMSO into a 96-well microplate (Costar 3359). The 5 μL of diluted compounds were added to 45μL of assay buffer (50 mM HEPES, 150 mM Li2SO4, 0.2 mM TCEP, 10% glycerol, pH 7.2, and 20 μM DHT) and the microplate was shaken at 600 rpm for 30 min at room temperature (IKA microtiter plate shaker). In 384-well microplates (Costar 3573) 20 μl of diluted compounds (1000-0.24 μM) were added to 20 μl of a protein cocktail (50 mM HEPES, 150 mM Li2SO4, 0.2mM TCEP, 10% glycerol, pH 7.2, 1 μM liganded AR-LBD and 0.01 μM fluorescent labeled peptide) yielding final compound concentrations of 500-0.12 μM and DMSO to 5%. The samples were allowed to equilibrate for 3 hours. Binding was measured using fluorescence polarization (excitation λ 485 nm, emission λ 530 nm) and given in polarization (mP). The mP value (milli-polarization level) is defined by: Polarization (mP) = 1000*(S−G*P)/(S+G*P), where S and P are background subtracted fluorescence count rates and G (grating) is an instrument and assay dependent factor.
PPARγ / DRIP2 Competition Assay
pET15b-PPAR-LBD expression plasmid, encoding the PPARγ-LBD (amino acids 173-475) was a generous gift from Gabor J. Tigyi, University of Tennessee, Memphis. PPARγ was expressed in BL21 (DE3) (Invitrogen), and the peptide DRIP2 (CKNHPMLMNLLKDNP) was labeled with Texas Red C2-maleimide (Invitrogen). The assay buffer was constituted of 20 mM TRIS (pH 7.50), 100 mM NaCl, 0.01% NP-40, 20 μM roziglitazone, 2 μM PPARγ-LBD, 10 nM DRIP-2 Texas Red, 5% DMSO.
VDR / SRC2-3 Competition Assay
This assay has been described in detail previously (32). MBP-VDR-LBDmt was expressed in BL21 (DE3) (Invitrogen), and the peptide SRC2-3 (CKKKENALLRYLLDKDDTKD) was labeled with Alexa Fluor 647 (Invitrogen). The assay buffer was constituted of 25 mM PIPES (pH 6.75), 50 mM NaCl, 0.01% NP-40, 6 μM LG190178, 1 μM MBP-VDR-LBD, 5 nM SRC2-3 Alexa Fluor 647, 5% DMSO.
TR / SRC2-2 Competition Assay
This assay has been described in detail previously (31). hTRβ-LBD (His6 T209-D461) was expressed in BL21 (DE3) (invitrogen), and the peptide SRC2-2 (CLKEKHKILHRLLQDSSSPV) was labeled with 5-iodoacetamidofluorescien (Molecular Probes). The assay buffer was constituted of 20 mM Tris (pH 7.0), 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, 0.01% NP-40, 1 μM T3, 1 μM hTRβ-LBD, 25 nM SRC2-2 fluorescein, 5% DMSO.
Cell based Assays
Transcription Reporter Assays. Androgen Receptor Assays in MDA-kb2 (33)
MDA-kb2 cells were cultured to 80% confluence, collected, re-suspended, and seeded at a concentration of 300 000 cells/mL in 96-well cell cultured treated microplates (Costar 3903) at 100 μl per well. The cells were allowed to attach at 37 °C for at least 15 hours, without CO2. Compounds were serially diluted in DMSO into a 96-well microplate (Costar 3359). 0.25 μL of compound solution and 0.25 μL of DHT solution were added to the cells yielding a final DMSO concentration of 0.5 %. After 20 hour incubation at 37°C and without CO2, cells were cooled at room temperature and 80 μL of Bright-Glo reagent (Promega) were added and luminescence was read directly.
Luciferase Inhibition Assay
MDA-kb2 cells at 80% confluence were incubated with 8nM DHT for 20 hours at 37°C and without CO2. After incubation, cells were lysed with a passive RIPA buffer (ThermoScientific), centrifuged at 1000g for 3 minutes, supernatant was collected, re-suspended in media, and dispensed in 96-well microplates (Costar 3903). 0.25 μL of compound solution was added to media and let incubate at room temperature for 1 hour. 80 μL of Bright-Glo reagent (Promega) were added and luminescence was read directly.
Cytotoxicity Assays
Rajii, HEPG2, BJ and Hek293 cells were grown to 80% confluence, collected, and re-suspended at a concentration of 50 000 cells/mL in 384-well microplates (Costar 3712) at 35 μL per well. Compounds were added to exponential growth phase cultured cells and incubated for 72 hours at 37°C. MDA-kb2 cells were grown to 80% confluence, collected, and re-suspended at concentrations of 300,000 cells/mL in 96 well microplates (Costar 3903) at 100 μL per well. The plates were incubated for 20 hours at 37 °C. The cytotoxicities were evaluated by using a Cell-Titer Glo reagent (Promega) and by reading luminescence. Compounds were tested at final concentrations of 40 μM and 1 μM. If more than 50% of the cells were killed at 40 μM, the compound was titrated in a dose response matter.
Real Time rt-PCR
MDA-kb2 cells were incubated at 37°C with tested compounds in the presence or absence of 8 nM DHT for 20 hours. Total RNA was isolated from cells using an RNAeasy kit (Qiagen). Genomic DNA was removed and cDNA was generated using equal amounts of RNA (QuantiTect® Reverse Transcription Kit, Qiagen). The cDNA was then diluted 50-fold and the QuantiFast SYBR Green PCR Kit (Qiagen) was used for the real time PCR following manufacturer's recommendations. Primers used in these studies are as follows: GAPDH Forward Primer 5'-accacagtccatgccatcac-3', Reverse Primer 5'-tccaccaccctgttgctgta-3'; 18S FP 5'-atcctcagtgagttctcccg-3', RP 5'- ctttgccatcactgccatta-3'; FKBP51 (FK506-binding immunophilin 51) FP 5'- ctgtgacaaggcccttgga-3', RP 5'-ctgggcttcacccctccta-3'. Real-time rt-PCR was carried out on a 7900HT Fast rt-PCR system (Applied Biosystems). We used the ΔΔCt method to measure the fold change in gene expression of target genes. Standard deviations were calculated from 3 biological independent experiments performed in triplicates.
Physicochemical Assays
Solubility Assay
30 μL of a DMSO stock of each compound (10 mM) was added to 600 μL of phosphate buffered saline (PBS) in a 96-well plate (Costar 3359) and mixed. The plate was sealed and incubated in room temperature for 18 hours. After mixing, 200 μL of sample suspension was transferred to a filter plate (catalog #110322, pION Inc., Woburn, MA) and pre-filtered. Another 200 μL of sample was filtered with the same filter plate. The filtrate was collected and injected to UPLC-MS (Waters, Milford, MA) and the concentration was determined according to UV absorbance standard curves.
PAMPA Assay
The PAMPA procedure was conducted using a published method (34–35). All liquid-handling steps for the PAMPA assay were performed on a Biomek FX Laboratory Automation Workstation (Beckman-Coulter) and analyzed by pION's (London, UK) PAMPA evolution 96 Command Software. The distribution of the compounds in the donor and acceptor buffers (100 μL aliquot) was determined by measuring the UV spectra from 200 to 500 nm using SpectraMax reader (Molecular Devices). Standards used were Verapamil (Pe = 1505 × 10−6 cm s−1) as a high permeability standard, Carbamazepine (Pe = 150 × 10−6 cm s−1) as a medium permeability standard, and Ranitidine (Pe = 2.3 × 10−6 cm s−1) as a low permeability standard.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children's Research Hospital (SJCRH), the NIH (DK58080) and the Department of Defense, Prostate Cancer Research Program (PC060344-W81XWH-07-1- 0073).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
REFERENCES
- 1.Chang C. Androgens and androgen receptor: mechanisms, functions, and clinical applications. Kluwer Academic Publishers ed.; Norwell: 2002. [Google Scholar]
- 2.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemppainen JA, Lane MV, Sar M, Wilson EM. Androgen receptor phosphorylation, turnover, nuclear transport, and transcriptional activation. Specificity for steroids and antihormones. J Biol Chem. 1992;267:968–974. [PubMed] [Google Scholar]
- 5.Amler LC, Agus DB, LeDuc C, Sapinoso ML, Fox WD, Kern S, Lee D, Wang V, Leysens M, Higgins B, Martin J, Gerald W, Dracopoli N, Cordon-Cardo C, Scher HI, Hampton GM. Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen- independent prostate cancer xenograft model CWR22-R1. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- 6.DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, Tibshirani R, Brown PO, Brooks JD. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nantermet PV, Xu J, Yu Y, Hodor P, Holder D, Adamski S, Gentile MA, Kimmel DB, Harada S, Gerhold D, Freedman LP, Ray WJ. Identification of genetic pathways activated by the androgen receptor during the induction of proliferation in the ventral prostate gland. J Biol Chem. 2004;279:1310–1322. doi: 10.1074/jbc.M310206200. [DOI] [PubMed] [Google Scholar]
- 8.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna NJ, O'Malley BW. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 10.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 11.Berrevoets CA, Doesburg P, Steketee K, Trapman J, Brinkmann AO. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor2) Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 12.Burd CJ, Morey LM, Knudsen KE. Androgen receptor corepressors and prostate cancer. Endocr Relat Cancer. 2006;13:979–994. doi: 10.1677/erc.1.01115. [DOI] [PubMed] [Google Scholar]
- 13.Estebanez-Perpina E, Moore JM, Mar E, Delgado-Rodrigues E, Nguyen P, Baxter JD, Buehrer BM, Webb P, Fletterick RJ, Guy RK. The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. J Biol Chem. 2005;280:8060–8068. doi: 10.1074/jbc.M407046200. [DOI] [PubMed] [Google Scholar]
- 14.Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- 15.Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A. 2007;104:16074–16079. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klotz L, Schellhammer P. Combined androgen blockade: the case for bicalutamide. Clin Prostate Cancer. 2005;3:215–219. doi: 10.3816/cgc.2005.n.002. [DOI] [PubMed] [Google Scholar]
- 17.Marques RB, Erkens-Schulze S, de Ridder CM, Hermans KG, Waltering K, Visakorpi T, Trapman J, Romijn JC, van Weerden WM, G. J. Androgen receptor modifications in prostate cancer cells upon long-term androgen ablation and antiandrogen treatment. Int J Cancer. 2005;117:221–229. doi: 10.1002/ijc.21201. [DOI] [PubMed] [Google Scholar]
- 18.Schellhammer P. An update on bicalutamide in the treatment of prostate cancer. Expert Opin Investig Drugs. 1999;8:849–860. doi: 10.1517/13543784.8.6.849. [DOI] [PubMed] [Google Scholar]
- 19.Gao W, Dalton JT. Expanding the therapeutic use of androgens via selective androgen receptor modulators (SARMs) Drug Discov Today. 2007;12:241–248. doi: 10.1016/j.drudis.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narayanan R, Mohler ML, Bohl CE, Miller DD, Dalton JT. Selective androgen receptor modulators in preclinical and clinical development. Nucl Recept Signal. 2008;6:e010. doi: 10.1621/nrs.06010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 22.Buchanan G, Yang M, Harris JM, Nahm HS, Han G, Moore N, Bentel JM, Matusik RJ, Horsfall DJ, Marshall VR, Greenberg NM, Tilley WD. Mutations at the boundary of the hinge and ligand binding domain of the androgen receptor confer increased transactivation function. Mol Endocrinol. 2001;15:46–56. doi: 10.1210/mend.15.1.0581. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb B, Beitel LK, Wu JH, Trifiro M. The androgen receptor gene mutations database (ARDB): 2004 update. Hum Mutat. 2004;23:527–533. doi: 10.1002/humu.20044. [DOI] [PubMed] [Google Scholar]
- 24.Jung F, Selvaraj S, Gargus JJ. Blockers of platelet-derived growth factor-activated nonselective cation channel inhibit cell proliferation. Am J Physiol. 1992;262:C1464–1470. doi: 10.1152/ajpcell.1992.262.6.C1464. [DOI] [PubMed] [Google Scholar]
- 25.Zhu W, Smith A, Young CY. A nonsteroidal anti-inflammatory drug, flufenamic acid, inhibits the expression of the androgen receptor in LNCaP cells. Endocrinology. 1999;140:5451–5454. doi: 10.1210/endo.140.11.7246. [DOI] [PubMed] [Google Scholar]
- 26.Bauman DR, Rudnick SI, Szewczuk LM, Jin Y, Gopishetty S, Penning TM. Development of nonsteroidal anti-inflammatory drug analogs and steroid carboxylates selective for human aldo-keto reductase isoforms: potential antineoplastic agents that work independently of cyclooxygenase isozymes. Mol Pharmacol. 2005;67:60–68. doi: 10.1124/mol.104.006569. [DOI] [PubMed] [Google Scholar]
- 27.Steckelbroeck S, Jin Y, Gopishetty S, Oyesanmi B, Penning TM. Human cytosolic 3alpha-hydroxysteroid dehydrogenases of the aldo-keto reductase superfamily display significant 3beta-hydroxysteroid dehydrogenase activity: implications for steroid hormone metabolism and action. J Biol Chem. 2004;279:10784–10795. doi: 10.1074/jbc.M313308200. [DOI] [PubMed] [Google Scholar]
- 28.Féau C, Arnold LA, Kosinski A, Guy RK. A High-Throughput Ligand Competition Binding Assay for the Androgen Receptor and other Nuclear Receptors. Journal of Biomolecular Screening. 2009;14:43–48. doi: 10.1177/1087057108326662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisson WH, Cheltsov AV, Bruey-Sedano N, Lin B, Chen J, Goldberger N, May LT, Christopoulos A, Dalton JT, Sexton PM, Zhang XK, Abagyan R. Discovery of antiandrogen activity of nonsteroidal scaffolds of marketed drugs. Proc Natl Acad Sci U S A. 2007;104:11927–11932. doi: 10.1073/pnas.0609752104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansch C, Fujita T. sigma-pi Analysis. A Method for the Correlation of Biological Activity and Chemical Structure. J. Amer. Chem. Soc. 1964;86:1616–1626. [Google Scholar]
- 31.Arnold LA, Estebanez-Perpina E, Togashi M, Shelat A, Ocasio CA, McReynolds AC, Nguyen P, Baxter JD, Fletterick RJ, Webb P, Guy RK. A high-throughput screening method to identify small molecule inhibitors of thyroid hormone receptor coactivator binding. Sci STKE. 2006;2006:pl3. doi: 10.1126/stke.3412006pl3. [DOI] [PubMed] [Google Scholar]
- 32.Teichert A, Arnold LA, Otieno S, Oda Y, Augustinaite I, Geistlinger TR, Kriwacki RW, Guy RK, Bikle DD. Quantification of the vitamin D receptor-coregulator interaction. Biochemistry. 2009;48:1454–1461. doi: 10.1021/bi801874n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson VS, Bobseine K, Lambright CR, Gray LE., Jr. A novel cell line, MDA-kb2, that stably expresses an androgen- and glucocorticoid-responsive reporter for the detection of hormone receptor agonists and antagonists. Toxicol Sci. 2002;66:69–81. doi: 10.1093/toxsci/66.1.69. [DOI] [PubMed] [Google Scholar]
- 34.Balimane PV, Han YH, Chong S. Current industrial practices of assessing permeability and P-glycoprotein interaction. Aaps J. 2006;8:E1–13. doi: 10.1208/aapsj080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu S, Konstantin T, Zhu Z, Graham T. Automation of a double-Sink PAMPA Permeability Assay on the Biomek→ FX Laboratory Automation Workstation. Pharmaceutical Discovery. 2005 [Google Scholar]
- 36.Febbo PG, Lowenberg M, Thorner AR, Brown M, Loda M, Golub TR. Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. J Urol. 2005;173:1772–1777. doi: 10.1097/01.ju.0000155845.44729.ba. [DOI] [PubMed] [Google Scholar]
- 37.Periyasamy S, Warrier M, Tillekeratne MP, Shou W, Sanchez ER. The immunophilin ligands cyclosporin A and FK506 suppress prostate cancer cell growth by androgen receptor-dependent and -independent mechanisms. Endocrinology. 2007;148:4716–4726. doi: 10.1210/en.2007-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.