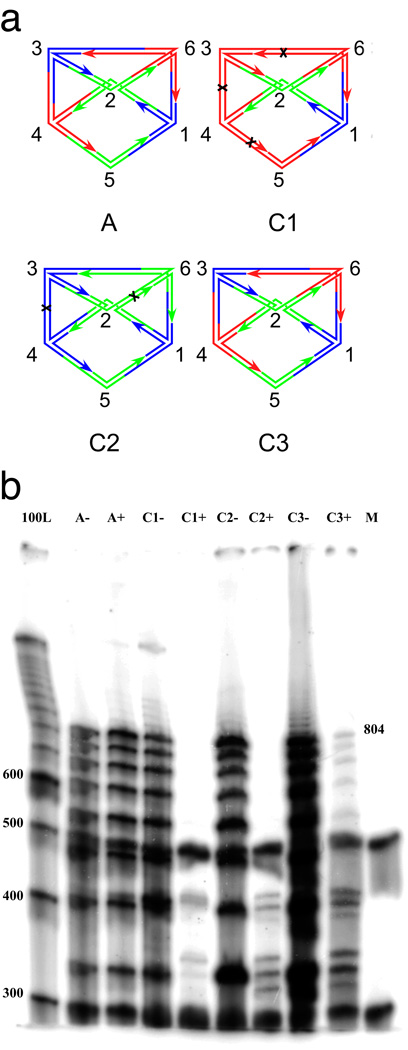
Figure 2. Denaturing gel electrophoresis.
(a) Schematic representations of the experiments reported here. The labels of the graphs correspond to the names of the experiments. Experiment A (for answer) contains only the branched junctions that produce the result. Experiment C1 (for one color) contains branched junctions corresponding to the two fixed vertices, vertex 1 (blue), vertex 2 (green) and all other vertices in red. Black crosses indicate the cleavage sites on the red edges. Experiment C2 (for two colors) contains branched junctions corresponding to all vertices in colors blue and green. The graph labeled C2 is only one possibility that can be assembled. Cleavage sites on homochromatic edges are indicated. Experiment C3 (for three colors) contains branched junctions corresponding to blue vertex 1, green vertex 2, and all colors for vertices 3–6. The correct undigested answer is shown. There are 81 (34) different molecules formed. Except for the answer illustrated, all other molecules contain at least one homochromatic edge and therefore are cleaved. (b) The gel showing the results of the experiments performed. From left to right, the lanes correspond to a 100 nucleotide marker, pairs of experiments containing the answer only (A), a one-color combination (C1), a two-color combination (C2) a three-color combination (C3) and a labeled marker (M) added in equal quantities to each experimental lane so that relative intensities of the lane's contents can be estimated; this prevents some lanes from being under-represented. Experiments after ligation but before enzyme restriction are indicated with a '−', and experiments after restriction are labeled with a '+'. The target reporter strand traverses every edge; seven edges once and two 64 bp edges twice. Its length, including the free thymidines is 804. Note that the target 804-mer reporter strand survives digestion in lane C3+, but not in lane C1+ nor in lane C2+.