Abstract
Purpose
The small leucine-rich repeat proteins (SLRPs) are involved in organizing the collagen fibrils of the sclera and vitreous. The shape of the eyeball is determined by the sclera and vitreous, so defects in SLRP family members may contribute to myopia. The purpose of this study was to test whether mutations in the two members of the class III SLRPs, opticin (OPTC) and dermatan sulfate proteoglycan 3 (EPYC), are responsible for high myopia.
Methods
DNA was prepared from venous leukocytes of 93 patients with high myopia (refraction of spherical equivalent ≤-6.00D) and 96 controls (refraction of spherical equivalent between -0.50D and +1.00D). The coding regions and adjacent intronic sequences of OPTC and EPYC were amplified by the polymerase chain reaction (PCR), and the products were then analyzed by cycle sequencing. The detected variations were further evaluated in normal controls and available family members by a heteroduplex-single strand conformation polymorphism (heteroduplex-SSCP) analysis or sequencing.
Results
Two substitutions in OPTC, including c.491G>T and c.803T>C, were identified. The c.491G>T mutation (p.Arg164Leu), a novel heterozygous variation, was detected in one of the 93 patients but in none of the 96 controls. The c.803T>C mutation (p.Pro267Leu), a known polymorphism, was detected in 22 of the 93 patients and in 15 of 48 controls. No variation was observed in EPYC.
Conclusions
Only one novel variation in OPTC was detected in a Chinese patient with high myopia. Our results imply that OPTC and EPYC are unlikely to play a major role in high myopia.
Introduction
Myopia is the most common human vision impairment all over the world, especially in Chinese people living in urban/suburban areas [1-6]. High myopia, an extreme form of myopia with refraction less than -6.0D (diopters), is a common cause of irreversible blindness due to its association with retinal degeneration and detachment, glaucoma, and other abnormalities. The exact molecular basis of high myopia and the genes that cause a predisposition to this disorder are still unknown, although abundant evidence has demonstrated that genetic factors play an important role in the development of high myopia [7].
The shape of the eye is largely determined by the sclera and the vitreous that are composed of abundant extracellular matrix (ECM) [8]. Abnormalities due to mutations in several ECM proteins or proteoglycans, such as COL2A1 and COL11A1 in Stickler syndrome and fibrillin-1 in Marfan syndrome, have been associated with syndromic high myopia [9,10]. The small leucine-rich repeat proteins (SLRPs), members of the ECM family, play important roles in organizing collagen fibrils [11]. Experiments in SLRP knockout mice have shown that a wide array of defects are caused by abnormal collagen fibrillogenesis, resulting in a longer eye axial length and high myopia [12]. Therefore, it would be reasonable to determine whether there are mutations in these SLRPs in human subjects with high myopia.
The OPTC gene (gene ID: 26254; OMIM 605127), located in chromosome 1q32.1, is a member of the class III SLRPs [13], and the OPTC protein is widely distributed in ocular tissue, including the cornea, iris, trabecular meshwork, ciliary body, retina, and optic nerve. OPTC is specifically expressed in the vitreous humor where the protein plays a key role in maintaining the gel structure of the vitreous humor [14-16]. The dermatan sulfate proteoglycan 3 gene (DSPG3/EPYC; GENE ID: 1833; OMIM 601657), located in chromosome 12q21 where MYP3 (OMIM 603221) mapped [17,18], is another member of the class III SLRPs [19]. The EPYC protein is predominantly expressed in cartilage [20,21] and is important for fibrillogenesis through the regulation of collagen fibrils.
In this study, the coding exons and the adjacent non-coding regions of OPTC and EPYC were analyzed in 93 Chinese patients with high myopia.
Methods
Subjects
The procedure for collecting the subjects and obtaining informed consent was the same as previously described [22]. The study was carried out following the tenets of the Declaration of Helsinki and was approved by the Institute Review Board of the Zhongshan Ophthalmic Center. Informed consent was obtained from the participating subjects before the study. Ophthalmological examinations were performed by ophthalmologists of the Zhongshan Ophthalmic Center (Q.Z. and X.G.). The recruitment of subjects as same as we described previously [23].
Mutation analysis
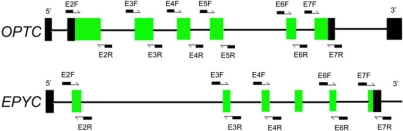
Genomic DNA was prepared from peripheral leukocytes as described previously [24]. Table 1 lists the primers used to amplify the coding exons and adjacent introns of OPTC (NCBI human genome build 36.3, NC_000001.10 for genomic DNA, NM_ 014359.3 for mRNA and NP_ 055174.1 for protein) and EPYC (NCBI human genome build 36.2, NC_000012.11 for genomic DNA, NM_004950.4 for mRNA and NP_004941.2 for protein; Figure 1). The nucleotide sequences of OPTC and EPYC were determined by cycle sequencing. The sequencing results from patients as well as the OPTC (NT_004487.19) and EPYC (NT_029419.12) consensus sequences were compared to identify variations. The variations were confirmed by bi-directional sequencing. The description of the variations followed the nomenclature recommended by the Human Genomic Variation Society (HGVS). The mutation we detected was further evaluated in 96 normal controls by single-strand conformational polymorphism (SSCP) using an extra pair of primers (Table 1), using the method we described previously [24]. The SIFT program (Sorting Intolerant From Tolerant) was used to predict whether an amino acid substitution was likely to affect the protein function [26].
Table 1. Primers used for the amplification and sequencing of OPTC and EPYC.
| Gene | Exon | Direction | Primer sequence | Size of PCR product (bp) | Annealing temperature (°C) |
|---|---|---|---|---|---|
|
OPTC |
E2 |
F |
5'-CGAGCCATTCCCACAACACT-3' |
439 |
64 |
| |
|
R |
5'-GCTGGAGACACCCGACTTT-3' |
||
| |
E3 |
F |
5'-TTCCCTCCTCCTCCTCCACA-3' |
387 |
62 |
| |
|
R |
5'-GTAGCCCTGACCCTCCAT-3' |
||
| |
E4 |
F |
5'-TGTACATGGTGCCCGTCAGT-3' |
425 |
62 |
| |
|
R |
5'-GAGAGCCAACAGGAAAGAGC-3' |
||
| |
E5 |
F |
5'-AGGTGGAGAGCTGGTTAGAA-3' |
433 |
64 |
| |
|
R |
5'-GGTGGTGGTGGAGGTGAGTG-3' |
||
| |
E6 |
F |
5'-GCGCATGGCTGAGGCTAAG-3' |
346 |
62 |
| |
|
R |
5'-CAATCAAGGAGACCCCACAG-3' |
||
| |
E7 |
F |
5'-TTGGCCCTAGATGCTTCAGT-3' |
425 |
62 |
| |
|
R |
5'-ATTAATGCCTGGGCTAAAGA-3' |
||
| |
SSCP |
F |
5'-CTGCGTGTGCCTCGGTTCCTCT-3' |
180 |
64 |
| |
|
R |
5'-ATGCCTGCCCCCACTCCTCTTT-3' |
||
|
EPYC |
E2 |
F |
5'-ACATCTGGCAGCAATGGTC-3' |
538 |
60 |
| |
|
R |
5'-TCCGGTATTAACCTCCAAAAG-3' |
||
| |
E3 |
F |
5'-CAAGCAGGATGGTCAAACTAT-3' |
451 |
62 |
| |
|
R |
5'-TGCTTCAAACCTAGCCTATTC-3' |
||
| |
E4 |
F |
5'-CTTGGTTAGTGGGAGACTCTA-3' |
371 |
62 |
| |
|
R |
5'-AACCAGGCTTTGGCATTCAA-3' |
||
| |
E5-1* |
F |
5'-ATTAACTGTGCATTGCTAAAT-3' |
473 |
44-68 |
| |
|
R |
5'-AAGCCTGGGTGACAGAGTGA-3' |
||
| |
E5-2* |
F |
5'-TAATGCCTGTATCTTTGACTAT-3' |
630 |
44-68 |
| |
|
R |
5'-AATTTATTTTCCCTACACGA-3' |
||
| |
E5-3* |
F |
5'-GTTGCTTTATGATGGATGAATG-3' |
979 |
44-46 |
| |
|
R |
5'-CCTAGGAAATATGGCGAAACC-3' |
||
| |
E5-4 [25] |
F |
5'-CGTGGTGTACCAATTTCTTTGCT-3' |
579 |
44-68 |
| |
|
R |
5'-ACTCAAGCCTGGGTGACAGAGTG-3' |
||
| |
E6 |
F |
5'-AAAACTAAGCTGCCTCAACT-3' |
353 |
60 |
| |
|
R |
5'-AACATGCCATTACAGACAAA-3' |
||
| |
E7 |
F |
5'-TTACCCCCATTGCTTAGAAA-3' |
433 |
60 |
| R | 5'-GAACACAATCTCAAAGTATCA-3' |
The asterisk indicates that in order to amplify exon 5 of EPYC, the PCR conditions, including DMSO concentration, and the enzymes used (rTaq, LATaq, hotMaster, and AmpiTaq Gold), were varied.
Figure 1.
The exon-intron structures of OPTC and EYPC. The black rectangles represent the untranslated region and the green rectangles represent the coding region.
Results
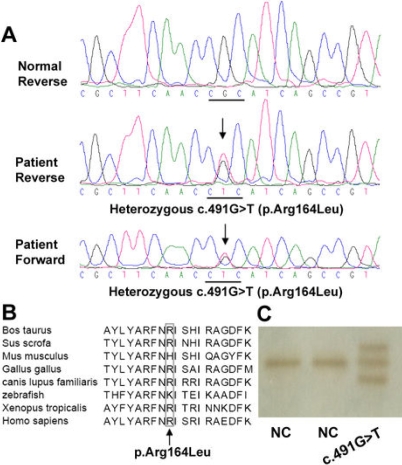
After sequencing the OPTC gene from 93 high myopia patients, two missense variations, c.491G>T and c.803T>C, were identified. The c.491G>T mutation is a novel heterozygous variation in which an arginine (a charged amino acid) would be replaced by a leucine (an aliphatic, hydrophobic amino acid) in the encoded protein (p.Arg164Leu; Figure 2A), with a residue weight of -2 (Blosum 62). The arginine at position 164 of OPTC is conserved among the six orthologs from Homo sapiens (NP_055174), Bos Taurus (NP_991339), Canis lupus familiaris (NP_001003056), Gallus gallus (NP_989804), Sus scrofa (NP_999173), and Xenopus tropicalis (NP_001016499; Figure 2B), but is not conserved in two orthologs from Mus Musculus (NP_473417) and Zebrafish (NP_001003583). The SIFT analysis predicted that the substitution at position 164 from arginine to leucine would to be tolerated with a score of 0.17. Substitutions with a score less than 0.05 are predicted to affect protein function [26]. This novel variant was identified only in a single patient, but not in the 96 controls, by SSCP analysis (Figure 2C). However, the c.803T>C mutation (p.Pro267Leu), a known polymorphism, was detected in 22 of the 93 patients with high myopia (18 heterozygous and 4 homozygous) as well as in 15 of the 48 controls (13 heterozygous and 2 homozygous).
Figure 2.
The novel variation identified in OPTC. A: The c.491G>T variant (indicated by an arrow) was sequenced in two directions and was compared to the wild type sequences. B: The missense variation p.Arg164Leu (c.491G>T; indicated by an arrow) changed a highly conserved residue from arginine to leucine. C: Variant c.491G>T (p.Arg164Leu) was not observed in the 96 normal controls by the heteroduplex-SSCP analysis.
No variation was identified in exons 2, 3, 4, 6, and 7 of EPYC. Our attempts to amplify part or all of exon 5 of EPYC failed, possibly due to special DNA structures in the region, such as those caused by a high AT content.
Discussion
OPTC is widely expressed in ocular tissue and was screened as a candidate gene in patients with high myopia, glaucoma, and age-related macular degeneration [27-30]. Recently, mutations in OPTC were suggested to be genetic risk factors underlying the pathogenesis of high myopia, where five variations were identified in five of 125 Caucasian patients with high myopia [30]. Co-segregation analyses were carried out in four families, where three variants (p.Arg325Trp, p.Arg330His, and p.Gly329Ser) were also present in relatives with normal refraction or moderate myopia. The p.Thr177Arg variation was only detected in the proband but not in the affected offspring. Incomplete penetrance was suggested as an explanation for the presence of the variation in unaffected family members [30].
In this study, only one novel variation (c.491G>T, p.Arg164Leu) was detected in one of the 93 patients with high myopia. The protein information analysis using the SIFT program indicated that the p.Arg164Leu variation was unlikely to affect protein function, and the effect of this variation was similar to the effects seen in Mus musculus (p.Arg164His) or Zebrafish (p.Arg164Lys). However, it is impossible to confirm or deny the association of this variation with high myopia based on the current evidence, especially because of our limited understanding of complex diseases. In addition to the novel variant, a known c.803T>C variation was detected in 22 of the 93 patients as well as in 15 of the 48 controls. The genotype and allele frequencies of this variation were similar between the patients and controls (χ2 test, p=0.6175, and p=0.3529 separately). This variation was previously detected in ten Caucasian patients with high myopia as well as in 27 normal controls [30], suggesting that this variation is more likely to be a polymorphism.
EPYC (DSPG3), located in the mapping interval of MYP3, has been suggested as a candidate gene for high myopia [17]. An analysis of four single nucleotide polymorphisms (SNPs: rs1135866, rs1920748, rs1920751, and rs1920752) in EPYC was performed for 120 patients with high myopia and 137 controls, but no association was detected [31]. However, an evaluation of the coding and adjacent intronic sequences of EPYC has not reported in patients with high myopia thus far. In this study, no variation was detected in the sequences of five of the six coding and adjacent intronic regions of EPYC in 93 Chinese patients with high myopia. The PCR amplification of exon 5 of EPYC was unsuccessful despite the use of several primer pairs and different amplification conditions. An extremely high AT content in this region may be the cause of the difficulty with the PCR amplification.
In summary, our results suggest that both OPTC and EPYC are unlikely to play a major role in high myopia. Additional studies will be necessary to clarify these results.
Acknowledgments
The authors thank all of the patients and controls for their participation. This study was supported in part by the National 863 Plan of China (Z19-01-04-02 to Q.Z.) and the National Science Fund for Distinguished Young Scholars (30725044 to Q.Z.).
References
- 1.Feldkamper M, Schaeffel F. Interactions of genes and environment in myopia. Dev Ophthalmol. 2003;37:34–49. doi: 10.1159/000072037. [DOI] [PubMed] [Google Scholar]
- 2.Saw SM. A synopsis of the prevalence rates and environmental risk factors for myopia. Clin Exp Optom. 2003;86:289–94. doi: 10.1111/j.1444-0938.2003.tb03124.x. [DOI] [PubMed] [Google Scholar]
- 3.Sperduto RD, Seigel D, Roberts J, Rowland M. Prevalence of myopia in the United States. Arch Ophthalmol. 1983;101:405–7. doi: 10.1001/archopht.1983.01040010405011. [DOI] [PubMed] [Google Scholar]
- 4.Choo V. A look at slowing progression of myopia. Lancet. 2003;361:1622–3. doi: 10.1016/S0140-6736(03)13315-6. [DOI] [PubMed] [Google Scholar]
- 5.He M, Zeng J, Liu Y, Xu J, Pokharel GP, Ellwein LB. Refractive error and visual impairment in urban children in southern china. Invest Ophthalmol Vis Sci. 2004;45:793–9. doi: 10.1167/iovs.03-1051. [DOI] [PubMed] [Google Scholar]
- 6.Fredrick DR. Myopia. BMJ. 2002;324:1195–9. doi: 10.1136/bmj.324.7347.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young TL. Molecular genetics of human myopia: an update. Optom Vis Sci. 2009;86:E8–22. doi: 10.1097/OPX.0b013e3181940655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hausman RE. Ocular extracellular matrices in development. Prog Retin Eye Res. 2007;26:162–88. doi: 10.1016/j.preteyeres.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Ammash NM, Sundt TM, Connolly HM. Marfan syndrome-diagnosis and management. Curr Probl Cardiol. 2008;33:7–39. doi: 10.1016/j.cpcardiol.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Bayazit YA, Yilmaz M. An overview of hereditary hearing loss. ORL J Otorhinolaryngol Relat Spec. 2006;68:57–63. doi: 10.1159/000091090. [DOI] [PubMed] [Google Scholar]
- 11.Iozzo RV. The biology of the small leucine-rich proteoglycans. Functional network of interactive proteins. J Biol Chem. 1999;274:18843–6. doi: 10.1074/jbc.274.27.18843. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti S, Paul J, Roberts L, Chervoneva I, Oldberg A, Birk DE. Ocular and scleral alterations in gene-targeted lumican-fibromodulin double-null mice. Invest Ophthalmol Vis Sci. 2003;44:2422–32. doi: 10.1167/iovs.02-0783. [DOI] [PubMed] [Google Scholar]
- 13.Le Goff MM, Bishop PN. Focus on molecules: opticin. Exp Eye Res. 2007;85:303–4. doi: 10.1016/j.exer.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Friedman JS, Ducharme R, Raymond V, Walter MA. Isolation of a novel iris-specific and leucine-rich repeat protein (oculoglycan) using differential selection. Invest Ophthalmol Vis Sci. 2000;41:2059–66. [PubMed] [Google Scholar]
- 15.Hindson VJ, Gallagher JT, Halfter W, Bishop PN. Opticin binds to heparan and chondroitin sulfate proteoglycans. Invest Ophthalmol Vis Sci. 2005;46:4417–23. doi: 10.1167/iovs.05-0883. [DOI] [PubMed] [Google Scholar]
- 16.Ramesh S, Bonshek RE, Bishop PN. Immunolocalisation of opticin in the human eye. Br J Ophthalmol. 2004;88:697–702. doi: 10.1136/bjo.2003.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurnberg G, Jacobi FK, Broghammer M, Becker C, Blin N, Nurnberg P, Stephani U, Pusch CM. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–38. [PubMed] [Google Scholar]
- 19.Zhou W, Shirabe K, Kuwada JY. Molecular cloning and expression of two small leucine-rich proteoglycan (SLRP) genes, dspg3l and optcl, in zebrafish. Gene Expr Patterns. 2006;6:482–8. doi: 10.1016/j.modgep.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Deere M, Dieguez JL, Yoon SJ, Hewett-Emmett D, de la Chapelle A, Hecht JT. Genomic characterization of human DSPG3. Genome Res. 1999;9:449–56. [PMC free article] [PubMed] [Google Scholar]
- 21.Kurita K, Shinomura T, Ujita M, Zako M, Kida D, Iwata H, Kimata K. Occurrence of PG-Lb, a leucine-rich small chondroitin/dermatan sulphate proteoglycan in mammalian epiphyseal cartilage: molecular cloning and sequence analysis of the mouse cDNA. Biochem J. 1996;318:909–14. doi: 10.1042/bj3180909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Li S, Xiao X, Jia X, Guo X. The 208delG mutation in FSCN2 does not associate with retinal degeneration in Chinese individuals. Invest Ophthalmol Vis Sci. 2007;48:530–3. doi: 10.1167/iovs.06-0669. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Xiao X, Li S, Xing Y, Guo X, Zhang Q. Evaluation of EGR1 as a candidate gene for high myopia. Mol Vis. 2008;14:1309–12. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Guo X, Jia X, Xiao X, Guo L, Li S. Penetrance of Leber hereditary optic neuropathy in Chinese individuals with mitochondrial DNA 11778 mutation. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2001;18:441–3. [PubMed] [Google Scholar]
- 25.Aldave AJ, Sonmez B, Bourla N, Schultz G, Papp JC, Salem AK, Rayner SA, Yellore VS. Autosomal dominant cornea plana is not associated with pathogenic mutations in DCN, DSPG3, FOXC1, KERA, LUM, or PITX2. Ophthalmic Genet. 2007;28:57–67. doi: 10.1080/13816810701351321. [DOI] [PubMed] [Google Scholar]
- 26.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman JS, Faucher M, Hiscott P, Biron VL, Malenfant M, Turcotte P, Raymond V, Walter MA. Protein localization in the human eye and genetic screen of opticin. Hum Mol Genet. 2002;11:1333–42. doi: 10.1093/hmg/11.11.1333. [DOI] [PubMed] [Google Scholar]
- 28.Acharya M, Mookherjee S, Bhattacharjee A, Thakur SK, Bandyopadhyay AK, Sen A, Chakrabarti S, Ray K. Evaluation of the OPTC gene in primary open angle glaucoma: functional significance of a silent change. BMC Mol Biol. 2007;8:21. doi: 10.1186/1471-2199-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Basavaraj MG, Gupta SK, Qamar I, Ali AM, Bajaj V, Ramesh TK, Prakash DR, Shetty JS, Dorairaj SK. Role of CYP1B1, MYOC, OPTN, and OPTC genes in adult-onset primary open-angle glaucoma: predominance of CYP1B1 mutations in Indian patients. Mol Vis. 2007;13:667–76. [PMC free article] [PubMed] [Google Scholar]
- 30.Majava M, Bishop PN, Hagg P, Scott PG, Rice A, Inglehearn C, Hammond CJ, Spector TD, Ala-Kokko L, Mannikko M. Novel mutations in the small leucine-rich repeat protein/proteoglycan (SLRP) genes in high myopia. Hum Mutat. 2007;28:336–44. doi: 10.1002/humu.20444. [DOI] [PubMed] [Google Scholar]
- 31.Wang IJ, Chiang TH, Shih YF, Hsiao CK, Lu SC, Hou YC, Lin LL. The association of single nucleotide polymorphisms in the 5'-regulatory region of the lumican gene with susceptibility to high myopia in Taiwan. Mol Vis. 2006;12:852–7. [PubMed] [Google Scholar]