Abstract
A gene, qid74, of mycoparasitic filamentous fungus Trichoderma harzianum and its allies encodes a cell wall protein that is induced by replacing glucose in the culture medium with chitin (simulated mycoparasitism conditions). Because no trace of this gene can be detected in related species such as Gibberella fujikuroi and Saccharomyces cerevisiae, the qid74 gene appears to have arisen de novo within the genus Trichoderma. Qid74 protein, 687 residues long, is now seen as highly conserved tandem repeats of the 59-residue-long unit. This unit itself, however, may have arisen as tandem repeats of the shorter 13-residue-long basic unit. Within the genus Trichoderma, the amino acid sequence of Qid74 proteins has been conserved in toto. The most striking is the fact that Qid74 shares 25.3% sequence identity with the carboxyl-terminal half of the 1,572-residue-long BR3 protein of the dipteran insect Chironomus tentans. BR3 protein is secreted by the salivary gland of each aquatic larva of Chironomus to form a tube to house itself. Furthermore, the consensus sequence derived from these 59-residue-long repeating units resembles those of epidermal growth factor-like domains found in divergent invertebrate and vertebrate proteins as to the positions of critical cysteine residues and homology of residues surrounding these cysteines.
When a distinctive body part is shared between certain organisms belonging to different phyla or classes, convergent evolution traditionally has been invoked as an explanation, implying unrelated mutations affecting different sets of gene loci resulted in the manifestation of that distinct body part. An oft invoked example of convergent evolution has been compound eyes of insects versus singular eyes of vertebrates. This proved to be a wrong example, however, because development of all metazoan eyes recently has been shown to be under the control of the same regulatory gene that encodes Pax-6 protein (1–4). In hindsight, this was no surprise, because nearly all the extant animal phyla emerged almost simultaneously during the early Cambrian period some 530 million years ago, and early Cambrian arthropods already included trilobites with compound eyes.
Similarly, in the case of proteins encoded by individual genes, the same or similar domains (e.g., epidermal growth factor-like domains) often are shared by divergent proteins belonging to different families. Such apparent convergence of a protein part, however, has been interpreted as consequences of domain exchanges between unrelated proteins because of exon shufflings (5, 6). Nevertheless, how, by exon shufflings, the same domain can be propagated to be included in a number of unrelated proteins has never been made clear. If a particular protein donated the domain in question to the other by an exon shuffling, that donor itself had to lose that domain, in return receiving an unrelated domain.
In the present paper, we present Qid74 protein of a mycoparasitic filamentous fungus Trichoderma harzianum and show its sequence conservation within the genus Trichoderma. Then we show a surprising identity shared between this fungal Qid74 and the previously determined sequence of BR3 protein of the dipteran insect Chironomus, thus raising the possibility of convergent evolution at the protein level.
MATERIALS AND METHODS
Fungal Materials and Growth Conditions.
Trichoderma harzianum CECT 2413, Trichoderma viride CECT 2423, Gliocladium virens CECT 2460 (reclassified as Trichoderma virens), Trichoderma longibrachiatum CECT 2606, Trichoderma koningii CECT 2412, Trichoderma reesei CECT2414, Gibberella fujikuroi CECT 2152, and Saccharomyces cerevisiae CECT1329 were obtained from the Colección Española de Cultivos Tipo, Burjasot, Valencia, Spain; T. harzianum IMI 206040 was obtained from the Imperial Mycological Institute, Kew, U.K. Hypocrea jecorina was a generous gift from C. Kubicek, University of Technology, Vienna. Glucose/agar/potato medium was used for the maintenance of cultures (7).
cDNA Library and Differential Screening.
A cDNA library was constructed from mRNA isolated from 33-h cultures as described (8). The library was constructed in the λgt11 SfiI-NotI vector by using the RiboClone kit (both from Promega). The library was screened by using first-strand cDNA synthesized on poly(A)+ mRNA populations as differential probes; they were obtained from control cultures (incubated with 10% glucose) and cultures induced with 1.5% chitin. The cDNA generated was labeled with [α-32P]dCTP (Amersham) by using a random primer-labeling kit from Boehringer Mannheim. The library was plated, transferred to nitrocellulose filters (Millipore), and hybridized as described (9). Plaques hybridizing only with cDNA from the induced culture were isolated and purified, and the cDNA was isolated according to the kit manufacturer’s instructions (Promega). The cDNA inserts were excised by digesting the recombinant phage DNA with EcoRI and NotI. These cDNAs were subcloned into pBluescript SK(+) (Stratagene), obtaining the recombinant plasmid pQID74.
PCR.
Total DNA from 4-day-old cultures was prepared according to Murray and Thomson (10) and was used as a template for PCR by using, as a sense primer, the oligonucleotide Q74up, 5′-ATGTTGCTTAAGCAGGTCCTTGTGGC-3′ (nucleotides 1–28 of the ORF) and, as antisense primer, the oligonucleotide Q74lo1, 5′-ATGTTCTTGGCACACGCACTTGTTGTTCTG-3′ or Q74lo2, 5′-TCAAGGATAGTTCATCTTACAAGTCTTCTT-3′ (2074–2100). To clone the 5′ region inverse PCR was carried out with the oligonucleotides IQ74up, 5′-GCAAGAACATCGGCCAAGTCTTTGAT-3′ (1832–1860) as sense primer and IQ74lo, 5′-GCCACAAGGACCTGCTTAAGCAACAT-3′ as antisense primer. The amplification protocol used was as follows: 95°C, 1 min; 60°C, 30 sec; 72°C, 1–4 min. This standard cycle was repeated 35 times. PCR experiments were always carried out with pFU polymerase from Stratagene.
DNA Sequencing and Data Analysis.
The recombinant plasmid (pQID74) was subjected to restriction analysis. Using the information obtained, constructions and nested deletions made from the original full cDNA were subjected to double-stranded DNA sequencing reactions by using the dideoxy chain termination method of Sanger et al. (11) and following the Sequenase protocol (United States Biochemical). Both strands were fully sequenced from overlapping constructs. The clones amplified by PCR were subcloned in pBluescript SK(+) (Stratagene), and nucleotide sequence was determined with an A.L.F. automatic DNA sequencer from Pharmacia. Sequence data and predictions of protein structure were analyzed by using programs developed by the University of Wisconsin Genetics Computer Group (12). Other computer analyses were performed by using programs developed by Altschul et al. (13).
RESULTS
Isolation of Genomic and cDNA Clones of Qid74 from T. harzianum.
As already noted, Qid74 protein is a cell wall component of T. harzianum, which is induced by replacing glucose in the culture medium with chitin as a sole carbon source (simulated mycoparasitism conditions). This permitted us to use a differential hybridization approach to single out cDNA copies of chitin-induced mRNAs. A λgt11-based cDNA expression library was prepared. Upon screening of approximately 2 × 104 plaques, 20 clones were isolated, all of them showing a strong hybridization signal with the chitin-induced probe and no signal with the noninduced probe. One of the clones that showed the strongest hybridization signal was λQID74, which was further purified. DNA from λQID74 was isolated and sequenced as described in Materials and Methods. From the cDNA sequence, oligonucleotides were designed that allowed the isolation of the genomic DNA corresponding to the ORF region and the promoter region from T. harzianum and from other Trichoderma species. To isolate these genomic clones, a PCR approach was followed (see Material and Methods). Under the experimental conditions described, a band of ca. 2.1 kbp was initially amplified from T. harzianum 2413 chromosomal DNA. This band was then subcloned and sequenced.
Sequence Analysis of qid74.
The complete nucleotide sequence for the QID74 cDNA and the genomic clone was determined. The most relevant characteristic of the sequence was its internal organization into tandemly repeating units. The genomic sequence of qid74 was identical to that of the QID74 cDNA, indicating a lack of introns in this gene. The full cDNA clone is 2,546 bp in length, with an ORF of 2,115 nucleotides. Such an ORF encoded a protein of 704 aa residues with a predicted molecular mass of 77,872 Da. The amino acid composition of the mature protein coded by qid74 was rather unusual, with a high proportion of lysine (13.5%), cysteine (12.8%), and glycine (9.6%). The codon usage of the T. harzianum qid74 gene resembles that of other Trichoderma genes that in the induced state demonstrated a very high transcription rate (14). Out of the 61 codons, 6 were not used at all whereas 24 were used 10 or more times. For example, of 76 glycine residues, 47 were encoded by the same triplet. With regard to serine, 40% of the residues also were encoded by the same triplet, in spite of the fact that there are six different serine codons available. The amino acid sequence of Qid74 also contains a potential signal peptide of 17 aa, which has an α-helical structure and is highly hydrophobic (15). This structure has been observed in secreted proteins such as cell wall proteins.
Internal Organization of qid74.
A dot matrix analysis of the nucleotide or amino acid sequence of the qid74 gene indicated a number of regions that contained internally repetitious sequences. Multiple alignment of randomly generated fragments was carried out to identify the nature of apparent repetitiousness. Such an analysis showed internal repetitiousness in qid74 at two hierarchical levels: (hereafter) major and minor repeating units.
Major repeating units.
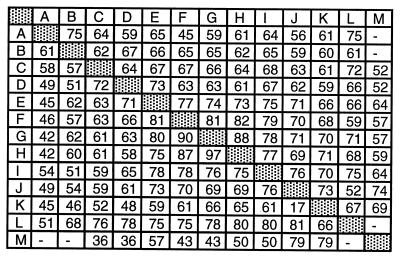
The protein consisted of five incomplete and nine complete copies of the 59-residue-long unit. Thus, almost the entire ORF of the gene (from the residue position 46–680) can be divided into these tandemly repeated copies, as shown in Fig. 1. The pattern of sequence conservation among 59-residue-long units follows a pattern characteristic of other proteins with internal repeats (16). The units from the central region of the protein are more similar to one another as compared with those from the terminal regions. In fact, although the overall sequence identity was 60% among the repeats, units G and H demonstrated 97% identity with G, 75% with E, 61% with C, and only 41% with repeat A (Fig. 2). The alignment of the sequence repeats indicates that the shorter amino acid sequences of units A, B, C, L, and M may be because of small deletions that had occurred in these copies.
Figure 1.
Alignment of the deduced Qid74 amino acid sequence of the 14 repetitions that are 59 aa in length. The sequence of the repetitive regions have been aligned to achieve maximal homology by using the pileup and bestfit programs of the Wisconsin Gene Computer Group package.
Figure 2.
Relationship among the sequence of repetitive regions of the qid74 gene. The lower middle section of matrix lists the average similarity for all pairwise comparisons between the individual repeats at amino acid levels. The upper middle section shows the average of mutations in the third position.
The observed tendency of copies in the central position remaining more conserved than those located in the periphery has been observed previously in all repeating DNA sequences, regardless of whether they are coding or noncoding (17). Yet, in the case of our Qid74, degeneracy most often affected the third base of codons, and even missense degeneracies were of the kind that led to conservative amino acid substitutions, e.g., glutamate to aspartate. Thus, degeneracy of repeating units in our case was under the strict surveillance by natural selection.
Cysteine is a highly reactive residue and has the tendency to form a disulfide bridge with another cysteine, thereby profoundly affecting a secondary structure of a protein. Not surprisingly, cys residues were conserved in fixed positions in all repeating units. Furthermore, they appeared in two motifs: CXC tripeptidic motif, where X is Val, Ser, or Ala and CXXXC pentapeptidic motifs, which shall be discussed later (Figs. 1 and 2). Motifs related to the above also have been found in other cell wall proteins such as the ice nucleation protein (18).
Minor repeating units.
In all repeating sequences, the larger repeating unit of today tends to be made of multiple copies of the smaller and more ancient repeating unit (19). This apparently was the case with Qid74. Observing Fig. 1, we note that the tridecapeptide KSKTCSCPGNQYW (in single-letter code) occupied the carboxyl-terminal position of the K copy. Furthermore, its apparent derivatives were found not only in the carboxyl half but also in the amino-terminal half of the C copy. In fact, two or more derivatives, at times truncated, of the above-noted tridecapeptide were found in nearly every copy of the 59-residue-long unit. Thus it would appear that the ultimate ancestor of the qid74 gene was tandem repeats of the 39-bp-long unit.
The qid74 Gene in Other Trichoderma Species.
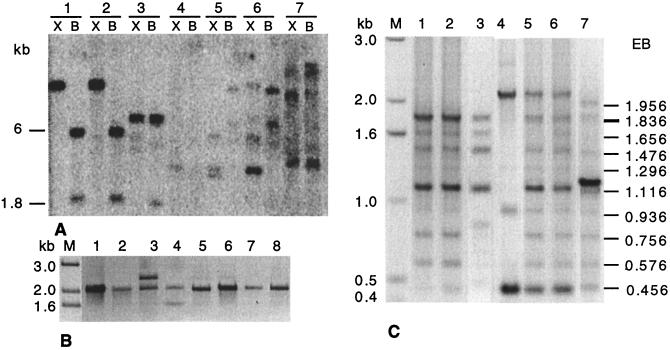
Southern blot and PCR analyses showed that the qid74 gene of the same ORF length was present in all the Trichoderma species tested. To determine a degree of sequence conservation among qid74 genes of various Trichoderma species, the study on a variation of tandemly repeating sequences was done. This analysis revealed almost the identical pattern shown by all the Trichoderma species tested. To verify the above finding based on PCR analyses by actual sequencing, a 1.1-kb band (Fig. 3C) from T. reesei and T. koningii and two 600-nt-long segments representing 5′ as well as 3′ ends of T. virens were sequenced. In addition, one 1-kb-long fragment corresponding to the upstream 5′ sequence of qid74 was cloned and sequenced on the strain T. harzianum 2413 and a species, T. reesei. To do so, inverse PCR using IQ74up and λQ74lo as sense and antisense primers was carried out.
Figure 3.
Detection of the qid74 gene in different species. (A) Southern blot analysis of genomic DNA, digested with BamHI (B) and XmaI (X). (B) PCR amplification of the ORF of qid74, using the 5′ (Q74up) and 3′ (Q74lo2) ends as sense and antisense primers (see Materials and Methods). (C) Variation of tandemly repeating sequences analysis. A single-copy sequence oligonucleotide (Q74up) was used as sense primer and, as antisense, a oligonucleotide (Q74lo1) was used that would hybridize with different melting temperatures to a number of nonidentical repeats. In this condition, 10 bands, which include from the ORF of repeat C to the ORF of repeat L, were expected in T. harzianum 2413. Hypocrea jecorina showed the same pattern of T. reesei (data not shown). Lanes: M, molecular weight marker; 1, T. harzianum 2413; 2, T. viride; 3, T. virens; 4, T. harzianum IMI; 5, T. longibrachiatum; 6, T. koningii; 7, T. reesei; 8, Hypocrea jecorina; T, bands expected in T. harzianum 2413; EB, expected bands in T. harzianum 2413.
As expected from PCR analyses, the ORFs of T. reesei, T. virens, and T. koningii indeed were identical with ORF of T. harzianum 2413. As to the 5′ upstream noncoding region, by contrast, 7 base substitutions per 600 bases separated T. harzianum 2413 from T. reesei. The promoter region apparently tolerated more base substitutions than the coding region. In addition to the conserved qid74 noted above, T. reesei was endowed with its redundant copy, which was rendered functionally defunct by an insertion of a 180-nt-long segment at the positions between 677 and 678. This pseudogene maintained only 65% identity with the real qid74 gene.
Although the qid74 gene apparently was conserved in toto within the genus Trichoderma, no trace of this gene was found in related fungi such as G. fugikuroi and S. cerevisiae (data not shown).
Qid74 and Homologous Proteins Found in Other Organisms.
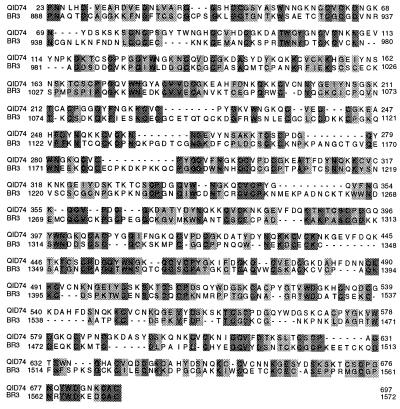
The blast program (13) has been used to perform a database search to find a protein or proteins that are reasonably homologous with Qid74. The maximal identity (p = 4.3e-44) with Qid74 was found in BR3 protein precursor of the dipteran insect Chironomus tentans (accession number Q03376) and its homolog of C. thumi and C. pallividitatus. BR3 protein is secreted in an enormous amount by salivary glands of an aquatic larvae of the midge Chironomus and it is used to weave a tube in which an aquatic larva houses itself. In preparations of Chironomus polytene salivary gland chromosomes, a gene encoding BR3 is readily recognized as an enlarged RNA puff (Balbiani Ring) reflecting its enormous transcriptional activity, and BR3 protein too is encoded by a coding sequence that is made of tandem repeats (6, 19). As shown in the alignments of Fig. 4, Qid74 and the carboxyl-terminal half of BR3 demonstrated 25.3% sequence identity.
Figure 4.
Alignment between Qid74 (704 residues) and BR3 (1,700 residues). The alignment was carried out using the program sim, which is available in the Molecular Biology Server Expasy.
Other proteins with lesser homology included an unknown protein of Caenorhabditis elegans (p = 1.1e-16), fibrillins, Notch, and others, all of which contained repeating epidermal growth factor-like domains. Another reasonably homologous protein was an oocyst wall protein of Chryptospodidium parvum (accession number A48456). The homology score between Qid74 and this oocyst wall protein was p = 8.9e-13. All these proteins noted above had a few characteristics in common. As indicated by the presence of a signal peptide at each protein’s N-terminal region, these proteins were meant to be secreted by the cell, they were made of repeating domains rich in cysteine residues, and they were also rich in charged residues presenting similar hydropathic profiles.
DISCUSSION
A major problem is encountered whenever an attempt is made to classify various strains of deuteromycetal fungi, because there is an inherent uncertainty in deciding whether they represented different strains belonging to the same species or they are, in fact, different species belonging to the same genus (20). In the case of the genus Trichoderma, however, species statuses of those presently studied were confirmed by internal transcribed spacers sequence comparisons (C. Kubicek, personal communication). Accordingly, only the very strict surveillance by natural selection can account for the presently observed virtual in toto conservation of the qid74 gene among various Trichoderma species. However, because we found no trace of the qid74 gene in either G. fugikuroi or S. cerevisiae, it would appear that this gene arose de novo in the ancestor of the teleomorphic genus Hypocrea whence the presently studied anamorphic genus Trichoderma sprung (21).
Let us now turn our attention to BR3 protein of the dipteran insect Chironomus with which Qid74 protein shared 25.3% sequence identity. Of the insect order Diptera, Chironomus together with mosquitoes belong to the suborder Nematocera, whereas the familiar Drosophila together with house flies belong to the other suborder Cyclorrhapha. Whereas larvae of the first suborder tend to lead aquatic lives, those of the second suborder are terrestrial. Indeed, no gene homologous to that encoding BR3 was found within the Drosophila genome (6, 19). Thus it would appear that BR3 gene also arose de novo either within the suborder Nematocera or much later within the genus Chironomus.
There can be no propinquity of descents between two genes separately created de novo in two different kingdoms: the kingdom Fungi and the kingdom of Animalia. Therefore, one is forced to conclude that the observed homology between the qid74 gene of mycoparasitic filamentous fungi Trichoderma species and BR3 gene of dipteran flies Chironomus species constituted a priori the case of convergent evolution of genes. The next question is: what was the reason behind this convergence? The ancestor of each gene created de novo has to be a noncoding DNA sequence that abounds in every eukaryotic genome. Of those noncoding sequences, only repetitious ones retain the potential to become a coding sequence, simply because of the 64 base triplets, 3 in the universal coding system are chain terminators. It follows that nonrepetitious noncoding sequences are too full of chain terminators in all three reading frames to supply a long enough ORF. In contrast, repetitious ones are either full of chain terminators or free of chain terminators. Accordingly, the later type of repetitious noncoding sequences are the only source of genes to be created de novo (22).
Repetitious sequences of different kinds abound in intergenic spacer regions of all eukaryotic genomes, and they invade even intragenic introns. At the simplest, they are repeats of base oligomers, which are 2–4 bases in length. Yet, even those simplest ones appeared to have yielded certain genes in the past. Involucrins are the major components that, by cross-linking with each other via Gln–Lys bonds, develop a physically as well as chemically resistant envelope that forms within each keratinocyte of vertebrate skin epidermis. The ultimate ancestry of these 410- to 630-residue-long Gln-rich proteins with the hexadecapeptidic periodicity was traced back to one of the simple CAG trimeric repeats (23). The more likely ultimate ancestors of Qid74 as well as BR3, however, are repeats derived from degenerate copies of a small 5S rRNA gene as well as from various tRNA genes, some of which came to be known as short interspersed nuclear elements and long interspersed nuclear elements (24). Because some of the repeating units are still equipped with the internal promoter for RNA polymerase III, multiple copies incorporated in various parts of the genome are still transcribed and, via reverse transcription, their DNA copies spread still further into the genome. One of those made of the 39-base-long repeating unit capable of encoding CXC tripeptide could have served as the ultimate ancestor of Qid74 of the fungal Trichoderma as well as BR3 of the diptera Chironomus. If so, the propinquity of descents was not between the genes but between the similar noncoding repeats whence two genes sprung quite independently of each other. In the end, the apparent convergence presently observed also was not really a good example of convergent evolution.
Acknowledgments
We thank Lorenzo Marquez and Beatriz Cubero for valuable discussion, Gabriel Gutierrez for helpful computer analysis, and Ramón Espejo for style correction. M.R. was the recipient of a postgraduate fellowship from the Ministerio de Educación y Ciencia. This work was supported by Grants BIO94-0289 and TS3-CT92-0140 from the Comision Interministerial de Ciencia y Tecnologia and the European Commission, respectively.
Footnotes
References
- 1.Glasser T, Watson D S, Mass R L. Science. 1990;250:823–827. [Google Scholar]
- 2.Hogan, B. L. M., Hirst, E. M. A., Horisburgh, G. A. & Hetherington, C. M. (1988) Development (Cambridge, U.K.) 103, Suppl., 115–119. [DOI] [PubMed]
- 3.Matsuo T, Osmi-Yamashita N, Noji S, Ohuchi H, Koyama E, Myokai F, Matsumoto N, Taniguichi S, Doi H, Iseki S, Ninoniya Y, Fujiware M, Watanabe T, Eto K. Nat Genet. 1993;3:299–304. doi: 10.1038/ng0493-299. [DOI] [PubMed] [Google Scholar]
- 4.Loosli F, Kmita-Cunisse M, Gehring W. Proc Natl Acad Sci USA. 1996;93:2658–2663. doi: 10.1073/pnas.93.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolittle R F. Trends Biochem Sci. 1985;10:233–337. [Google Scholar]
- 6.Paulsson G, Lendahl U, Galli J, Ericsson C, Wieslander L. J Mol Biol. 1990;211:331–349. doi: 10.1016/0022-2836(90)90355-P. [DOI] [PubMed] [Google Scholar]
- 7.Dawson C, Belloch C, García-López M D, Uruburu F. Catalogue of Strains, Spanish Type Culture Collection. Valencia, Spain: Univ. of Valencia; 1990. [Google Scholar]
- 8.Lora J M, Cruz J, Benítez T, Llobell A, Pintor-Toro J A. Mol Gen Genet. 1994;242:461–466. doi: 10.1007/BF00281797. [DOI] [PubMed] [Google Scholar]
- 9.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Murray M G, Thomson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanger F, Nicklen S, Coulson R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Vasseur V, Van Montagu M, Goldman G H. Microbiology. 1995;141:767–774. doi: 10.1099/13500872-141-4-767. [DOI] [PubMed] [Google Scholar]
- 15.von Heijne G. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 16.Patthy L. Cell. 1985;41:657–663. doi: 10.1016/s0092-8674(85)80046-5. [DOI] [PubMed] [Google Scholar]
- 17.Smith G P. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 18.Green R L, Warren G J. Nature (London) 1985;317:645–648. [Google Scholar]
- 19.Höög C, Daneholt B, Wieslander L. J Mol Biol. 1988;200:655–664. doi: 10.1016/0022-2836(88)90478-0. [DOI] [PubMed] [Google Scholar]
- 20.Samuels G J. Mycol Rev. 1996;100:923–935. [Google Scholar]
- 21.Kuhls K, Lieckfeldt E, Samuels G J, Kovacs W, Meyer W, Petrini O, Gams W, Borner T, Kubicek C P. Proc Natl Acad Sci USA. 1996;93:7755–7760. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno S. Proc Natl Acad Sci USA. 1996;93:8475–8478. doi: 10.1073/pnas.93.16.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green H, Djian P. Mol Biol Evol. 1992;9:977–1017. doi: 10.1093/oxfordjournals.molbev.a040775. [DOI] [PubMed] [Google Scholar]
- 24.Hwu H R, Roberts J W, Davidson E H, Britten R J. Proc Natl Acad Sci USA. 1986;83:3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]