Abstract
Nosiheptide (NOS), belonging to the e series of thiopeptide antibiotics that exhibit potent activity against various bacterial pathogens, bears a unique indole side ring system and regiospecific hydroxyl groups on the characteristic macrocyclic core. Here, cloning, sequencing and characterization of the nos gene cluster from Streptomyces actuosus ATCC 25421 as a model for this series of thiopeptides has unveiled new insights into their biosynthesis. Bioinformatics-based sequence analysis and in vivo investigation into the gene functions show that NOS biosynthesis shares a common strategy with recently characterized b or c series thiopeptides for forming the characteristic macrocyclic core, which features a ribosomally synthesized precursor peptide with conserved posttranslational modifications. However, it apparently proceeds via a different route for tailoring the thiopeptide framework, allowing the final product to exhibit the distinct structural characteristics of e series thiopeptides, such as the indole side ring system. Chemical complementation supports the notion that the S-adenosylmethionine (AdoMet)-dependent protein NosL may play a central role in converting Trp to the key 3-methylindole moiety by an unusual carbon side chain rearrangement, most likely via a radical-initiated mechanism. Characterization of the indole side ring-opened analog of NOS from the nosN mutant strain is consistent with the proposed methyltransferase activity of its encoded protein, shedding light into the timing of the individual steps for indole side ring biosynthesis. These results also suggest the feasibility of engineering novel thiopeptides for drug discovery by manipulating the NOS biosynthetic machinery.
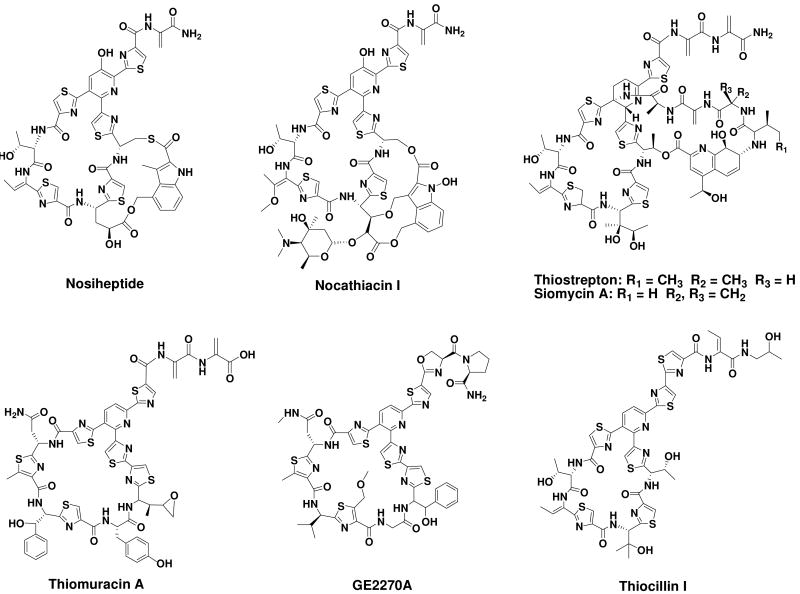
Thiopeptides are a growing class of sulfur-rich, highly modified heterocyclic peptides (1). Despite overall structural diversity, they share a characteristic macrocyclic core, consisting of a nitrogen-containing, 6-membered ring central to multiple thiazoles and dehydroamino acids (Figure 1). Nosiheptide-like members, classified as e series thiopeptides according to a central 2,3,5,6-tetrasubstituted pyridine domain, possess an indolic acid ring system that is appended to the side chains of the Ser/Cys and hydroxylated Glu residues of the macrocyclic core via at least two carboxylic ester linkages (e.g. O- and S- linkages for nosiheptide, and two O- linkages for nocathiacins), and in some cases, (an) attached sugar unit(s) (2, 3). There is significant clinical interest in thiopeptides due to their potent activity against various bacterial pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), penicillin-resistant Streptococcus pneumoniae (PRSP), and vancomycin-resistant enterococci (VRE). For example, the nocathiacins have been investigated as drug leads for developing broad-spectrum antibiotics by chemical modifications, aiming at the generation of biologically comparable and water-soluble compounds to fight against progressively emergent bacterial resistance to traditional chemotherapies (4–8).
Figure 1.
Structures of the e series thiopeptides nosiheptide (NOS) and nocathiacin I, b series thiostrepton (TSR)/siomycin A (SIO-A), and d series thiomuracin A (TMR-A), GE2270A and thiocillin I (TCL-I).
Nosiheptide (NOS), produced by Streptomyces actuosus ATCC 25421, is one of the oldest known thiopeptides and has been widely used as a feed additive for animal growth (9, 10). The structure and stereochemistry of NOS was ultimately confirmed by X-ray crystallography (11), following extensive analysis of chemically hydrolyzed fragments by NMR spectroscopic methods (12, 13). While the total synthesis of NOS has not been achieved, NOS, as the model molecule in the e series, was one of the first thiopeptides to be investigated biosynthetically by incorporation of isotope-labeled precursors, along with thiostrepton (TSR), a representative of b series that contains a distinct quinaldic acid side ring system appended to the characteristic thiopeptide macrocyclic core (14–17). All moieties of the peptidyl backbones of NOS and TSR were shown to originate exclusively from proteinogenic amino acids, including dehydroamino acids (from the Ser or Thr residues undergoing the anti elimination of water), thiazoles (from the Cys residues with cyclodehydration followed by deoxygenation), and the central 6-membered nitrogen heterocycle (produced by cyclization between two corresponding dehydroalanine acids with incorporation of an adjacent carbonyl group). Interestingly, the Trp residue was confirmed as a common precursor for the side ring systems in both NOS and TSR biosynthesis (14, 16), in spite of the difference in structures of the resulting motifs (i.e. the indolic acid moiety for NOS and the quinalidic acid moiety for TSR) and their linkages to the thiopeptide macrocyclic core (Figure 1).
It had long been controversial whether the thiopeptides are biosynthesized via a ribosome-dependent route of maturation of short peptides to complex, highly functionalized molecules, such as lantibiotics (18), bacteriocins (19) and cyanobactins (20), or in a manner similar to peptide antibiotics vancomycin and cyclosporin, whose peptidyl backbones are assembled by non-ribosomal peptide synthetases (NRPSs) (21, 22). Very recently, we and other research groups cloned, sequenced and characterized the biosynthetic gene clusters of the bicyclic b series thiopeptides TSR and siomycin A (SIO-A), and monocyclic d series thiocillins (TCLs), GE2270A and thiomuracins (TMRs), uncovering a common paradigm for the characteristic macrocyclic core biosynthesis that features conserved posttranslational modifications on a ribosomally synthesized precursor peptide (23–26). Given the similarities in structures and precursor-labeling patterns, the biosynthesis of NOS likely shares a conserved strategy with those of above thiopeptides to form the thiopeptide macrocyclic core. However, the tailoring of the elementary framework into the e series-specific member, particularly for the indolic acid moiety formation and attachment via a route distinct from the quinaldic acid pathway in TSR biosynthesis, was unclear. To exploit the genetic basis for fulfilling the knowledge gap, we now report the localization of the nos biosynthetic gene cluster from S. actuosus ATCC 25421 by cloning the thiopeptide-specific cyclodehydratase gene nosG using our recently developed PCR approach. The sequence analysis of the entire nos gene cluster allows for assignment of functions to the deduced gene products, setting the stage to propose the NOS biosynthetic pathway. While the finding of the ribosomal origin of NOS along with conserved posttranslational modifications again validates the generality of thiopeptide biosynthesis, in vivo functional investigations of genes involved in the indole side ring formation have revealed new insights into the biosynthesis of the e series-specific thiopeptides, including a novel strategy for the carbon side chain rearrangement to convert the Trp residue into the key 3-methylindole moiety.
Results and Discussion
NOS biosynthetic gene cluster
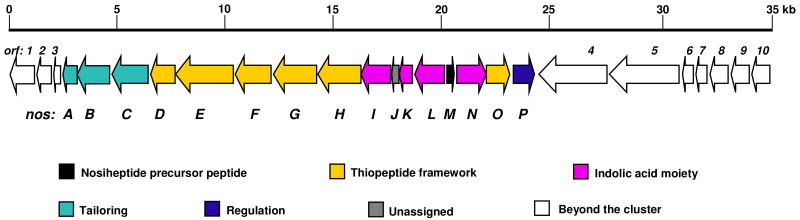
The enzymes for the formation of the thiopeptide-characteristic framework are highly conserved in and unique to thiopeptide biosynthesis (24), as exemplified by the thiopeptide cyclodehydratases. These enzymes exhibit high sequence homology to each other, but are distinct from other proteins with similar functions, including the polythiazole synthetases in both bacteriocin and cyanobactin biosynthesis (27). We decided to localize the NOS biosynthetic gene cluster by cloning the NOS cyclodehydratase gene using a specific PCR approach developed recently (24). With the genomic DNA of S. actuosus as the template, a distinct product with the expected size of 0.7 kb was readily amplified and subsequently cloned into pMD19-T vector for sequencing. Analysis of randomly selected clones confirmed the identity of the PCR product (P1), the deduced amino acid sequence of which is highly similar to known thiopeptide cyclodehydratases. To determine the relevance of P1 to NOS production, we set out to inactivate the target allele by gene disruption in S. actuosus. As anticipated, the P1 allele (namely nosG) mutant strain SL4001 completely lost the ability to produce NOS, confirming that the cloned locus encodes NOS biosynthesis (Figure S1). Consequently, using P1 as a probe, approximately 2,000 clones from the S. actuosus genomic library were screened, resulting in the identification of 3 overlapping cosmids that span a 70 kb region on the chromosome.
The DNA region represented by the cosmid pSL4001 was selected for sequencing, yielding a 34,713 bp contiguous sequence with the overall GC content of 71.6%, characteristic of Streptomyces DNA. Bioinformatics-based analysis of the sequenced region revealed 26 open reading frames (orfs), 15 of which, including 14 structural genes and 1 regulatory gene, were proposed to constitute the nos gene cluster according to functional assignments to the deduced products (Figure 2 and Table 1). The boundaries of the nos gene cluster were judged by identification of the immediately flanking orfs (at the both ends) that encode DNA-directed RNA polymerase subunits and various ribosomal proteins for the construction of the bacterial ribosome. Attempts to inactivate these genes were not successful, consistent with their essential nature to the survival of S. actuosus (data not shown). Considering the antibacterial mechanism of NOS, with the mode of action on the ribosome to inhibit protein synthesis by occupying a 23S ribosomal RNA domain that was known for the binding of the ribosomal protein L11 (28), the direct insertion of the nos gene cluster into the ribosomal protein encoding locus is interesting. Whether the resulting ribosome in S. actuosus is resistant to the action of NOS remains to be determined.
Figure 2.
Organization of the NOS biosynthetic genes. The deduced functions of which are labeled in color and summarized in Table 1.
Table 1.
Deduced functions of orfs in the nosiheptide biosynthetic gene cluster.
| Gene | Sizea | Protein Homologb and Origin | Identity / Similarity, % | Proposed Function |
|---|---|---|---|---|
| orf1 | 324 | SCO4661 (NP_628821), from S. coelicolor A3(2) | 91/96 | elongation factor G |
| orf2 | 156 | SCO4660 (NP_628820), from S coelicolor A3(2) | 99/100 | 30S ribosomal protein S7 |
| orf3 | 123 | SCO4659 (NP_628819), from S coelicolor A3(2) | 99/100 | 30S ribosomal protein S12 |
| nosA | 151 | Francci3_4114 (YP_483191), from Frankia sp. CcI3 | 55/67 | hypothetical protein |
| nosB | 455 | Sare_3149 (YP_001537948), from Salinispora arenicola CNS-205 | 32/47 | cytochrome P450 |
| nosC | 408 | SACE_1426 (YP_001103673), From Saccharopolyspora erythraea NRRL 2338 | 39/55 | cytochrome P450-like enzyme |
| nosD | 345 | TsrK (ACN80673), from S. laurentii ATCC 31255 | 32/54 | dehydratase |
| nosE | 920 | TsrJ (ACN80672), from S. laurentii ATCC 31255 | 29/50 | dehydratase |
| nosF | 549 | TsrM (ACN80675), from S. laurentii ATCC 31255 | 29/51 | dehydrogenase |
| nosG | 656 | TsrO (ACN80677), from S. laurentii ATCC 31255 | 36/58 | cyclodehydratase |
| nosH | 654 | TsrN (ACN80676), from S. laurentii ATCC 31255 | 22/37 | hypothetical protein |
| nosI | 419 | SAMR0921 (CAJ88630), from S. ambofaciens ATCC 23877 | 29/39 | AMP-dependent acyl-CoA synthetase/ligase |
| nosJ | 79 | -- | -- | hypothetical protein |
| nosK | 264 | BCE_2449 (NP_978758), from Bacillus cereus ATCC 10987 | 29/43 | α/β fold family hydrolase/acyltransfer ase |
| nosL | 400 | ThiH (AAD48429), from Salmonella typhimurium | 26/41 | Radical S-AdoMet: Biotin and thiamine synthesis associated |
| nosM | 50 | -- | -- | NOS precursor peptide |
| nosN | 395 | Tlm Orf11 (ABL74954), from Streptoalloteichus hindustanus | 32/48 | S-AdoMet-dependent oxidase or methyl transferase |
| nosO | 370 | TsrL (ACN80674), from S. laurentii ATCC 31255 | 28/49 | hypothetical protein |
| nosP | 323 | pSLA2-L_p071(NP_851493), from S. rochei | 51/66 | pathway specific regulatory protein |
| orf4 | 1299 | SAV_4915 (NP_826092), from S. avermitilis MA-4680 | 96/98 | DNA-directed RNA polymerase subunit β′ |
| orf5 | 1181 | SAV_4914 (NP_826091), from S. avermitilis MA-4680 | 95/98 | DNA-directed RNA polymerase subunit β |
| orf6 | 129 | SCO4653 (NP_628814), from S. coelicolor A3(2) | 81/86 | 50S ribosomal protein L7/L12 |
| orf7 | 176 | SSEG_01531 (YP_002208236), from S. sviceus ATCC 29083 | 89/96 | 50S ribosomal protein L10 |
| orf8 | 272 | SCO4651 (NP_628812), from S. coelicolor A3(2) | 65/78 | lipoprotein |
| orf9 | 297 | SSEG_01533 (YP_002208238), from S. sviceus ATCC 29083 | 55/70 | lipoprotein |
| orf10 | 241 | SCO4649 (NP_628810), from S. coelicolor A3(2) | 92/97 | 50S ribosomal protein L1 |
Numbers are in amino acids.
NCBI accession numbers are given in parentheses.
Ribosomal origin of NOS
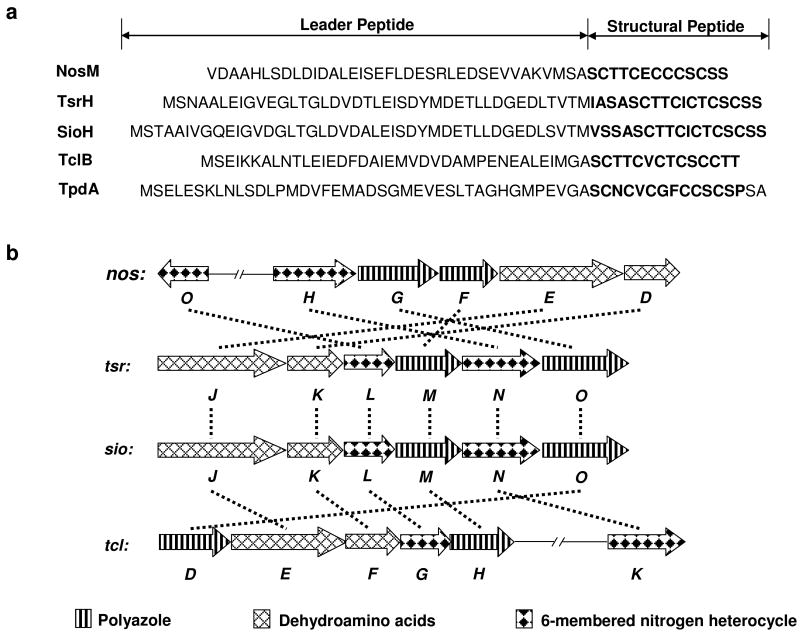
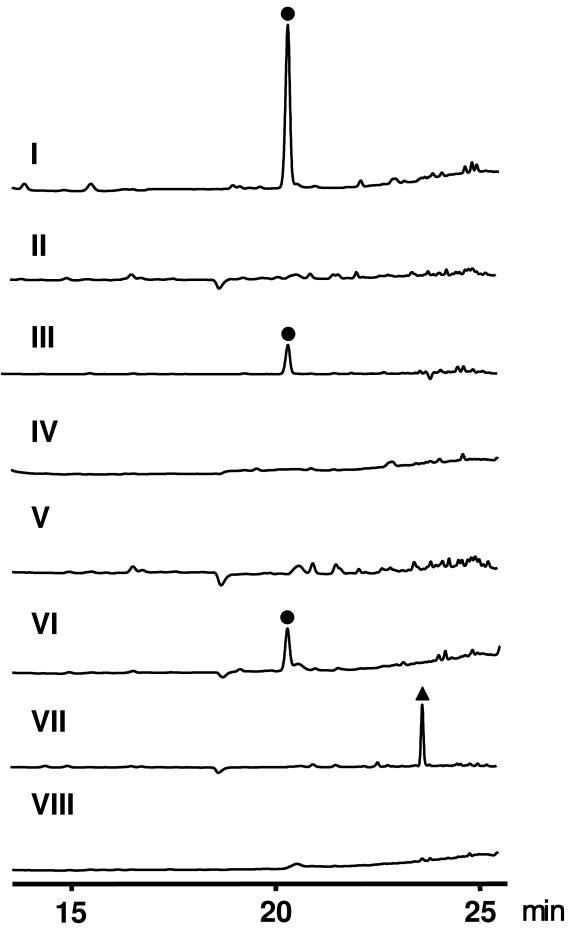
Within the predicted nos gene cluster, nosM encodes a 50-aa peptide that contains a 37-aa leader peptide (LP) and a 13-aa, Cys and Ser/Thr-rich structural peptide (SP) (Figure 3a). The SP (SCTTCECCCSCSS) is in complete agreement with the peptide sequence of the NOS backbone, with the exception of the C-terminal Ser residue that can be removed (except for the amino group) in the tailoring process. The LP sequence of NosM is rich in acidic amino acids, similar to those of thiopeptide precursor peptides such as TsrH for TSR and TclB for TCLs, but no overall significant homology in sequence was identified among them. This indicates that the Glu and Asp residues of LPs might be commonly important to substrate recognitions for posttranslational modifications that, however, may take place specifically on the cognate precursor peptide in each thiopeptide biosynthesis. To confirm that nosM is indispensable in NOS biosynthesis, we inactivated and complemented it in vivo. As shown in Figure 6 (II), the resulting in-frame deletion mutant strain SL4002 completely lost the ability to produce NOS, providing experimental evidence to support the origin of NOS from a ribosomally synthesized precursor peptide. Furthermore, the NOS production can be partially restored in the recombinant strain SL4003, where nosM was expressed in trans in SL4002 (Figure 6, III), implying a promising way to access the structural diversity of NOS by sequence permutations to the precursor peptide.
Figure 3.
Genetic features for the thiopeptide framework formation. (a) Peptide precursors for NOS (NosM), TSR (TsrH), SIO-A (SioH), TCLs (TclB), and GE2270A (TpdA). The SP sequences are labeled in bold. (b) Organization of the thiopeptide framework-forming genes identified from the producers of NOS (nos), TSR (tsr), SIO-A (sio), and TCLs (tcl). Their deduced functions are labeled in pattern, and homologies in sequence are indicated by dashed lines.
Figure 6.
HPLC analysis of the production of NOS or its analog, in the wild type strain S. actuosus ATCC 25421 (I), nosM mutant SL4002 (II), recombinant strain SL4003 that derives from SL4002 by expressing nosM in trans (III), nosH mutant SL4004 (IV), nosL mutant SL4005 (V), nosL mutant SL4005 complemented by chemically feeding 3-methylindole-2-carboxylic acid (8) (VI), nosN mutant SL4006 (VII), and nosP mutant SL4007 (VIII). Solid dot indicates NOS. Solid triangle indicates the NOS analog 4.
Conserved posttranslational modifications
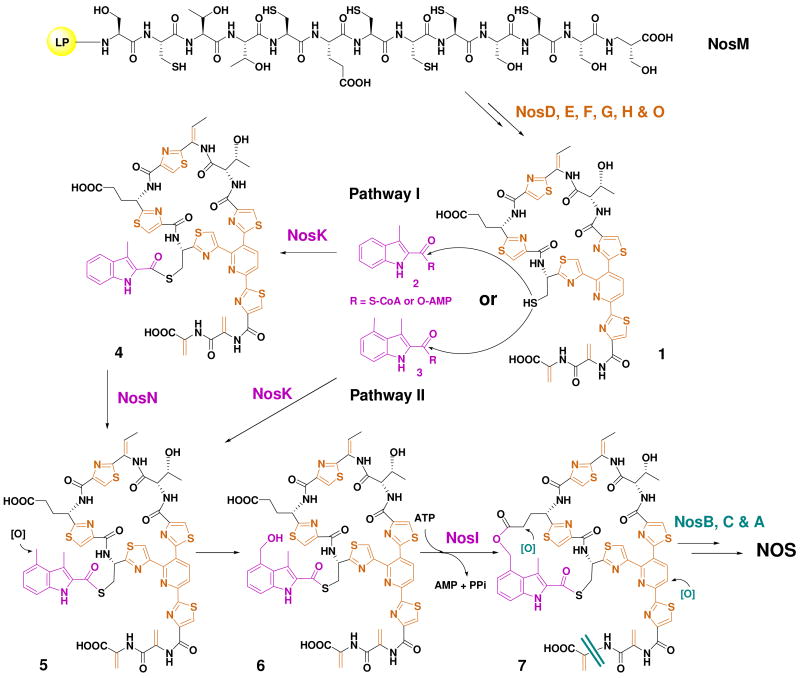
Central to the nos gene cluster are five orfs, nosDEFGH, the deduced products of which are highly homologous to the conserved posttranslational modification enzymes for affording the thiopeptide-characteristic macrocyclic core of previously characterized thiopeptides (Figure 3b). They encode a cyclodehydratase/dehydrogenase complex (NosG and NosF, 36% and 29% identity to TsrO and TsrM, respectively), a dehydratase pair (NosE and NosD, 29% and 32% identity to TsrJ and TsrK, respectively), and NosH, a TsrN-like unknown protein (27% identity). In addition, nosO, which encodes a protein similar to TsrL (28% identity) that was proposed to be involved in the 6-membered nitrogen heterocycle formation along with TsrN, was identified in the downstream region of the gene cluster. The presence of these genes, along with nosM that encodes the ribosomally synthesized precursor peptide (whose SP is enriched by Cys and Ser/Thr residues), is in perfect agreement with the criteria for a thiopeptide biosynthetic machinery and support the newly unveiled common paradigm to furnish the family-specific framework (24). Thus, these encoded enzymes may act on NosM and catalyze 1) the polythiazole formation by nucleophilic addition of each Cys side chain to the proceeding carbonyl group followed by dehydration and dehydrogenation (with one exception of Cys45, whose –SH group is reserved for appending the indolic acid moiety); 2) multiple dehydrations of Ser and Thr residues to generate dehydroamino acids (except for Thr40, presumably due to the regiospecific activity); and 3) intramolecular cyclization to afford the 6-membered heterocyclic ring and complete the biosynthesis of the 26-membered macrocyclic system, yielding the intermediate 1 for further modifications. It should be noted that the resulting 6-membered nitrogen heterocycle may undergo an elimination of the 37-aa LP after dehydration to form the central aromatic pyridine domain, distinct from a reductive reaction required for the dehydropiperidine formation in TSR or SIO-A biosynthesis (24, 25). To support this proposed modification pathway, selected orfs were inactivated to validate their indispensability. As exemplified by the in-frame deletion of nosH, the resulting mutant strain SL4004 completely lost the ability to produce NOS (Figure 6, IV), thereby clearly confirming its involvement in NOS biosynthesis.
Unusual indole side ring system formation
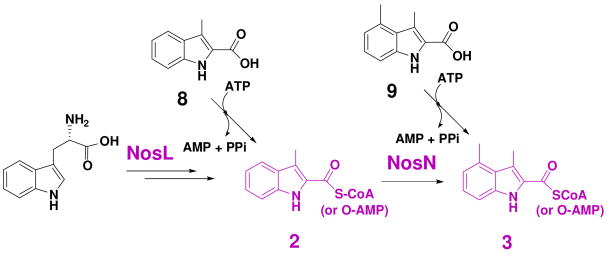
Functional assignment of the remaining orfs in the nos gene cluster revealed four genes, nosIKLN, whose deduced products could serve as the candidates responsible for the biosynthesis and attachment of the indolic acid moiety to the thiopeptide macrocyclic ring system of 1 (Figures 4 and 5). NosL, containing a conserved CxxxCxxC motif essential for binding a [4Fe-4S] cluster, shows sequence similarity to various ThiH-like proteins (e.g. ThiH from Salmonella typhimurium, 26% identity) that are S-adenosylmethionine (AdoMet)-dependent and catalyze the cleavage of the Cα-Cβ of Tyr initiated by a radical Ado intermediate in thiamine biosynthesis (29, 30). Since previous isotope-labeling experiments had confirmed Trp as the precursor of the indolic acid moiety (14), this conversion should involve an unusual carbon side chain rearrangement on Trp. In a manner similar to the ThiH-catalyzed reaction, NosL may activate the substrate Trp by a radical-forming mechanism, and further carbon side chain reconstruction eventually result in the key intermediate 3-methylindolyl derivative (2) (Figure 4). To validate the above proposal, we first inactivated nosL by in-frame deletion. As anticipated, the resulting mutant strain SL4005 completely lost the ability to produce NOS (Figure 6, V). Next, feeding of the chemically synthesized 3-methylindole-2-carboxylic acid (8) into the fermentation culture of SL4005 partially restored NOS production (Figure 6, VI), strongly supporting the central role of NosL in the formation of the indolic acid moiety. Since the conversion of Trp into 2 is novel and requires multiple reactions that, to our knowledge, are not similar to those catalyzed by known enzymes, the identification of NosL in NOS biosynthesis presents an excellent opportunity to investigate its novel chemistry and enzymology.
Figure 4.
Proposed biosynthetic pathway of the indolic acid moiety of NOS.
Figure 5.
Biosynthetic hypothesis for conversion of the thiopeptide framework into NOS. Color coding indicates the posttranslational modifications on the peptide precursor NosM (orange), attachment of the indolic acid moiety to the macrocyclic core (purple), and tailoring steps including hydroxylations on Glu43 at the γ-position and central pyridine domain at the C-5 position, and final cleavage of an acrylic acid to afford NOS (blue).
NosN shares high sequence similarity to various putative methyltransferases with a radical S-AdoMet-dependent manner, including Tlm-Orf11 (32% identity) in tallysomycin biosynthesis (31). To determine its involvement and corresponding function in NOS biosynthesis, we inactivated nosN by in-frame deletion. As shown in Figure 6, the resulting mutant strain SL4006 indeed lost the ability of NOS production (Figure 6, VII) and, intriguingly, accumulated a new compound with ultraviolet (UV) absorptions at λmax 254 nm, 280 nm and 350 nm, characteristic for all thiopeptides. HR-ESI-MS analysis showed its positive [M + Na]+ ion at m/z = 1270.1528 (± 0.003), establishing the molecular formula as C53H45N13O12S6Na (1270.1257 calculated). Although it is too labile for complete structural elucidation by NMR spectroscopic methods, this compound, upon extensive tandem MS spectrometry analysis, was deduced to be the intermediate 4 (Figure 7), a side ring-opened NOS analog that contains the 3-methylindolyl moiety appended to the –SH group of Cys45 on the thiopeptide framework via a single thioester linkage. These findings not only support that NosN functions as a methyltransferase and acts on the 3-methylindolyl group at the C-4 position for furnishing the 3,4-dimethylindolyl moiety, but also shed light into the timing of the individual steps for indole side ring biosynthesis. As shown in Figure 5, the intermediate 4 can be resulted from the thioesterification between 2 and Cys45 of 1 (pathway I). NosK, containing a putative acyltransferase domain at the C-terminus, may contribute to this enzymatic process. The NosN-catalyzed methylation may occur on 4 to yield the intermediate 5, and the subsequent hydroxylation of the resulting C-4 methyl group could give the intermediate 6. The priority of the S- linkage formation to the methyl group hydroxylation in NOS biosynthesis is consistent with previous studies that 8 and 3,4-dimethylindole-2-carboxylic acid 9, but not 4-(hydroxylmethyl)-3-methylindole-2-carboxylic acid, can be activated as the adenylate derivatives and incorporated into NOS (14). However, it does not exclude another possibility of the substrate specificity of NosN. It may act on 2 (instead of the 3-dimethylindolyl moiety-attached compound 4) to generate the 3,4-dimethylindolyl derivative 3 (Figure 4), which could then be appended directly onto the thiopeptide framework via the NosK-catalyzed thioesterification to give the intermediate 6 (pathway II, Figure 5). Finally, 6 may undergo an intramolecular esterification, presumably catalyzed by NosI (belonging to an AMP-dependent CoA ligase family), to form the O- linkage between the indole moiety and Glu43 on the macrocyclic core, close the 19-membered side ring system and give the product 7 for further tailoring (Figure 5).
Figure 7.
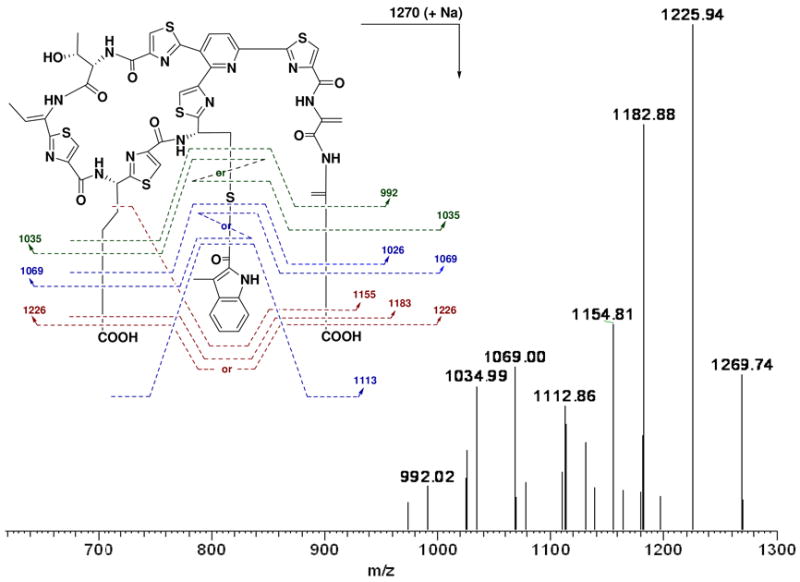
MS/MS spectrum of the [M + Na]+ ion at m/z = 1269.74 corresponding to the NOS analog 4. The obtained fragments clearly indicate a permutation and combination pattern (labeled in color) for cleavage on the free carboxylic group(s) of the C-terminal dehydroalanine acid and/or side chain of Glu43, and/or on the linkage of the 3-methylindole moiety (2) to Cys45 of the macrocyclic core.
Tailoring to NOS and regulation
To eventually produce NOS, post-modifications on 7 are postulated to proceed with a set of tailoring enzymes encoded by nosA, nosB and nosC as outlined in Figure 5. NosB and NocC, belonging to a P450 hydroxylase family, may oxidatively act on Glu43 at the γ-position and the central pyridine domain at the C-5 position, giving the hydroxylation pattern characteristic to all known e series thiopeptides. The final formation of NOS could be resulted from the removal of an acrylic acid group to form the C-terminal amide moiety, and NosA serves as a candidate to catalyze this reaction. This proposed mechanism is distinct from that in TSR biosynthesis, which employs an asparagine synthetase-like protein TsrC for the terminal amidotransfer (25).
The only putative regulatory gene identified within the nos gene cluster was nosP (Figure 2). It encodes a protein that contains an effector domain of a response regulator at the N-terminus and a transcriptional activator domain at the C-terminus. Inactivation of nosP by in-frame deletion completely abolished NOS production (Figure 6, VIII), validating that NosP functions as a pathway specific regulator to positively control the NOS biosynthesis in S. actuosus.
In summary, we have uncovered the biosynthetic machinery of nosiheptide by cloning, sequencing and characterizing the nos gene cluster in S. actuosus ATCC 25421. Our findings extend the current paradigm for thiopeptide biosynthesis into the e series members with distinct features. Thus, consistent with the similarities in structures to the b or c series thiopeptides whose biosynthetic pathways were established, NOS biosynthesis shares the common strategy for forming the characteristic thiopeptide macrocyclic core that features a ribosomally synthesized precursor peptide and conserved posttranslational modifications. However, the NOS biosynthetic machinery is unique, apparently proceeding via a different route for tailoring the intermediate into the final product by installing an unusual indole side ring system and two regiospecific hydroxyl groups, characteristics of e series thiopeptides. Although Trp serves as a common precursor for the indolic acid moiety of NOS and the quinaldic acid moiety of TSR, the distinct pathways for biosynthesis and attachment onto the thiopeptide macrocyclic cores lead to major differences in the side ring systems. The conversion of Trp into the key 3-methylindolyl moiety (2) requires an unusual carbon side chain rearrangement, in which the S-AdoMet-dependent protein NosL may play a central role with a radical-initiated mechanism. NOS biosynthesis involves many novel enzymes and reactions, and the characterization of these will contribute new chemistry and enzymology to thiopeptide biosynthesis. Since all e series members contain these structurally similar features (e.g. the indole side ring system and the hydroxylation pattern) (1), their biosyntheses may employ similar approaches in affording the NOS-like scaffold, but with additional oxidoreduction(s) and glycosylation(s) for tailoring into the other individual members, such as nocathiacins. Finally, the success in identification of the novel NOS analog 4 by gene inactivation continuously inspires attempts to apply this knowledge to metabolic engineering for structural derivation in the pharmaceutical development of thiopeptide-like antibiotics.
Methods
Bacterial Strains, Plasmids, and Reagents
Bacterial strains and plasmids used in this study are summarized in Table S1. Biochemicals and media were purchased from Sinopharm Chemical Reagent Co., Ltd. and Oxoid Ltd. unless otherwise stated. Restriction enzymes were purchased from TaKaRa Biotechnology Co., Ltd.
DNA Isolation, Manipulation, and Sequencing
DNA isolation and manipulation in Escherichia coli and Streptomyces were performed according to standard methods (32, 33). PCR amplifications were carried out in an Eppendorf AG 22331 thermal cycler using LA Taq DNA polymerase (TaKaRa). Primer synthesis and DNA sequencing were performed at Shanghai Invitrogen Biotech Co., Ltd. and the Chinese National Human Genome Center. For cloning the cyclodehydratase gene from S. actuosus, PCR amplification was carried out by using the degenerate primers ThioF (5′-TAC GAG ACC TCC AAY GGN TGY GCN-3′) and ThioR (5′-GTG GCC RAA SGT CAT NGG-3′) described previously (24). Gene inactivations and complementation in this study are described in the Supporting Information.
Construction of the Genomic Library
The genomic library of S. actuosus ATCC 25421 was constructed in pOJ446. Briefly, the total DNA of S. actuosus was randomly sheared to approximately 30–40 kb fragments (judged by electrophoresis at 4 °C), which were blunt-ended by the treatment with the End-Repair Enzyme Mix (EPICENTRE Biotechnologies). The resultant DNA fragments were then ligated into the EcoRV site of pOJ446. DNA packaging and transfection were carried out by using the Packagene Lambda DNA Packaging System (Promega) and E. coli LE392 according to the manufacture's instructions.
Sequence Analysis
ORFs were deduced from the sequence with the assistance of FramePlot 4.0beta program (http://nocardia.nih.go.jp/fp4/). The corresponding deduced proteins were compared with other known proteins in the databases by available BLAST methods (http://www.ncbi.nlm.nih.gov/blast/). Amino acid sequence alignments were performed with CLUSTALW from BIOLOGYWORKBENCH 3.2 software (http://workbench.sdsc.edu).
Fermentation, Isolation and Analysis of NOS
S. actuosus wild-type and recombinant strains were grown on ISP2 agar plates at 30 °C for sporulation. To produce NOS, 50 μl of S. actuosus or recombinant strains spores were inoculated into a 250-ml flask containing 50 ml of seed medium (sucrose 2.0%, corn steep liquor 3.0%, peptone 0.5% and CaCO3 0.5%) and incubated at 28 °C and 250 rpm for 24 hrs. Ten ml of seed culture was then transferred into 100 ml of fermentation medium (Pharmamedia cotton meal 1.0%, NaCl 0.3%, glucose 3.0%, 2 × trace elements solution 0.5% (32) and CaCO3 0.3%, pH 7.0) in a 500-ml flask and incubated at 28 °C for 4 days. For chemical feeding of the synthetic 3-methylindole-2-caboxylic acid (8) to restore NOS production in the nosL mutant SL4005, 8 was added into the fermentation culture of SL4005 on day 2 at a final concentration of 20 μg ml-1. For NOS isolation, each 100 ml of culture broth was centrifuged for 10 minutes at 6,000 rpm. After removing the supernatant, the precipitate was soaked with 100 ml of ethyl acetate for 6 hrs. The extract was then concentrated in vacuum and resolved in 1 ml of methanol for further analysis. HPLC analysis was carried out on an Agilent ZORBAX SB-C18 column (4.6×250 mm, part number 880975-902, S/N USCL024998), which was equilibrated with 85% solvent A (H2O containing 0.1% TFA) and 15% B (CH3CN containing 0.1% TFA), and developed with the following program: 0 to 3 min, constant 85% A/15% B; 3 to 6 min, a linear gradient from 85% A/15% B to 60% A/40% B; 6 to 12 min, constant 60% A/40% B; 12 to 19 min, a linear gradient from 60% A/40% B to 45% A/55% B; 19 to 22 min, a linear gradient from 45% A/55% B to 15% A/85% B; 22 to 28 min, constant 15% A/85% B; and 28 to 32 min, a linear gradient from 15% A/85% B to 85% A/15% B. This was carried out at a flow rate of 1 ml min-1 and UV detection at 254 nm using an Agilent 1100 HPLC system (Agilent Technologies). The identity of NOS was confirmed by HPLC-ESI-MS analysis (for HPLC, Agilent 1100 used; and for ESI-MS, Thermo Fisher LTQ Fleet used) performed under the same conditions. NOS showed an [M+H]+ ion at m/z 1222.36 consistent with the molecular formula C51H43N13O12S6 (1221.15 calculated).
Tandem Mass Spectrometry Analysis of the NOS analog 4
Production and isolation of 4 was carried out according to the method described previously (24). Tandem Mass Spectrometry analysis of 4 was performed on a LCQ Fleet Ion Trap MSn tandem mass spectrometer (Thermo Fisher Scientific) (Figure 7).
Synthesis of 3-Methylindole-2-Carboxylic Acid (8)
The synthesis of 8 was achieved by following the previously described method (34). 1HNMR assignments in CDCl3 at 300 MHz are as follows (s means singlet, d means doublet, and t means triplet): δ8.85 (s, 1H), δ7.69 (d, 1H, J = 7.8 Hz), δ7.40 (d, 1H, J = 9.3 Hz), δ7.35 (t, 1H, J = 8.7 Hz), δ7.16 (t, 1H, J = 7.2 Hz), δ2.67 (s, 3H). ESI-MS m/z = 174.0 (M - H+).
Supplementary Material
Acknowledgments
We thank H.G. Floss, Department of Chemistry, University of Washington, USA, for providing of S. actuosus ATCC 25421 and his pioneer works on NOS biosynthesis. This work was supported in part by grants from NIH of U.S (CA094426 to B.S.), the National Natural Science Foundation of China (30525001, 90713012 and 20832009), the Ministry of Science and Technology of China (2006AA022185), the Chinese Academy of Science (KJCX2-YW-H08), and the Science and Technology Commission of Shanghai Municipality (09QH1402700) (all to W.L.).
Footnotes
Supporting Information Available: This material is available free of charge via the internet at http://pubs.acs.org.
Nucleotide Sequence Accession Number: The sequence reported in this paper has been deposited in GenBank under the accession number FJ438820.
References
- 1.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 2.Leet JE, Li W, Ax HA, Matson JA, Huang S, Huang R, Cantone JL, Drexler D, Dalterio RA, Lam KS. Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. II. Isolation, characterization, and structure determination. J Antibiot (Tokyo) 2003;56:232–242. doi: 10.7164/antibiotics.56.232. [DOI] [PubMed] [Google Scholar]
- 3.Li W, Leet JE, Ax HA, Gustavson DR, Brown DM, Turner L, Brown K, Clark J, Yang H, Fung-Tomc J, Lam KS. Nocathiacins, new thiazolyl peptide antibiotics from Nocardia sp. I. Taxonomy, fermentation and biological activities. J Antibiot (Tokyo) 2003;56:226–231. doi: 10.7164/antibiotics.56.226. [DOI] [PubMed] [Google Scholar]
- 4.Naidu BN, Sorenson ME, Zhang Y, Kim OK, Matiskella JD, Wichtowski JA, Connolly TP, Li W, Lam KS, Bronson JJ, Pucci MJ, Clark JM, Ueda Y. Nocathiacin I analogues: synthesis, in vitro and in vivo biological activity of novel semi-synthetic thiazolyl peptide antibiotics. Bioorg Med Chem Lett. 2004;14:5573–5577. doi: 10.1016/j.bmcl.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Huang S, Liu X, Leet JE, Cantone JL, Lam KS. N-Demethylation of nocathiacin I via photo-oxidation. Bioorg Med Chem Lett. 2008;18:4051–4053. doi: 10.1016/j.bmcl.2008.05.112. [DOI] [PubMed] [Google Scholar]
- 6.Naidu BN, Sorenson ME, Matiskella JD, Li W, Sausker JB, Zhang Y, Connolly TP, Lam KS, Bronson JJ, Pucci MJ, Yang H, Ueda Y. Synthesis and antibacterial activity of nocathiacin I analogues. Bioorg Med Chem Lett. 2006;16:3545–3549. doi: 10.1016/j.bmcl.2006.03.079. [DOI] [PubMed] [Google Scholar]
- 7.Naidu BN, Sorenson ME, Bronson JJ, Pucci MJ, Clark JM, Ueda Y. Synthesis, in vitro, and in vivo antibacterial activity of nocathiacin I thiol-Michael adducts. Bioorg Med Chem Lett. 2005;15:2069–2072. doi: 10.1016/j.bmcl.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 8.Pucci MJ, Bronson JJ, Barrett JF, DenBleyker KL, Discotto LF, Fung-Tomc JC, Ueda Y. Antimicrobial evaluation of nocathiacins, a thiazole peptide class of antibiotics. Antimicrob Agents Chemother. 2004;48:3697–3701. doi: 10.1128/AAC.48.10.3697-3701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benazet F, Cartier JR. Effect of nosiheptide as a feed additive in chicks on the quantity, duration, prevalence of excretion, and resistance to antibacterial agents of Salmonella typhimurium; on the proportion of Escherichia coli and other coliforms resistant to antibacterial agents; and on their degree and spectrum of resistance. Poult Sci. 1980;59:1405–1415. doi: 10.3382/ps.0591405. [DOI] [PubMed] [Google Scholar]
- 10.Benazet F, Cartier M, Florent J, Godard C, Jung G, Lunel J, Mancy D, Pascal C, Renaut J, Tarridec P, Theilleux J, Tissier R, Dubost M, Ninet L. Nosiheptide, a sulfur-containing peptide antibiotic isolated from Streptomyces actuosus 40037. Experientia. 1980;36:414–416. doi: 10.1007/BF01975121. [DOI] [PubMed] [Google Scholar]
- 11.Pascard C, Ducruix A, Lunel J, Prange T. Highly modified cysteine-containing antibiotics. Chemical structure and configuration of nosiheptide. J Am Chem Soc. 1977;99:6418–6423. doi: 10.1021/ja00461a039. [DOI] [PubMed] [Google Scholar]
- 12.Prange T, Ducruix A, Pascard C, Lunel J. Structure of nosiheptide, a polythiazole-containing antibiotic. Nature. 1977;265:189–190. doi: 10.1038/265189a0. [DOI] [PubMed] [Google Scholar]
- 13.Mocek U, Chen LC, Keller PJ, Houck DR, Beale JM, Floss HG. 1H and 13C NMR assignments of the thiopeptide antibiotic nosiheptide. J Antibiot (Tokyo) 1989;42:1643–1648. doi: 10.7164/antibiotics.42.1643. [DOI] [PubMed] [Google Scholar]
- 14.Mocek U, Knaggs AR, Tsuchiya R, Nguyen T, Beale JM, Floss HG. Biosynthesis of the modified peptide antibiotic nosiheptide in Streptomyces actuosus. J Am Chem Soc. 1993;115:7557–7568. [Google Scholar]
- 15.Mocek U, Zeng Z, O'Hagan D, Zhou P, Fan LDG, Beale JM, Floss HG. Biosynthesis of the modified peptide antibiotic thiostrepton in Streptomyces azureus and Streptomyces laurentii. J Am Chem Soc. 1993;115:7992–8001. [Google Scholar]
- 16.Priestley ND, Smith TM, Shipley PR, Floss HG. Studies on the biosynthesis of thiostrepton: 4-(1-hydroxyethyl)quinoline-2-carboxylate as a free intermediate on the pathway to the quinaldic acid moiety. Bioorg Med Chem. 1996;4:1135–1147. doi: 10.1016/0968-0896(96)00126-5. [DOI] [PubMed] [Google Scholar]
- 17.Smith TM, Priestley ND, Knaggs AR, Nguyen T, Floss HG. 3,4-Dimethylindole-2-carboxylate and 4-(1-hydroxyethyl)quinoline-2-carboxylate activating enzymes from the nosiheptide and thiostrepton producers, Streptomyces actuosus and Streptomyces laurentii. J Chem Soc Chem Commun. 1993:1612–1614. [Google Scholar]
- 18.Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 19.Lee SM, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 22.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 23.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci U S A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 26.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 27.Walsh CT, Nolan EM. Morphing peptide backbones into heterocycles. Proc Natl Acad Sci U S A. 2008;105:5655–5656. doi: 10.1073/pnas.0802300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson DN. Antibiotics and the inhibition of ribosome function In Protein Synthesis and Ribosome Structure. Wiley-VCH; Weinheim: 2004. [Google Scholar]
- 29.Martinez-Gomez NC, Robers M, Downs DM. Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily. J Biol Chem. 2004;279:40505–40510. doi: 10.1074/jbc.M403985200. [DOI] [PubMed] [Google Scholar]
- 30.Kriek M, Martins F, Challand MR, Croft A, Roach PL. Thiamine biosynthesis in Escherichia coli: identification of intermediate and by-product derived from tyrosine. Angew Chem Int Ed. 2007;46:9223–9226. doi: 10.1002/anie.200702554. [DOI] [PubMed] [Google Scholar]
- 31.Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, Coughlin JM, Shen B. The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465-94 ATCC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics. Mol Biosyst. 2007;3:60–74. doi: 10.1039/b615284h. [DOI] [PubMed] [Google Scholar]
- 32.Kieser T, Bibb M, Butter M, Chater KF, Hopwood DA. Practical Streptomyces Genetics. The John Innes Foundation; Norwich: 2001. [Google Scholar]
- 33.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- 34.Jones CD. Preparation of 2-substituted indole sulfonamides and subsequent conversion to indole-2-carboxylic acids, indole-2-carbonitriles, and 2-acylindole. J Org Chem. 1972;37:3624–3625. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.