Abstract
Purpose
Previous gene expression profiling studies of breast cancer have focused on the entire genome to identify genes differentially expressed between estrogen receptor alpha (ER)-positive and ER-alpha-negative cancers.
Experimental Design
Here we used gene expression microarray profiling to identify a distinct kinase gene expression profile that identifies ER-negative breast tumors and subsets ER-negative breast tumors into 4 distinct subtypes.
Results
Based upon the types of kinases expressed in these clusters, we identify a cell cycle regulatory subset, a S6 kinase pathway cluster, an immunomodulatory kinase expressing cluster, and a MAPK pathway cluster. Furthermore, we show that this specific kinase profile is validated using independent sets of human tumors, and is also seen in a panel of breast cancer cell lines. Kinase expression knockdown studies show that many of these kinases are essential for the growth of ER-negative, but not ER-positive, breast cancer cell lines. Finally, survival analysis of patients with breast cancer shows that the S6 kinase pathway signature subtype of ER-negative cancers confers an extremely poor prognosis, while patients whose tumors express high levels of immunomodulatory kinases have a significantly better prognosis.
Conclusions
This study identifies a list of kinases that are prognostic and may serve as druggable targets for the treatment of ER-negative breast cancer.
Introduction
The genomic era has produced an exponential increase in our understanding of cancer biology and has greatly accelerated cancer drug development. With the advent and implementation of microarray expression profiling, it is now possible to evaluate gene expression in tumors on a genome-wide basis. Gene expression analysis is now extensively used to subtype cancers, predict prognosis and disease free survival, and determine optimal treatment (1–7).
Estrogen receptor alpha (ER)-positive breast cancers account for 60–70% of breast cancers, but the remaining 30–40% of breast cancers are ER-negative and are poorly responsive to traditional therapies (8). Selective estrogen receptor modulators (SERMs), such as tamoxifen and raloxifene, and aromatase inhibitors are currently used to treat ER-positive breast cancer and have been shown to reduce ER-positive breast cancer recurrence by approximately 50% (9). These agents, however, are not effective in treating ER-negative breast cancer. Currently, chemotherapy is used to treat ER-negative tumors (10). Such therapy is generally toxic and is not specifically targeted to ER-negative breast cancer.
A major goal of current breast cancer research has been to identify targets that are unique to cancer cells and to identify drugs that kill only cancerous cells without affecting normal tissue. While achieving this goal has been difficult, there are several examples of effective targeted therapies, including development of the monoclonal antibodies trastuzumab (targeting the HER2/neu receptor) and bevacizumab (targeting vascular epithelial growth factor) which have been shown to be effective in treating breast cancer (11, 12). Other successes include the development of small molecule tyrosine kinase inhibitors including gefitinib and erlotinib (both of which target the epidermal growth factor receptor), and lapatinib (a dual kinase inhibitor targeting both the epidermal growth factor receptor and the HER2/neu receptor) (13–16). Despite these advances, such therapies are effective only in the 10–15% of patients whose tumors overexpress HER2. To develop targeted therapies for the remaining ER-negative breast cancers, including the aggressive ER-negative, PR-negative, HER2-negative (“triple-negative”) breast cancers, we have used expression microarray analysis to identify molecules that play a role in breast cancer development and progression. Subsequent validation of these findings, along with the development of specific targeted inhibitors of these molecules, will certainly broaden treatment options and improve patient survival.
The purpose of this study was to identify the kinases that are over-expressed in ER-negative breast cancer and which may serve as “druggable targets” for the treatment of ER-negative breast cancer, and in particular, “triple-negative” breast cancer. We have used transcriptional profiling data to evaluate the expression of the human kinome and have identified a set of kinases which are differentially expressed and are critical for the growth of ER-negative breast cancer. Our results also demonstrate that ER-negative breast cancer can be subdivided into four separate subgroups based on their kinase expression profile. These kinases represent promising targets for the treatment of ER-negative breast cancers.
Materials and Methods
Study Population and Design
All ER-negative and ER-positive tumors were collected by Dr. Jenny Chang through IRB-approved, neoadjuvant studies to investigate gene expression changes in human tumors following drug treatment. Diagnostic core needle biopsies were taken first, then several (up to 6) additional cores were taken for biomarker studies. These additional cores were taken before treatment, placed immediately in liquid nitrogen, and used to prepare RNA, DNA, and protein. Immunohistochemical (IHC) staining for ER and HER2/neu expression was done on these sets of tumor samples as previously described (17). The tumor set comprised of pre-treatment specimens from studies of docetaxel (18), cyclophosphamide (19), docetaxel and cyclophosphamide (unpublished data), and trastuzumab (20). All studies were conducted with approval from the Institutional Review Boards at Baylor College of Medicine and participating sites.
Affymetrix microarray experiments
Total RNA from these tumor samples was isolated using Qiagen’s RNeasy kit, double-stranded cDNA synthesized, and reverse transcription carried out followed by biotin labeling. RNA was isolated from tumors that were not microdissected but tumor cellularity was confirmed to be greater than 40% in all tumor samples by IHC analysis. Additionally, about 250-fold linear amplification and phenol-chloroform cleanup was done as previously published (1). From each biopsy, 15 micrograms of biotin-labeled cRNA was hybridized onto an Affymetrix HGU133A GeneChip™, which comprise around 22,000 genes (www.affymetrix.com). The experiments were all done using the microarray core facility at the Lester and Sue Smith Breast Center at Baylor College of Medicine. Statistical analysis was done with dChip (www.dchip.org) and BRB ArrayTools software packages developed by Dr. Richard Simon and Amy Peng Lam. (http://linus.nci.nih.gov/BRB-ArrayTools.html). Gene expression was estimated with dChip software using Invariant Set normalization and Perfect Match (PM) only model (21). Comparison of ER-negative vs. ER-positive groups was done with BRB Array Tools, using t-test and computing permutation P-values (22). Hierarchical clustering was also done using dChip with rows standardized by subtracting the mean and dividing by the standard deviation. Pearson’s correlation and centroid linkage was used to generate the trees on Log2 transformed expression data with PM/MM difference background subtraction.
Gene ontology analysis
All gene ontology enrichment analyses were initially done using a Pathway Architect™ software package developed by Stratagene. Genes found to be overexpressed at least 2 fold with a permutation P-value score of <.05 were used as the input list and compared against the human kinome. Follow-up and confirmatory analysis was done using Gene Ontology Tree Maker (GOTM) and EASE software (23).
Selection of Genes for further study
After completing all microarray experiments and performing statistical analysis genes having a minimum of 2-fold or greater expression in E R-negative vs. ER-positive tumors with a permutation P-value <.05 were selected for further study. This selection yielded 52 kinases.
Z-transform test in multiple datasets
To validate that the differentially expressed kinases identified in this analysis were also differentially expressed in other publically available datasets of human breast cancer we employed the Z-transform test described by Whitlock (24). Briefly, this method allows for the combining of individual P-values and has proven superior to Fisher’s combined probability test. The Z-transform test takes advantage of the one-to-one mapping of the standard normal curve to the P-value of a one-tailed test. As Z goes from negative infinity to infinity, P will go from 0 to 1, and any value of P will uniquely be matched with a value of Z and vice versa. The Z-transform test converts the one-tailed P-values, Pi, from each of k independent tests into standard normal deviates Zi. The sum of these Zi's, divided by the square root of the number of tests, k, has a standard normal distribution if the common null hypothesis is true. The equation: was used in the calculation of summed z-scores, which were then related to the reported P-values.
RNA isolation and Quantitative RT-PCR (Q-RT-PCR)
Total RNA was isolated using the RNeasy RNA isolation kit (QIAGEN). Quantitative RT-PCR assays of transcripts were carried out using gene-specific double fluorescence-labeled probes in an ABI PRISM 7500 Sequence Detector (Applied Biosystem). The PCR reaction mixture consisted of 300nM each of the forward and reverse primers, 100nM probe, 0.025 units/µl of Taq Polymerase (Invitrogen), 125µM each of dNTP, 5mM MgCl2, and 1X Taq Polymerase buffer. Cycling conditions were 95°C for 30 seconds, followed by 40 cycles at 95°C for 5 seconds and 60°C for 30 seconds 6-Carboxy fluorescein (FAM) was used as the 5’ fluorescent reporter and black hole quencher (BHQ1) was used at the 3’end quencher. All reactions were performed using triplicate RNA samples. Standard curves for the quantification of each transcript were generated using a serially diluted solution of synthetic templates. Results were reported as average expression ± standard error of the mean.
siRNA transfection
siRNAs for all kinases (see supplementary table) were purchased from Sigma Aldrich. siRNA transfection was performed using DharmaFECT™ 1 (Dharmacon), according to the manufacture’s instruction. MDA-MB-468, MDA-MB-231, T47D, and MCF-7 cells were plated in 100 mm dishes and grown to 60% confluence before being transfected with Dharmacon siRNA dilution buffer (mock-transfection), 20 ng of kinase specific siRNA constructs, or with scrambled siRNA as a control. 36 hours after transfection, cells were replated in 96 well plates at a density of 2000 cells per well. RNA and protein were also harvested at this time (as described previously), as well as on days two and four, to confirm sufficient knockdown of kinase expression by Q-RT-PCR and western blotting, respectively. After replating in 96 well plates, growth was measured by MTS assay every 2 days for a total of 5 days.
Cell proliferation assays
Cell growth was measured using the CellTiter 96™ Aqueous Non-Radioactive Cell Proliferation assay (MTS assay, Promega) according to the manufacturer’s instructions. Briefly, cells were plated in 96-well plates at 2000 cells per well. Every 24 hours, a solution containing 20:1 ratio of M T S ( 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) and PMS (phenazine methosulfate) was added to the cells. Plates were incubated at 37°C for 2 h and absorption at 550nm was determined. Each data point was performed in heptuplicate, and the results were reported as average absorption ± standard deviation. The data is reported as percentage of growth compared to mock transfected controls for each cell line. Experiments were repeated at least twice and the percentage growth inhibition is the average of the experiments.
Kaplan-Meier Survival Analysis
Gene expression profiling and survival data generated by Wang et al. and van de Vijver et al. was used to evaluate prognostic import of the kinase clusters in this data set (3, 25). Data was obtained from GEO and hierarchical clustering performed only on the ER-negative samples from the Wang and van de Vijver data sets. MeV and R software package were used for data and statistical analysis. For hierarchical clustering, the expression values of the kinases identified as being over expressed in ER-negative tumors were extracted from the data sets using Affymetrix probe IDs. The expression values were mean centered and hierarchical clustering based on Pearson’s correlation with complete linkage again identified our 4 subsets of ER-negative kinase clusters. Figure of merit scoring showed that these four clusters were stable against reclustering in both datasets. Using this information each tumor sample was classified as falling into one of the 4 kinase clusters (cell cycle checkpoint, S6 kinase, MAPK signaling, or immunomodulatory). After classification of tumors, Kaplan-Meier analysis using the survival data from the ER-negative tumors in the data sets was performed using R (http://www.r-project.org) and survival curves were generated. Chi squared scores were calculated to determine significance.
Immunohistochemical Analysis of Immune Infiltration
Representative tumors from each of the four groups identified by gene expression profiling (cell cycle regulatory, S6 kinase, immunomodulatory, and MAPK) were selected and slides were prepared and stained with hematoxolyin and eosin. Each slide was analyzed and scored by a trained pathologist in a blinded fashion. Scoring was from 0–3 for neutrophil infiltration and 0–3 for lymphocyte infiltration in the entire section of tissue at both low and high power magnification. Scores of 0 corresponded to no evidence of infiltration, 1 for minimal foci of infiltration (<1% of tumor mass), 2 for moderate amounts of immune cell infiltration (2–15% of tumor mass) and 3 for considerable amounts of immune cell infiltration (>15% of tumor mass). Scores for the neutrophil and lymphocyte scoring were added to give a composite score (0–6).
Results
To identify kinases that are differentially expressed in ER-negative breast cancers, we designed a study to compare kinase expression levels in ER-positive and ER-negative human breast tumor samples. A summary of the study design is outlined in Supplementary Figure S1.
Patient Population
A total of 102 patients with invasive breast cancer were recruited through IRB-approved, neoadjuvant studies to investigate gene expression in human tumors before and after drug treatment. Breast biopsies using a core needle were taken before initiation of any treatment and were used in this study. Because the patients did not receive systemic adjuvant or neoadjuvant therapy prior to the biopsy, the results from the gene expression analysis represent basal gene expression in these breast cancers. For these gene expression profiling experiments, 102 breast tumors were studied, 58 of which were ER-positive and 44 ER-negative by IHC-staining (24 of which were confirmed as “triple-negative”). The tumors were all stage III or IV from pre- and post-menopausal women, with all tumors showing >40% cellularity. The women were from several racial groups (as shown in Table 1) and the majority had no palpable nodes at baseline. Most of the women were premenopausal and presented with relatively large tumors (ranging from 2.5 to 25 cm). The clinical and demographic features of these tumors are summarized in Table 1.
Table 1. Clinical characteristics of the patients and tumor samples used in the study.
Characteristics of 102 patients with breast cancer. Tumors from these patients were used for gene expression profiling to identify overexpressed kinases and kinase-associated genes in ER-negative breast tumors
| Characteristic | Tumor Set N=102 (%) |
|---|---|
| Age | |
| Mean | 48.1 |
| Range | (32–72) |
| Race | |
| Caucasian | 50 (57%) |
| Hispanic | 7 (8%) |
| African-American | 23 (27%) |
| Asian | 7 (8%) |
| Menopausal Status | |
| Pre | 49 (62%) |
| Post | 30 (38%) |
| BMI | |
| Mean | 29.7 |
| Range | (16.1–48.3) |
| Baseline Tumor Size, cm | |
| Mean | 6.3 |
| Range | (2.5–25.0) |
| Palpable Nodes at Baseline | |
| Yes | 20 (21%) |
| No | 77 (79%) |
| ER | |
| Positive | 57 (56%) |
| Negative | 45 (44%) |
| Unknown | 0 (0%) |
| PR | |
| Positive | 37 (36%) |
| Negative | 47 (46%) |
| Unknown | 18 (18%) |
| HER2/Neu | |
| Positive | 27 (26%) |
| Negative | 58 (57%) |
| Unknown | 17 (17%) |
Affymetrix Gene Expression Profiling Identified Kinases Overexpressed in Human ER-Negative Breast Tumors
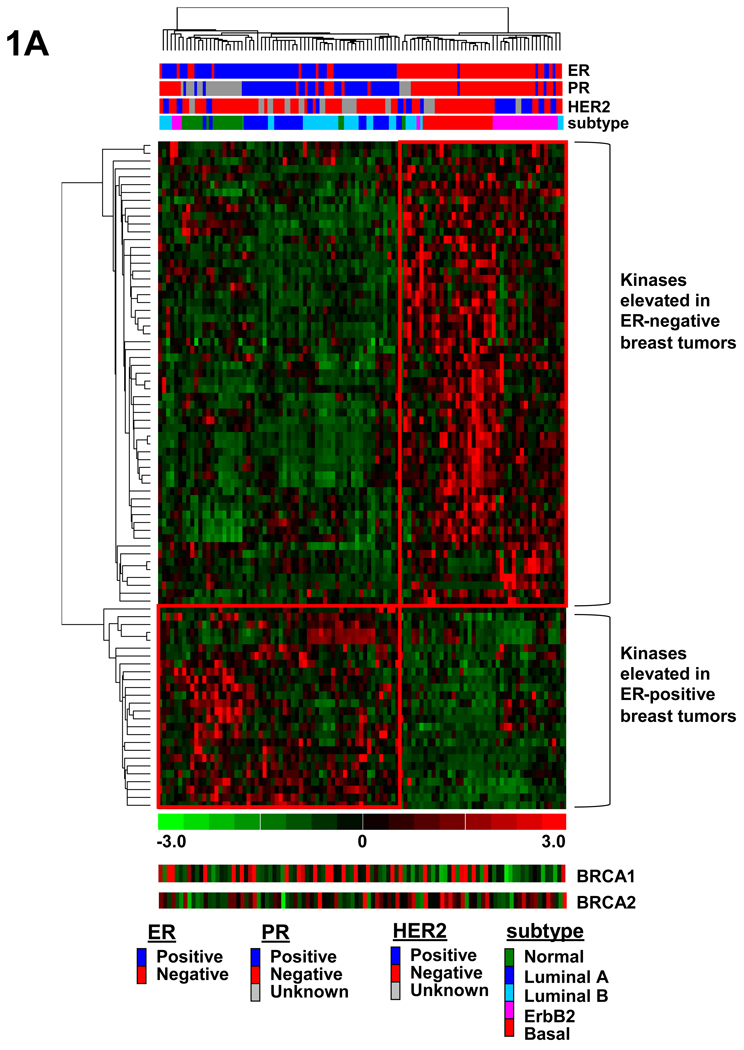
To identify signaling molecules that are differentially expressed in ER-negative breast cancers, we performed Affymetrix gene expression profiling to compare human ER-negative and ER-positive breast tumors. Data analysis and clustering for this study was limited to the known kinome with interrogation of the 779 known and putative human protein, nucleotide, and lipid kinases as well as kinase-interacting proteins and regulatory subunits as previously described (26–28). These kinases and kinase-associated genes are listed in Supplementary Table S1. We first performed analysis to identify those kinases that were differentially expressed in ER-positive and ER-negative breast tumors. Our analysis revealed a significant difference (permutation P-value< 0.05, hereafter referred to as P-value) in the expression of 86 kinases or kinase-associated genes between ER-negative and ER-positive tumors (see Supplementary Table 2 and Table 3). To visualize the clustering of the ER-positive and ER-negative tumors, hierarchical clustering analysis was done using only those kinases identified as being differentially expressed between the two groups (Fig. 1A). Hierarchical clustering showed that these 86 kinases or kinase-associated genes were able to segregate ER-positive and ER-negative tumors and that the highest percentage of the HER2-positive tumors were ER-negative. As BRCA1 and BRCA2 mutations have been shown to be enriched in ER-negative tumors, especially basal-like tumors, we show relative BRCA1 and BRCA2 gene expression values in figures 1A. BRCA1 and BRCA2 gene expression was generally lower in the ER-negative samples than in the ER-positive samples, and in the tumors which were basal-like in their gene expression profile. Additionally, ER, PR, and HER2 status is listed above the figure, as is the subtype of breast cancer identified based on the application of the intrinsic gene list identified by previous groups to these breast tumor samples (Fig. 1A). Upon further analysis, 52 of these 86 differentially expressed genes were expressed at least 2 fold higher with a P-value <.05 in the ER-negative breast tumors as compared to ER-positive tumors. These 52 genes were selected for further study.
Fig. 1.
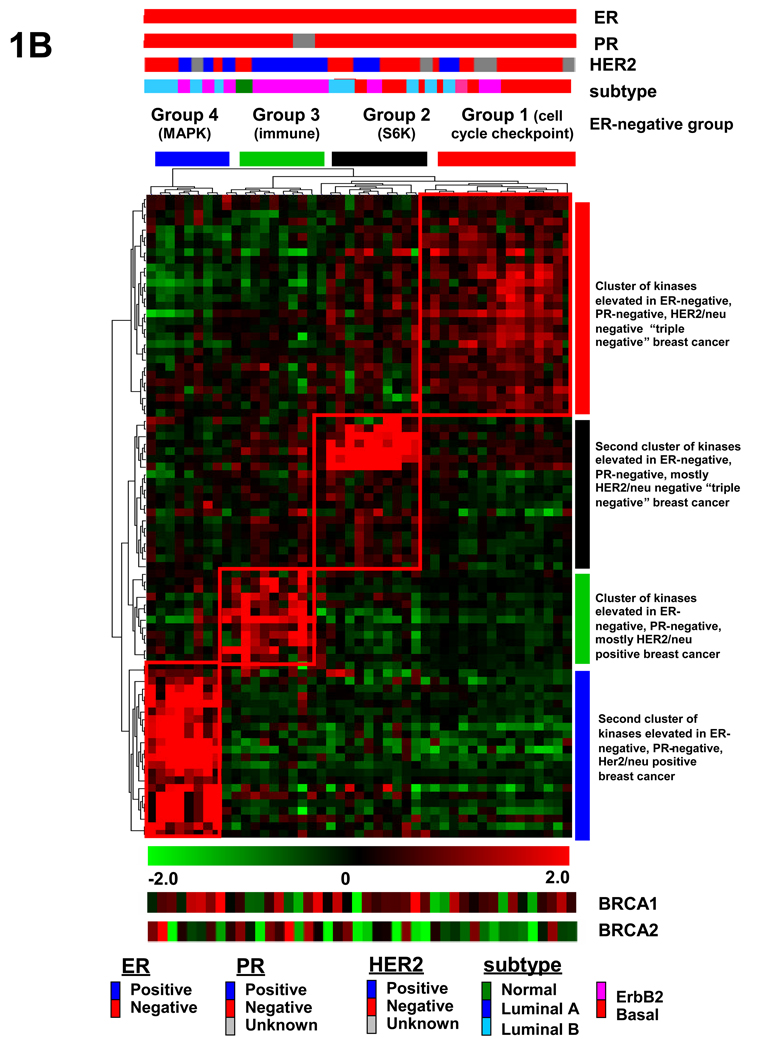
Supervised hierarchical clustering identifies different subsets of ER-negative breast cancer. (A) Hierarchical clustering analysis of kinases that distinguish ER-positive from ER-negative human breast tumors. Gene expression analysis of 102 human breast tumors reveals 86 kinases and kinase-associated genes that are differentially expressed between ER-negative and ER-positive human breast tumors with a permutation P-value <.05. (B) Unsupervised hierarchical clustering analysis of only ER-negative tumors using kinases and kinase-associated genes overexpressed in ER-negative breast cancers reveals 4 distinct subsets of ER-negative breast cancer. These four subset are defined by kinases that are involved in cell cycle checkpoint control (group 1), S6 kinase signaling (group 2), immunomodulatory (group 3), or paracrine signaling involving many MAPKs (group 4). Subtype refers to the breast cancer subtypes identified by Sotiriou et al. (46). BRCA1 and BRCA2 relative gene expression has been included for comparison.
Unsupervised Clustering Analysis Revealed Four Distinct Subtypes of ER-negative Breast Cancer
We next determined whether this list of 52 kinases or kinase-associated genes overexpressed in ER-negative breast cancers could subcluster the ER-negative tumors in an unbiased manner. We performed unsupervised clustering analysis using only the ER-negative breast cancer samples and found that these tumors clustered broadly into 4 distinct subtypes of ER-negative breast cancer (Fig. 1B, labeled as groups 1–4). Upon further inspection of these four subsets of tumors, there was one subset of tumors defined by kinases involved in cell cycle checkpoint control and mitogenesis, including CHK1, BUB1, TTK, and AK2 (group 1 termed the “cell cycle checkpoint group”). Another tumor subset was defined by kinases involved in the S6 kinase signaling pathway and includes RPS6KA3, SMG-1, and RPS6KA1 kinases (group 2 termed the “S6 kinase group”). Of the two other ER-negative clusters, one is defined by kinases that are involved in modulating the immune system (IRAK1, TLR1, LCK, and LYN) (group 3, termed the “immunomodulatory group”). The fourth group is defined by kinases that govern paracrine growth signaling and include mitogen activated protein kinases MAP4K2, MAP4K4, and MAPK1 (group 4 termed the “MAPK group”). In these later two groups, many of the tumors were HER2-positive by IHC staining. HER2 itself did not act as a cluster driver because despite being more highly expressed in the ER-negative samples, it did not meet our strict P-value criterium and thus was not included in the list of kinases and kinase-associated genes that were used in the unsupervised clustering of the ER-negative samples.
Immunohistochemical Analysis Shows No Significant Increase in Lymphocytic Infiltrate
With the identification of 4 distinct subsets of ER-negative disease, and with one of those subsets being marked by high expression of many immunomodulatory genes, we investigated whether the gene expression signature from these tumors was derived from the epithelial tumor compartment or from infiltrating immune cells. To address this we characterized the proportion of infiltrating immune cells in the tumors (prior to treatment) by examining the hemotoxylin and eosin (H&E) stained slides from the tumors used in this study. The histological appearance and lymphocytic infiltration was scored by a trained pathologist in a blinded fashion. Comparison of the four groups (cell cycle checkpoint, S6 kinase, MAPK-associated, and immunomodulatory) showed no increase in lymphocytic infiltration in the immunomodulatory tumors (Supplementary Fig. S2). Thus, this data argues that the immunomodulatory gene expression profile associated with this subtype of ER-negative breast cancer is not just the result of immune cell infiltration into the tumor tissue.
Gene Ontology Analysis
To gain insight into the function of kinases highly expressed in ER-negative breast cancer, we performed gene ontology (GO) enrichment analysis using EASE and found that several classes of biological function were highly enriched in our selected sets (Table 2). We observed enrichment for kinases involved in the regulation of metabolism (P-value <10−14), cell cycle checkpoint control (P-value <10−12), DNA damage checkpoint control (P-value <10−11), cell-to-cell signaling (P-value <10−9), and apoptosis regulation (P-value <10−9). Many of these kinases fell in linear pathways, for example TTK, CHK1, and BUB1 kinases (group 1), all of which play a role via sequential phosphorylation and activation in regulating G2/M transitioning as well as DNA damage checkpoint control pathways, and PIK3CB, RPS6KA1, and SMG-1 kinases (group 2), that mark tumors with activated cytoplasmic kinases involved in mitogenesis and signal transduction.
Table 2. Kinases identified in analysis as most highly overexpressed in ER-negative tumors.
52 overexpressed kinases in ER-negative breast cancer fall into 4 distinct subsets with varying biological functions. Gene ontology analysis shows that these kinases have varying biological functions, but most regulate growth, metabolism, affect cell cycle, or are involved in DNA damage sensing and repair.
| Cell cycle checkpoint cluster | Gene Bank Assession | Kinase function |
|---|---|---|
| BUB1 | NM_001211 | cell cycle checkpoint |
| CHK1 checkpoint homolog | NM_001274 | cell cycle checkpoint |
| TTK protein kinase | NM_003318 | cell cycle checkpoint |
| serum/glucocorticoid regulated kinase | NM_005627 | cell cycle checkpoint |
| SFRS protein kinase 1 | NM_003137 | cell cycle checkpoint |
| maternal embryonic leucine zipper kinase | NM_014791 | cell cycle checkpoint |
| RYK receptor-like tyrosine kinase | NM_001005861 | positive regulation of proliferation |
| vaccinia related kinase | NM_006296 | anti-apoptosis |
| phosphoglycerate kinase 1 | NM_000291 | metabolism |
| selenophosphate synthetase 1 | NM_004226 | metabolism |
| uridine-cytidine kinase 2 | NM_012474 | metabolism |
| UDP-glucose pyrophosphorylase 2 | NM_006759 | metabolism |
| adenylate kinase 2 | NM_001625 | metabolism |
| aurora kinase B | NM_004217 | cell cycle checkpoint |
| cell division cycle 2 | NM_001786 | cell cycle checkpoint |
| cell division cycle 7 homolog | NM_003503 | cell cycle checkpoint |
| S6 kinase pathway cluster | ||
| ribosomal protein S6 kinase, 90kDa, polypeptide 1 | NM_001006665 | positive regulation of proliferation |
| PI-3-kinase-related kinase SMG-1 | NM_015092 | DNA repair |
| EPH receptor B4 | NM_004444 | positive regulation of proliferation |
| serine/threonine kinase 38 like (NDR2) | NM_015000 | positive regulation of proliferation |
| PI3K catalytic subunit beta | NM_006219 | positive regulation of proliferation |
| death-associated protein kinase 1 | NM_004938 | anti-apoptosis |
| pim-1 oncogene | NM_002648 | anti-apoptosis |
| LIM domain kinase 2 | NM_001031801 | cell adhesion |
| phosphoribosyl pyrophosphate synthetase 1 | NM_002764 | metabolism |
| EPH receptor B6 | NM_004445 | nervous system development |
| EPH receptor B2 | NM_017449 | maintainance of polarity |
| EPH receptor A2 | NM_004431 | signal transduction |
| ribosomal protein S6 kinase, 90kDa, polypeptide 3 | NM_001006665 | positive regulation of proliferation |
| MAPK cluster | ||
| mitogen-activated protein 4K4 | NM_004834 | response to stress |
| mitogen-activated protein kinase kinase 6 | NM_002758 | DNA damage, cell cycle arrest |
| mitogen-activated protein kinase 1 (ERK2) | NM_002745 | positive regulation of proliferation |
| mitogen-activated protein 4K2 | NM_004579 | positive regulation of proliferation |
| mitogen-activated protein 3K5 | NM_005923 | regulation of apoptosis |
| mindbomb homolog 1 (14-3-3) | NM_020774 | receptor mediated endocytosis |
| v-raf-1 murine leukemia viral oncogene homolog | NM_002880 | anti-apoptosis |
| protein kinase, X-linked | NM_005044 | unknown |
| PTK7 protein tyrosine kinase 7 | NM_002821 | cell adhesion |
| myelin protein zero-like 1 | NM_003953 | cell to cell signaling |
| phosphofructokinase, platelet | NM_002627 | metabolism |
| epidermal growth factor receptor | NM_005228 | positive regulation of proliferation |
| MET proto-oncogene | NM_000245 | activation of MAPK activity |
| Immunomodulatory cluster | ||
| toll-like receptor 1 | NM_003263 | immune system modulation |
| LYN | NM_002350 | positive regulation of proliferation |
| MALT lymphoma translocation gene 1 | NM_006785 | anti-apoptosis |
| serine/threonine kinase 17b | NM_004226 | anti-apoptosis |
| interleukin-1 receptor-associated kinase 1 | NM_001569 | positive regulation of transcription |
| chemokine (C-X-C motif) ligand 10 | NM_001565 | immune system modulation |
| lymphocyte-specific protein tyrosine kinase | NM_001042771 | immune system modulation |
| chemokine (C-C motif) ligand 4 | NM_002984 | cell to cell signaling |
| pyridoxal (pyridoxine, vitamin B6) kinase | NM_003681 | metabolism |
| v-yes-1 | NM_005433 | positive regulation of proliferation |
Differentially Expressed Kinases Validated Using Publicly Available Data Sets
Having identified differentially expressed kinases using gene expression profiling in a test set of tumors, we next investigated whether these kinases were differentially expressed in an independent set of human breast tumors. To demonstrate that these kinases are indeed overexpressed in ER-negative tumors compared to ER-positive tumors, we analyzed gene expression data from 12 additional publically available data sets. This data set from multiple investigators includes over 1800 additional tumor samples (556 ER-negative and 1282 ER-positive tumors) for which there is gene expression profiling data and is the most comprehensive breast tumor set available (3, 25, 29–37). Data for analysis was compiled and downloaded from GEO and used for analysis. Additionally, as verification, we used calculated P-values from the Oncomine™ repository for use in the Whitlock Z-transforms test. To utilize the power of such a large combined dataset, we employed a technique recently described by Whitlock that relies on a weighted Z-method to combine P-values (24). This robust approach, superior to Fisher’s combined probability test, revealed that all of the selected 52 kinases validated as being significantly more highly expressed (with extremely high z-scores and low P-values) in ER-negative breast tumors as compared to ER-positive tumors in an effective sample size of over 1800 tumors (Supplementary Table S4).
Validation of Kinase Overexpression in an Independent Set of Human Breast Tumors
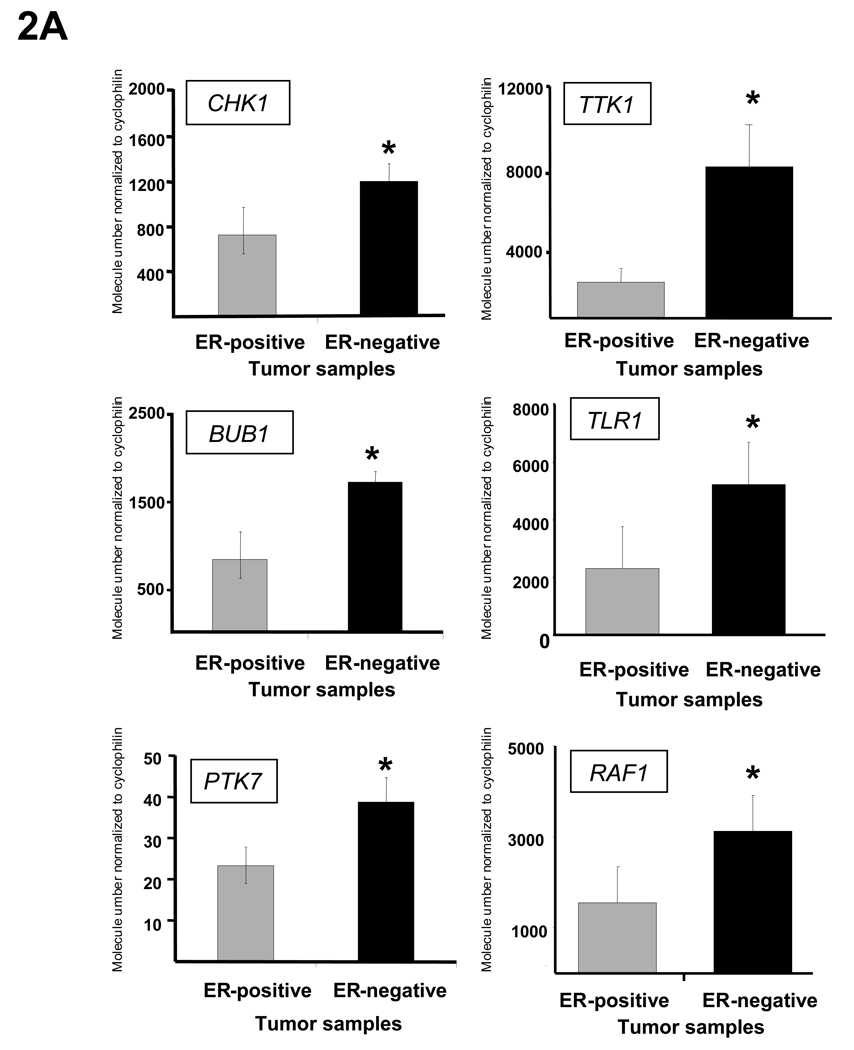
To confirm that the overexpression identified using Affymetrix gene expression profiling could be validated using another technique, we used an independent set of 60 human breast tumors from the tumor bank at Baylor College of Medicine for further validation. After identifying equal numbers of ER-positive and negative samples, we used quantitative RT-PCR (Q-RT-PCR) to confirm the overexpression of the kinases identified in the array profiling. 34 of the 34 kinases assayed were significantly more highly expressed in ER-negative human breast tumors than ER-positive tumors in this additional tumor set (P-value <0.05). Representative results from these experiments showing expression of six kinases (CHK1, BUB1, PTK7, TTK1, TLR1, and RAF1) are displayed in Figure 2A.
Fig. 2.
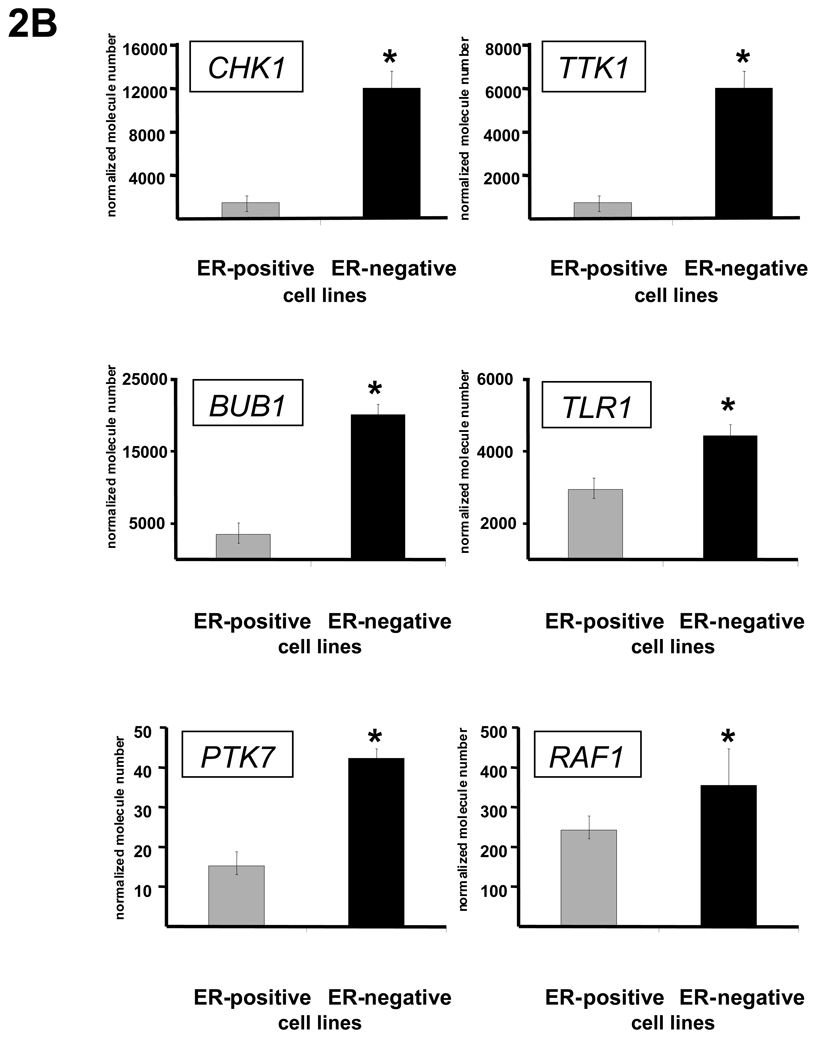
Kinase overexpression validated in independent human tumor sample data sets and in a panel of breast cancer cell lines. (A) The expression of 34 of 34 kinases and kinase-associated genes identified in the array profiling were validated as being more highly expressed in ER-negative tumors compared to ER-positive tumors as measured by Q-RT-PCR in an independent set of breast tumors. Expression data for 6 representative kinases (CHK1, BUB1, PTK7, TTK, TLR1, and RAF1) are shown. Asterisks indicate P-value <0.01. Data are represented as mean ± SEM. (B) The expression of 42 of 42 kinases was significantly higher in ER-negative breast cancer cell lines as compared to ER-positive cell lines. Again, expression data as measured by Q-RT-PCR, this time in a panel of breast cancer cell lines, for 6 representative kinases (CHK1, BUB1, PTK7, TTK, TLR1, and RAF1) are shown. Asterisks indicate P-value <0.01. Data are represented as mean ± SEM. Validation of kinase overexpression was also done in 12 human breast tumor datasets (Table S4).
Validation of Kinase Overexpression in Breast Cancer Cell Lines
To conduct further in vitro experimentation in cell lines, we investigated whether the kinases identified in ER-negative human breast tumors were also overexpressed in ER-negative breast cancer cell lines. Twelve ER-positive or ER-negative breast cancer cell lines were chosen and the expression of the identified kinases was measured under basal growth conditions. Of 42 kinases evaluated to date, all 42 were found to be statistically significantly increased (P-value <.05) in this panel of ER-negative breast cancer cell lines as compared to ER-positive cell lines using Q-RT-PCR. Representative results for several of these kinases (CHK1, BUB1, PTK7, TTK1, TLR1, and RAF1) are shown in Figure 2B.
Cluster of Human Breast Cancer Cell Lines using the Kinase Profile
To determine whether the 52 kinases and kinase-associated genes could accurately subgroup breast cancer cell lines, we used available expression data from 51 breast cancer cell lines. Recent work by Neve et al. showed that the recurrent genomic and transcriptional characteristics of breast cancer cell lines mirror those of primary breast tumors (38). These investigators performed Affymetrix gene expression profiling on a set of 51 ER-positive and ER-negative breast cancer cell lines and used hierarchical clustering to show that the cell lines clustered into three main groups: basal A, basal B, and luminal (38). We used this expression information from breast cancer cell lines to determine whether our list of 52 genes would group these cell lines into the similar groups as with the profiling of human tumors (cell cycle checkpoint control, S6 kinase, immunomodulatory, and MAPKs groups). When hierarchical cluster analysis was performed on the expression data from these 51 cell lines using the list of 52 genes identified here, the cell lines were accurately clustered into ER-positive or ER-negative groups (Supplementary Fig. S3A). Furthermore, the overexpressed kinases were able to subset ER-negative breast cancer cell lines into 4 groups in an unsupervised manner (Supplementary Fig. S3B). These results indicate that the expression profile of the identified kinases is sufficiently robust to accurately discriminate between ER-positive and ER-negative breast cancer cell lines and may serve as a reliable diagnostic tool to categorize human tumors in the future.
Kinase Knockdown Exerts Differential Growth Effects on ER-negative and ER-positive Breast Cancer Cell Lines
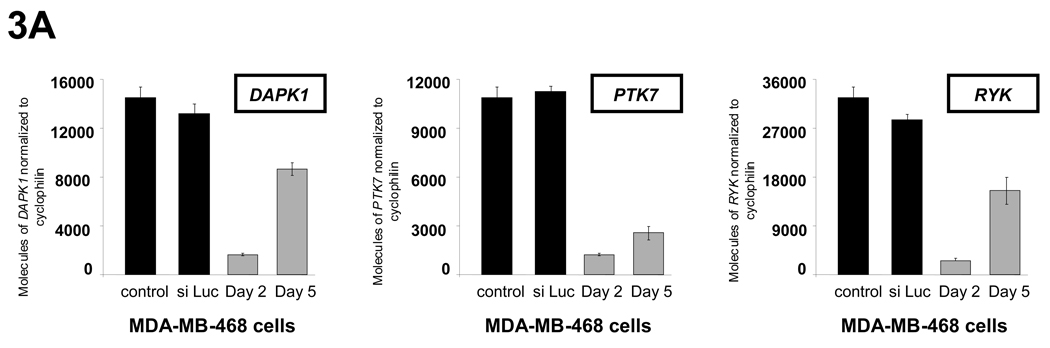
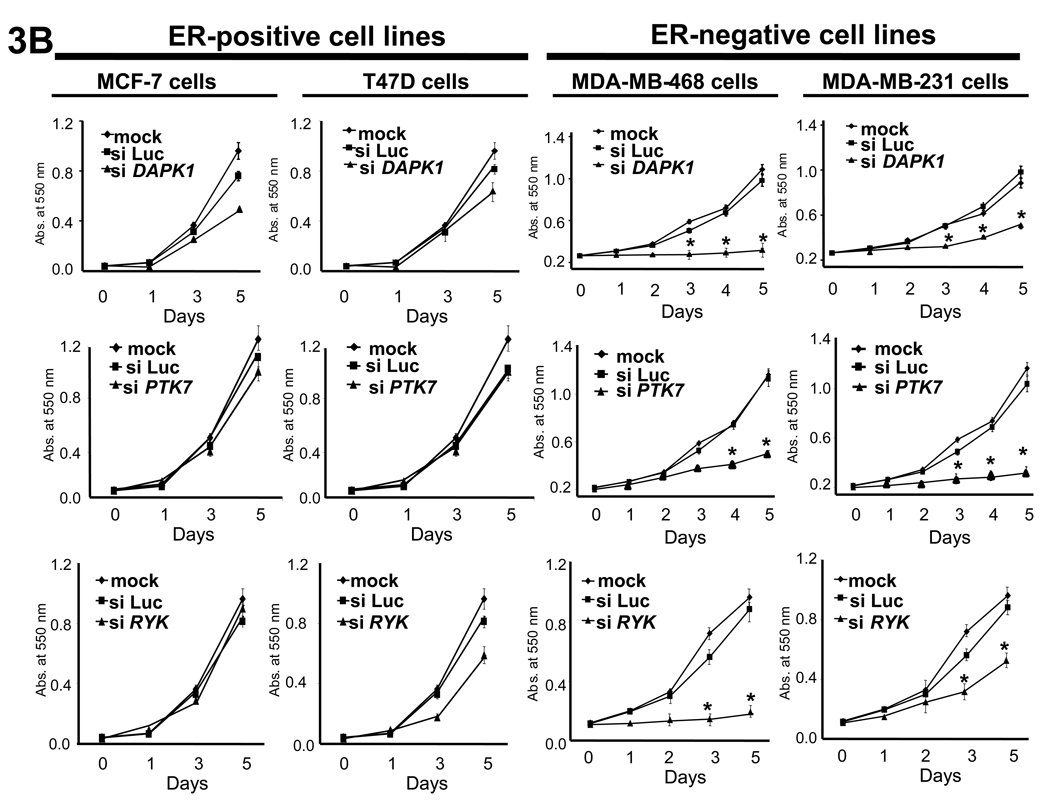
While the expression array profiling data allowed us to evaluate which kinases were differentially expressed, we investigated whether these kinases are critical for the growth of ER-negative breast cancer. To do this we performed siRNA knockdown studies to determine the effect of individual kinase knockdown on breast cancer cell proliferation. ER-positive (MCF-7 and T47D) and ER-negative (MDA-MB-468 and MDA-MB-231, both “triple-negative”) cells were transfected with siRNAs for 20 of the 52 genes identified in our screen. All siRNA constructs used in the study showed at least 70% knockdown of target kinase expression for 4 days after transfection (representative examples are shown in Fig. 3A).
Fig. 3.
Effect of siRNA knockdown on the growth of ER-negative and ER-positive breast cancer cells. (A) Knockdown of target kinase expression was achieved using siRNA against the identified kinases, with representative data of DAPK1, PTK7, and RYK knockdown in MDA-MB-468 cells shown. Knockdown was confirmed by Q-PCR at day 2 and day 5 and was >70% in all experiments. (B) DAPK1, PTK7, and RYK knockdown inhibited growth in the ER-negative breast cancer cell lines MDA-MB-468 and MDA-MB-231 but not in the ER-positive breast cancer cell lines MCF-7 and T47D. Asterisk denotes significant difference in curves between kinase of interest knockdown and siLuc transfected growth curves, P-value < 0.05. A complete summary of results of the kinase inhibition cell growth studies are shown in figure S4 and figure S5. Data are represented as mean ± SD.
Of the 20 kinases evaluated, 14 were critical for the growth of ER-negative breast cancer. Knockdown of 9 (EPHB4, LIMK2, DAPK1, YES1, RYK, VRK2, PTK7, RAF1, UCK2) had a significant growth-inhibitory effect on ER-negative breast cancer (MDA-MB-468 and MDA-MB-231) but had little or no effect on ER-positive breast cancer cells. An additional 5 of 20 kinases (BUB1, CHK1, IRAK1, CCL4, TTK) inhibited growth of all breast cancer cell lines. Knockdown of 5 of the 20 kinases (STK38L, PIM1, SFRS1, PKXL, TLR1) had no effect on the growth of any breast cancer cell lines, while knockdown of 1 of 20 kinases (MPZL1) had a significant growth-stimulatory effect on all breast cancer cell lines examined. Representative growth curves from these knockdown experiments are shown in Fig. 3B.
Knockdown of many of the genes in the “cell cycle checkpoint” cluster of ER-negative breast cancer had a profound inhibitory effect on ER-negative breast cancer cell growth but no effect on ER-positive breast cancer, while knockdown of certain genes in the “immunomodulatory” cluster inhibited the growth of all breast cancer cell lines examined. A summary of results is shown in Supplementary Fig. S5, with bolded genes exhibiting a differential growth phenotype between ER-negative and ER-positive breast cancer cell lines. These results indicate that many of the kinases found to be highly expressed in ER-negative breast cancers are indeed critical for breast cancer cell growth (actual percentage of growth inhibition values can be found in Supplementary Fig. S4).
S6 kinase Subtype of ER-negative Breast Cancer Predicts Poor Metastasis-Free Survival
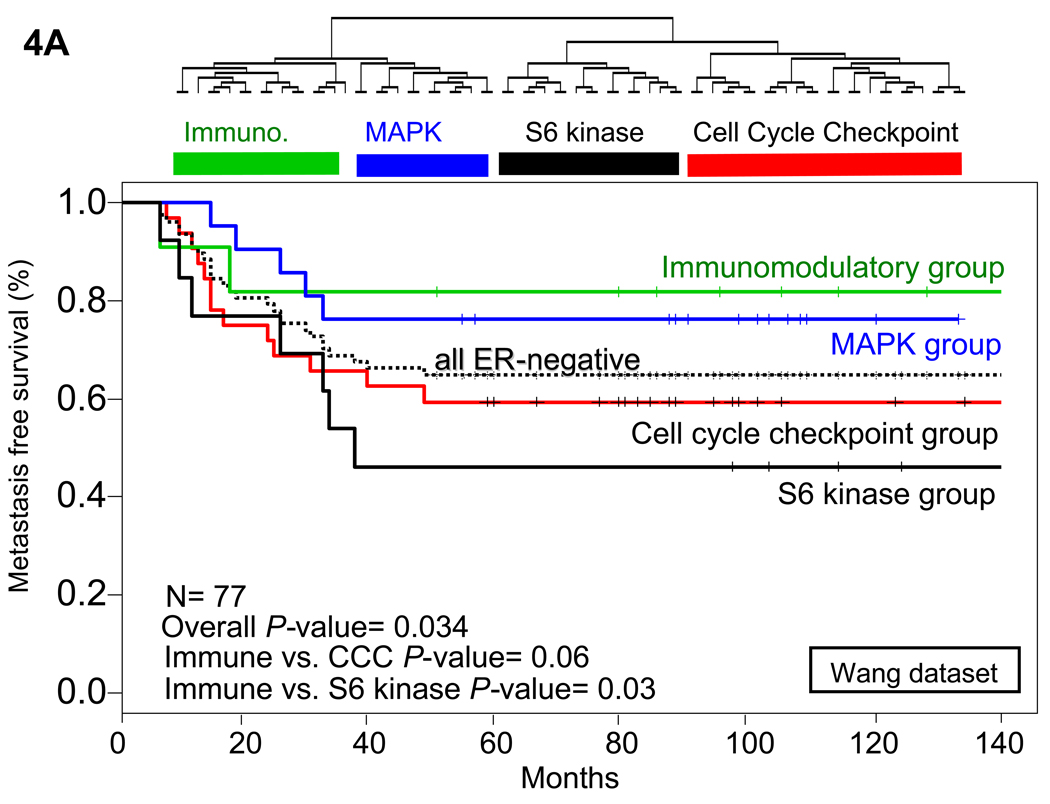
To determine whether the identified list of differentially-expressed kinases provided prognostic information, we analyzed the survival data from the Wang (25) and van de Vijver (3) data sets using the genes that we identified as being overexpressed in ER-negative breast tumors. The Wang dataset was obtained using breast cancer samples from patients with lymph-node negative breast cancer who were treated with breast conserving surgery or modified radical mastectomies from 1980–95. These patients also received radiotherapy when indicated, but did not receive systemic chemotherapy or hormonal adjuvant therapy. This time period was also prior to the development of the anti-HER2 therapy, trastuzumab (Herceptin), and these patients were not treated with trastuzumab (Herceptin). 219 patients had undergone breast-conserving surgery and 67 modified radical mastectomy. Radiotherapy was given to 248 patients (87%), and metastasis-free survival was tracked in all patients. In this data set, we first determined whether our list of 52 kinases and kinase-associated genes overexpressed in ER-negative breast tumors could subcluster the Wang dataset tumors into the 4 subtypes of ER-negative tumors identified in our analysis. Hierarchical clustering of the ER-negative tumors from the Wang dataset using expression values of the genes identified in this analysis did indeed identify 4 groups of ER-negative tumors (Fig. 4A). Figure of merit analysis showed that these four groups were stable against reclustering. Furthermore, these 4 clusters were similar in their kinase expression profiles to those previously identified, again identifying an S6 kinase signature cluster, a cell cycle checkpoint cluster, an immunomodulatory cluster, and a MAPK cluster.
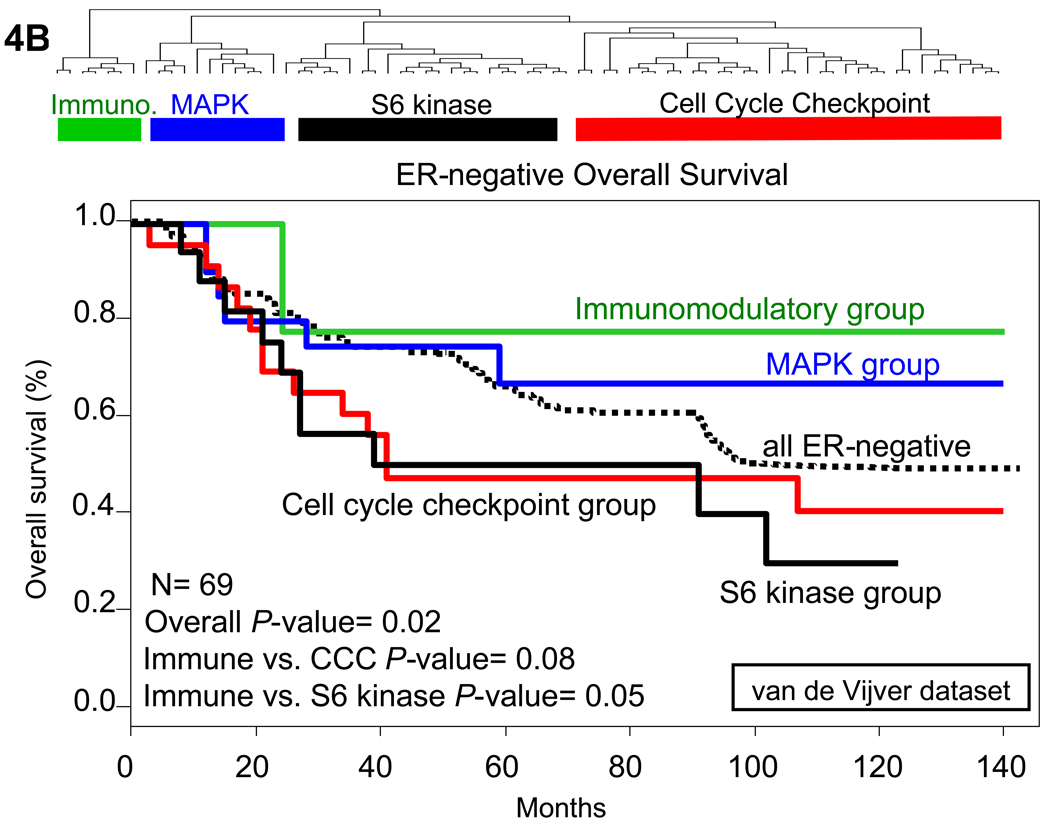
Fig. 4.
Hierarchical clustering and Kaplan-Meier metastasis free and overall survival analysis of ER-negative tumors in multiple datasets. (A) Hierarchical clustering of only ER-negative tumors identified the 4 clusters of ER-negative breast tumors in the Wang data set (25). The tumors were classified based on the expression level of the kinases identified in the analysis. Tumors that fell into the immunomodulatory cluster had a decreased risk of metastasis, and tumors in the cell cycle regulatory and S6 kinase clusters had a substantially elevated risk of metastasis at 5 years. (B) Similar results were found when hierarchical clustering was done in the van de Vijver data set (3). Overall survival was substantially higher in the immunomodulatory group than in the S6 kinase or cell cycle checkpoint groups. Overall P-value was calculated based on the assumption that there would be no difference between any of the survival curves and was initially used to determine whether any one of the curves were significantly different. Further P-values were calculated comparing the designated two groups with the calculation of Chi square values. Immune refers to immunomodulatory group, CCC to the cell cycle checkpoint group, and S6 kinase to the S6 kinase group.
Kaplan-Meier analysis of the metastasis-free survival between the different subgroups of ER-negative tumors from the Wang dataset shows that women with the S6 kinase signature-expressing tumors have a much worse prognosis than the other groups, while women with breast cancers expressing the immunomodulatory kinases have a much better prognosis (Fig. 4B). To provide additional validation of these results, we performed the same analysis in the van de Vijver (3) data set. In this dataset, all patients had stage I or II breast cancer and were younger than 53 years old; 151 had lymph-node–negative disease, and 144 had lymph node–positive disease. Ten of the 151 patients who had lymph-node–negative disease and 120 of the 144 who had lymph-node–positive disease had received adjuvant systemic therapy consisting of chemotherapy (90 patients), hormonal therapy (20), or both (20). As with the Wang dataset, hierarchical clustering of the ER-negative tumors identified the four groups of ER-negative tumors, which were again stable against reclustering.
Patients whose tumors had high expression of the immunomodulatory genes had a significantly better overall survival than those with high expression of the S6 kinase and cell cycle checkpoint clusters. These data suggest that our list of differentially-expressed kinases can be used to identify distinct subtypes of ER-negative breast tumors, and that the tumor clusters defined by the expression of these kinases have either a good prognosis (immunomodulatory group) or a particularly poor prognosis (S6 kinase signature group) based on their kinase expression profile.
Discussion
In this report we show that Affymetrix gene expression profiling of human breast tumors is able to identify kinases that are differentially-expressed in ER-negative breast cancers as compared to ER-positive breast cancers. Our analysis also revealed that ER-negative tumors can be clustered into 4 distinct groups, depending on the specific kinases and kinase-associated genes expressed and the level of their expression. Analysis of publicly available breast tumor data sets confirmed that these identified kinase and kinase-associated genes are indeed upregulated in ER-negative breast cancer. Studies in which knockdown of selected kinases using siRNA demonstrated which of the identified kinases are critical for ER-negative, including “triple-negative”, breast cancer growth. Finally, analysis of kinase and kinase-associated gene expression in human breast tumors demonstrated that the individual subtypes of ER-negative breast cancer identified by their kinase profile here have widely different outcomes. Specifically, women whose ER-negative tumors highly express the S6 kinase group kinases have a particularly bad prognosis, while women whose tumors highly express immunomodulatory genes have a relatively good prognosis. Such results suggest that characterization of human tumors based on kinase expression can be used to select patients appropriate for novel therapies. In addition, this study identifies potential targets for the treatment of ER-negative breast cancer, including the aggressive “triple-negative” form of breast cancer.
This is the first report to show that ER-negative breast cancers can be subdivided into biologically distinct groups based on expression levels of specific kinases. Our data indicate that ER-negative breast tumors can be subdivided into 4 distinct groups, of particular importance are group 2 (S6 kinase group) and group 3 (immunomodulatory group), and that patients whose tumors express these kinases have very different prognoses. The immunomodulatory group (group 3) identified in this report has recently become of a focus of increasing scientific inquiry. In this report, we show differential expression of these immunomodulatory genes in the epithelial compartment (as demonstrated by high expression in breast cancer cell lines grown in vitro). There remains a question of whether these kinases and kinase-associated genes are also expressed in the non-epithelial cells present in breast tumors, specifically in infiltrating immune cells. Three lines of evidence suggest that this difference is predominantly due to gene expression in epithelial cells. First, recent work by Neve et al. (38) validates the differential expression identified in this report (including the immunomodulatory cluster) in ER-negative breast cancer epithelial cell lines as compared to their ER-positive cell line counterparts. Their experiments were conducted using a purified, homogenous population of breast cancer epithelial cell lines that show the same differential expression we note in our human tumor studies. Secondly, the siRNA knockdown experiments reported herein also show that knockdown of these immunomodulatory kinases in vitro in epithelial breast cancer cell lines have a differential effect on cell growth. Finally, to definitively address this issue, we examined H&E slides taken from the tumors at the time of diagnostic biopsy. These slides were analyzed in a blinded fashion by a trained pathologist and scored as to their histology and level of lymphocytic infiltration. There was no increase in lymphocytic infiltration in the tumors that comprise the “immunomodulatory” subtype as compared to the other subsets of ER-negative breast cancer (Supplementary Fig. S2).
The role of the immune system in cancer has historically investigated how the immune system itself responds to the “foreign” cancer as the primary focus. It is now being appreciated that the tumor itself may act autonomously to influence the stromal microenvironment and evade recognition by the immunosurveillance machinery. It is possible that the immune-regulatory genes expressed by the epithelial cancer cells affect this local immune response to these tumors. Recent work by Teschendorff et al. supports our findings (39). This group also identified an immunomodulatory profile in ER-negative breast cancer which was shown to confer better prognosis (39). It will be interesting to investigate in the future whether modulation of intrinsic gene expression by the tumor is an important mechanism by which cancer cells can avoid immunosurveillance, including the proper controls meant to keep aberrant growth in check (39, 40).
These studies provide a large number of promising new targets for the treatment of ER-negative breast cancer. ER-positive breast cancers are now routinely treated using SERMs and aromatase inhibitors, and these cancers are now even prevented using such pharmacologic intervention (9). Recent studies have shown that intrinsic breast cancer subtypes differ depending on the ethnicity of the patient from whom the tumor is obtained. Carey et al. refined an IHC-based assay to categorize the prevalence of varying breast cancer subtypes in different populations (8). It was shown that the prevalence of the basal-like subtypes was strongly influenced by race and menopause status. The highest prevalence of basal-like tumors was noted in premenopausal African American breast cancer patients (8, 41). Basal-like tumors, which are almost uniformly ER-negative, PR-negative, and HER2 negative (“triple-negative”), are more aggressive, carry a higher proliferative capacity, occur at a younger age, and carry a particularly bad prognosis (41, 42). This work provides the rationale for targeted therapy using specific kinase inhibitors to treat this type of breast cancer more prevalent among a traditionally underserved population.
One particularly promising agent for the treatment of triple-negative breast cancer is the multiple kinase inhibitor dasatinib. Dasatinib is an oral kinase inhibitor that blocks BCR/Abl and c-Src and is currently approved for the treatment of chronic myelogenous leukemia (CML) and acute lymphoblastic leukemia (ALL) (43). Intriguingly, this kinase inhibitor blocks the activity of many of the kinases identified in this analysis including YES1, EPHB4, and EPHA2. Additionally, dasatinib blocks the activity of many other kinases identified in this analysis as being overexpressed in ER-negative tumors, including Abl, c-Src, and KIT, though these kinases did not meet our strict P-value inclusion criteria and thus are not part of our restricted list of 52 kinases. These kinases have, however, been shown to be elevated in other datasets. There is increasing pre-clinical and clinical data to suggest that this multi-kinase inhibitor may be an effective treatment for triple-negative breast cancer. Initial experiments in both prostate and breast cancer cell lines demonstrated that dasatinib significantly inhibited breast cancer cell line growth (44). Further in vitro experimentation shows that dasatinib is especially efficacious at inhibiting basal-like and post-EMT ER-negative breast cancer cell line growth and these studies led to the identification potential biomarkers of response (45). Clinical trials are currently being conducted using dasatinib in women with ER-negative breast cancer in the metastatic setting.
The results reported here demonstrate that genomic profiling of human breast cancers can identify subtypes of ER-negative breast cancer, but even more importantly, can also identify new targets for effective treatment of these aggressive breast cancers. Given the current difficulty in treating ER-negative breast cancer, and particularly the triple-negative form of breast cancer, the identification of the kinases that are critical for the growth of these cancers represents the first step towards effective individualized targeted therapy for these poor prognosis ER-negative breast cancers.
Translational Relevance
Estrogen receptor alpha (ER)-negative breast cancers remain a very difficult cancer to treat. There are few effective treatments for such cancers which are generally more aggressive, rapidly growing, and are often not cured by traditional chemotherapy. Furthermore, the signaling pathways that govern ER-negative cancer growth are poorly described. This study identifies critical growth regulatory molecules in ER-negative breast cancer that represent novel targets for the treatment of ER-negative breast cancer, specifically the aggressive ER-negative, PR-negative, HER2-negative or “triple negative” breast cancer, using gene expression profiling and siRNA knock-down studies. This is the first study to specifically evaluate differential expression of kinases in ER-negative breast cancer in human tumors. In the studies reported in this manuscript, we used gene expression profiling and siRNA knockdown to identify specific kinases that are required for the growth of ER-negative, but not ER-positive breast cancer cells. These kinases represent potential “druggable” targets for the treatment of these aggressive ER-negative tumors. In addition, our results divide these aggressive human ER-negative tumors into 4 groups, including one group which has a relatively good outcome (immunomodulatory) and another group that has an extremely poor outcome (S6 kinase). Thus, kinase gene expression profiling of ER-negative breast cancers could be used to identify patients who will respond very poorly to standard therapy. The kinases identified as overexpressed in this poor prognosis group of tumors may be targeted to more effectively treat these aggressive “triple-negative” breast cancers.
Supplementary Material
Effects of siRNA knockdown on the growth of ER-positive and ER-negative breast cancer cell lines. Individual kinases were knockdown in 4 breast cancer cell lines and growth was monitored using MTS assay. Effect of growth inhibition was based on the percentage of growth at day 4 compared to siLuc transfected controls. Cut-off percentages for inhibition, slight inhibition, no effect, and enhanced growth are shown in figure S5. Bolded genes exhibited a differential growth phenotype between ER-negative and ER-positive breast cancer cell lines. Actual percentage values can be found in figure S4.
Overview of the study design and results
H&E of immune infiltration in all subsets of ER-negative breast cancer. Scoring was from 0–3 for neutrophil infiltration and 0–3 for lymphocyte infiltration in the entire section of tissue at both low and high power magnification. Scores of 0 corresponded to no evidence of infiltration, 1 for minimal foci of infiltration (<1% of tumor mass), 2 for moderate amounts of immune cell infiltration (2–15% of tumor mass) and 3 for considerable amounts of immune cell infiltration (>15% of tumor mass). Scores for the neutrophil and lymphocyte scoring were added to give a composite score (0–6).
List of kinases validates in an independent data set of human breast cancer cell lines. (A) Publically available breast cancer cell line expression data was clustered in an unsupervised manner using only the 52 kinase genes identified in our analysis. Unsupervised hierarchical clustering using the 52 kinases identified as being at least 2 fold more highly expressed clusters ER-positive from ER-negative breast cancer cell lines and identifies the luminal, basal A, and basal B subtypes. (B) Clustering only of ER-negative breast cancer cell lines using the 52 kinases identifies 4 subsets of ER-negative breast cancer cell lines.
Effects of siRNA knockdown on the growth of ER-positive and ER-negative breast cancer cell lines. Individual kinases were knockdown in 4 breast cancer cell lines and growth was monitored using MTS assay. Data is shown as percentage of growth at day 4 compared to mock transfected controls.
List of the kinases overexpressed in ER-negative breast tumors that had at least 2-fold higher expression with a P-value <.01 in ER-negative tumors compared to ER-positive tumors.
List of kinases overexpressed in ER-positive breast tumors that had at least 2-fold higher expression with a P-value <.01 in ER-positive tumors compared to ER-negative tumors.
Kinase overexpression validated in independent human tumor sample data sets. Data analysis of an additional 12 publically available human breast tumor datasets shows the 52 kinases identified as being overexpressed in this study are also significantly overexpressed in ER-negative breast tumors in the other datasets. Z-scores were calculated using the Z-transform test and are listed with their correlating P-value.
List of kinases and kinase associated genes used in the kinome gene expression profiling. This list is similar to those used in other studies evaluating “kinome” gene expression.
Acknowledgment
We would like to thank all the patients enrolled in the clinical trials whose tumors samples were used to generate this data. Additionally we would like to thank the research nurses who were instrumental in accruing patients in these trials and obtaining tumor specimens. We would also like to thank Dr. Suzanne Fuqua and the microarray core at Baylor College of Medicine for assistance with the microarray experiments. Finally, we would like to thank Arlene Zamora for providing administrative help. This work was supported by grants from the NIH/NCI Breast Cancer Special Project of Research Excellence (SPORE) #P50CA58183, DOD predoctoral fellowship #W81XWH-06-1-0715, Susan G. Komen Breast Cancer Foundation Promise Grant, and the Breast Cancer Research Foundation (BCRF). The authors declare no conflict of interest in these studies.
References
- 1.Chang JC, Wooten EC, Tsimelzon A, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 2.Chang JC, Wooten EC, Tsimelzon A, et al. Patterns of resistance and incomplete response to docetaxel by gene expression profiling in breast cancer patients. J Clin Oncol. 2005;23:1169–1177. doi: 10.1200/JCO.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 3.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Expression profiling predicts outcome in breast cancer. Breast Cancer Res. 2003;5:57–58. doi: 10.1186/bcr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 9.Swaby RF, Sharma CG, Jordan VC. SERMs for the treatment and prevention of breast cancer. Rev Endocr Metab Disord. 2007;8:229–239. doi: 10.1007/s11154-007-9034-4. [DOI] [PubMed] [Google Scholar]
- 10.Tan AR, Swain SM. Adjuvant chemotherapy for breast cancer: an update. Semin Oncol. 2001;28:359–376. doi: 10.1016/s0093-7754(01)90130-7. [DOI] [PubMed] [Google Scholar]
- 11.Miller K, Wang M, Gralow J, et al. Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med. 2007;357:2666–2676. doi: 10.1056/NEJMoa072113. [DOI] [PubMed] [Google Scholar]
- 12.Viani GA, Afonso SL, Stefano EJ, De Fendi LI, Soares FV. Adjuvant trastuzumab in the treatment of her-2-positive early breast cancer: a meta-analysis of published randomized trials. BMC Cancer. 2007;7:153. doi: 10.1186/1471-2407-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowell JE, Minna JD. EGFR mutations and molecularly targeted therapy: a new era in the treatment of lung cancer. Nat Clin Pract Oncol. 2006;3:170–171. doi: 10.1038/ncponc0476. [DOI] [PubMed] [Google Scholar]
- 14.Herbst RS, Fukuoka M, Baselga J. Gefitinib--a novel targeted approach to treating cancer. Nat Rev Cancer. 2004;4:956–965. doi: 10.1038/nrc1506. [DOI] [PubMed] [Google Scholar]
- 15.Minna JD, Dowell J. Erlotinib hydrochloride. Nat Rev Drug Discov. 2005 Suppl:S14–S15. doi: 10.1038/nrd1612. [DOI] [PubMed] [Google Scholar]
- 16.Dancey J, Sausville EA. Issues and progress with protein kinase inhibitors for cancer treatment. Nat Rev Drug Discov. 2003;2:296–313. doi: 10.1038/nrd1066. [DOI] [PubMed] [Google Scholar]
- 17.Chang J, Clark GM, Allred DC, Mohsin S, Chamness G, Elledge RM. Survival of patients with metastatic breast carcinoma: importance of prognostic markers of the primary tumor. Cancer. 2003;97:545–553. doi: 10.1002/cncr.11083. [DOI] [PubMed] [Google Scholar]
- 18.Tham YL, Gomez LF, Mohsin S, et al. Clinical response to neoadjuvant docetaxel predicts improved outcome in patients with large locally advanced breast cancers. Breast Cancer Res Treat. 2005;94:279–284. doi: 10.1007/s10549-005-9020-z. [DOI] [PubMed] [Google Scholar]
- 19.Cleator S, Tsimelzon A, Ashworth A, et al. Gene expression patterns for doxorubicin (Adriamycin) and cyclophosphamide (cytoxan) (AC) response and resistance. Breast Cancer Res Treat. 2006;95:229–233. doi: 10.1007/s10549-005-9009-7. [DOI] [PubMed] [Google Scholar]
- 20.Mohsin SK, Weiss HL, Gutierrez MC, et al. Neoadjuvant trastuzumab induces apoptosis in primary breast cancers. J Clin Oncol. 2005;23:2460–2468. doi: 10.1200/JCO.2005.00.661. [DOI] [PubMed] [Google Scholar]
- 21.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura K, Bowman ED, Simon R, et al. Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking- and prognosis-related molecular profiles. Cancer Res. 2002;62:3244–3250. [PubMed] [Google Scholar]
- 23.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 26.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 27.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 28.Draviam VM, Stegmeier F, Nalepa G, et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 29.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 31.Yu K, Ganesan K, Miller LD, Tan P. A modular analysis of breast cancer reveals a novel low-grade molecular signature in estrogen receptor-positive tumors. Clin Cancer Res. 2006;12:3288–3296. doi: 10.1158/1078-0432.CCR-05-1530. [DOI] [PubMed] [Google Scholar]
- 32.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 35.Ginestier C, Cervera N, Finetti P, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006;12:4533–4544. doi: 10.1158/1078-0432.CCR-05-2339. [DOI] [PubMed] [Google Scholar]
- 36.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 38.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amini RM, Aaltonen K, Nevanlinna H, et al. Mast cells and eosinophils in invasive breast carcinoma. BMC Cancer. 2007;7:165. doi: 10.1186/1471-2407-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furberg H, Millikan R, Dressler L, Newman B, Geradts J. Tumor characteristics in African American and white women. Breast Cancer Res Treat. 2001;68:33–43. doi: 10.1023/a:1017994726207. [DOI] [PubMed] [Google Scholar]
- 42.Porter PL, Lund MJ, Lin MG, et al. Racial differences in the expression of cell cycle-regulatory proteins in breast carcinoma. Cancer. 2004;100:2533–2542. doi: 10.1002/cncr.20279. [DOI] [PubMed] [Google Scholar]
- 43.Padmanabhan S, Ravella S, Curiel T, Giles F. Current status of therapy for chronic myeloid leukemia: a review of drug development. Future Oncol. 2008;4:359–377. doi: 10.2217/14796694.4.3.359. [DOI] [PubMed] [Google Scholar]
- 44.Nam S, Kim D, Cheng JQ, et al. Action of the Src family kinase inhibitor, dasatinib (BMS-354825), on human prostate cancer cells. Cancer Res. 2005;65:9185–9189. doi: 10.1158/0008-5472.CAN-05-1731. [DOI] [PubMed] [Google Scholar]
- 45.Finn RS, Dering J, Ginther C, et al. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/"triple-negative" breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 46.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of siRNA knockdown on the growth of ER-positive and ER-negative breast cancer cell lines. Individual kinases were knockdown in 4 breast cancer cell lines and growth was monitored using MTS assay. Effect of growth inhibition was based on the percentage of growth at day 4 compared to siLuc transfected controls. Cut-off percentages for inhibition, slight inhibition, no effect, and enhanced growth are shown in figure S5. Bolded genes exhibited a differential growth phenotype between ER-negative and ER-positive breast cancer cell lines. Actual percentage values can be found in figure S4.
Overview of the study design and results
H&E of immune infiltration in all subsets of ER-negative breast cancer. Scoring was from 0–3 for neutrophil infiltration and 0–3 for lymphocyte infiltration in the entire section of tissue at both low and high power magnification. Scores of 0 corresponded to no evidence of infiltration, 1 for minimal foci of infiltration (<1% of tumor mass), 2 for moderate amounts of immune cell infiltration (2–15% of tumor mass) and 3 for considerable amounts of immune cell infiltration (>15% of tumor mass). Scores for the neutrophil and lymphocyte scoring were added to give a composite score (0–6).
List of kinases validates in an independent data set of human breast cancer cell lines. (A) Publically available breast cancer cell line expression data was clustered in an unsupervised manner using only the 52 kinase genes identified in our analysis. Unsupervised hierarchical clustering using the 52 kinases identified as being at least 2 fold more highly expressed clusters ER-positive from ER-negative breast cancer cell lines and identifies the luminal, basal A, and basal B subtypes. (B) Clustering only of ER-negative breast cancer cell lines using the 52 kinases identifies 4 subsets of ER-negative breast cancer cell lines.
Effects of siRNA knockdown on the growth of ER-positive and ER-negative breast cancer cell lines. Individual kinases were knockdown in 4 breast cancer cell lines and growth was monitored using MTS assay. Data is shown as percentage of growth at day 4 compared to mock transfected controls.
List of the kinases overexpressed in ER-negative breast tumors that had at least 2-fold higher expression with a P-value <.01 in ER-negative tumors compared to ER-positive tumors.
List of kinases overexpressed in ER-positive breast tumors that had at least 2-fold higher expression with a P-value <.01 in ER-positive tumors compared to ER-negative tumors.
Kinase overexpression validated in independent human tumor sample data sets. Data analysis of an additional 12 publically available human breast tumor datasets shows the 52 kinases identified as being overexpressed in this study are also significantly overexpressed in ER-negative breast tumors in the other datasets. Z-scores were calculated using the Z-transform test and are listed with their correlating P-value.
List of kinases and kinase associated genes used in the kinome gene expression profiling. This list is similar to those used in other studies evaluating “kinome” gene expression.