Abstract
BACKGROUND
The ooplasm plays a central role in forming the paternal pronucleus, and subsequently in regulating the expression of paternally inherited chromosomes. Previous studies in mice have revealed genetic differences in paternal genome processing by ooplasm of different genotypes. Ooplasm donation coupled to intracytoplasmic sperm injection (ICSI) has been used in human assisted reproductive technology (ART). This procedure exposes the developing paternal pronucleus to ‘foreign’ ooplasm, which may direct aberrant epigenetic processing. The potential effects of the foreign ooplasm on epigenetic information in the paternal pronucleus are unknown; however, some human progeny from ooplasm donation procedures display abnormalities.
METHODS
In this study, we employed inter-genotype ooplasm transfer followed by ICSI using two mouse strains, C57BL/6 and DBA/2, to explore the influence of foreign ooplasm on paternal pronucleus function. In order to assay for effects on the paternal genome without masking effects of the maternal genome, we examined ooplasm effects in diploid androgenones, which are produced by pronuclear transfer to contain exclusively two paternal sets of chromosomes, in combination with ICSI.
RESULTS
There was no significant effect of intra-strain ooplasm transfer among androgenones made with either C57BL/6 or DBA/2 oocytes. There was a significant negative effect on androgenone blastocyst development with inter-genotype transfer (10% volume) of DBA/2 ooplasm to C57BL/6 oocytes (P < 0.05). The reciprocal inter-genotype ooplasm transfer had no significant effect.
CONCLUSIONS
Thus, inter-genotype ooplasm transfer in conjunction with ICSI can alter the function of the paternal genome. However, the effect of foreign ooplasm is restricted to a negative effect, with no evidence of a positive effect. This study provides important new information about the possible consequences of ooplasm donation in human ART.
Keywords: ooplasm donation, androgenone, parthenogenone, IVF, ICSI
Introduction
The oocyte cytoplasm plays a fundamental role in creating the embryonic genome from two gamete genomes, and preparing it for transcriptional activation and execution of the developmental program. A key part of this process involves epigenetic modifications during the first few hours after fertilization, when parental genomes are still separated in the distinct pronuclei (PN). These modifications appear to modify or extend epigenetic modifications (e.g. imprinting) established in the gametes, and both kinds of modification (pre- and post-fertilization) are subject to genetic variation (Latham, 1999).
Strain-specific differences exist between C57BL/6 and DBA/2 (denoted as B6 and D2 hereafter) mouse strains with respect to embryonic genome modification. Nuclear transfer (NT) studies have revealed that the oocytes of these two strains differentially modify paternal PN after fertilization, so that androgenetic embryos (two paternal PN, achieved by pronuclear transfer) display very different developmental abilities (Latham and Solter, 1991; Latham, 1994; Latham and Sapienza, 1998). Genetic studies have revealed specific loci (Egm1 and Egm2) that control these processes. Other studies with other mouse strains have revealed other strain-specific effects on genome function in the early embryo (Latham and Solter, 1991; Sapienza et al., 1992; Latham, 1994, 1999; Latham and Sapienza, 1998; Pardo-Manuel De Villena et al., 2000; Pickard et al., 2001). These results are consistent with a model in which the different strains of mice differentially modify the paternal genome after fertilization, perhaps to make it compatible with the oocyte epigenotype (Latham, 1999). Disruption of this process could result in altered epigenetic modification of the paternal pronucleus, and an incompatibility with the epigenetic state of the maternal pronucleus, and subsequent epigenetic defects in development.
Studies in mice have reported that the ooplasm genetic strain of origin can exert subtle effects on blastomere fragmentation and that the ooplasm likely interacts with the paternal genome (Han et al., 2008). Other studies have revealed subtle negative effects of ooplasm donation on development in early stage fertilized embryos; however, long-term effects on growth were not manifested (Cheng et al., 2009). These results collectively indicate that the paternal genome may be especially susceptible to epigenetic alteration by foreign ooplasm.
Novel microsurgical procedures in the human, including ooplasm transfer, have been developed and applied for clinical treatment of infertility (Cohen et al., 1997, 1998; Zhang et al., 1999; Barritt et al., 2001a, b; Hawes et al., 2002). One of the innovations involves the augmentation of recipient ooplasm of patients who have repeatedly failed to achieve pregnancy, with ooplasm from a supposedly ‘healthy’ donor oocyte (Cohen et al., 1997, 1998; Barritt et al., 2001a, b). With this procedure, the donor ooplasm is microinjected simultaneously with the sperm (cytoplasm transfer-coupled intracytoplasmic sperm injection, CT-ICSI). A more extreme version of the procedure involves the total ooplasm replacement via germinal vesicle transfer or NT (Zhang et al., 1999). The basic premise for these procedures is that the ooplasm obtained from supposedly ‘healthy’ donor oocytes can augment and restore functionally important properties to the ‘unhealthy’ oocyte of the patient. However, neither the safety nor efficacy of this method has been adequately investigated. Mitochondrial heteroplasmy in the blood of children conceived using ooplasm donation has recently been described (Barritt et al., 2001a), and potential problems associated with heteroplasmy have been discussed (Hawes et al., 2002; Bredenoord et al., 2008). Additionally, both procedures (GV transfer and CT-ICSI) result in an altered environment for pronucleus formation.
Given the results from mouse studies that reveal genetically diverse effects of ooplasm on the epigenetic state of the paternal genome (Latham and Solter, 1991; Latham et al., 1995; Latham and Sapienza, 1998; Latham, 1999), the changes introduced by ooplasm transfer in the pronucleus-forming environment establish the potential for altered epigenetic modification of the paternal genome. If epigenetic compatibility with the maternal genome is disrupted by such changes, this could subsequently affect the epigenetic control of development.
Because ooplasm donation to address human infertility exposes the emerging paternal genome to foreign ooplasm which is presumably genetically different from the recipient oocyte, we wished to test whether foreign ooplasm can exert effects specifically on the paternal genome. To do this, we used ooplasm transfer and ICSI together to manipulate androgenetic embryos and then monitored their phenotypes in order to observe any adverse effects of ooplasm transfer on the paternal genomes. We found that ooplasm transfer could exert negative effects on androgenone development, but could not positively affect developmental potential. These results indicate that the advantages of ooplasm donation may be minimal, although undesirable negative epigenetic effects may arise.
Materials and Methods
Animals and reagents
Male (C57BL/6 × DBA/2)F1 mice (B6D2) at 10–14 weeks of age (National Cancer Institute, Frederick, MD, USA) were used for spermatozoa collection. Inbred strains B6 (Harlan Aprague-Dawley, Indianapolis, IN, USA) and D2 (The Jackson Laboratory, Bar Harbor, ME, USA) female mice at 8–12 weeks of age were used as a source of MII stage oocytes. Studies adhered to procedures consistent with the National Research Council Guide for the Care and Use of Laboratory Animals. All chemicals used in this study were purchased from Sigma-Aldrich Company (St Louis, MO, USA) unless otherwise noted.
Oocyte collection, embryo construction and embryo culture
Adult B6 and D2 females were superovulated by sequential administration of 5 IU equine chorionic gonadotrophin (Calbiochem, San Diego, CA, USA) and human chorionic gonadotrophin (hCG) 48 h apart. Oocytes were isolated at ∼14–15 h post-hCG injection, and cumulus cells were removed by hyaluronidase treatment in HEPES-buffered M2 medium as described (Chung et al., 2002).
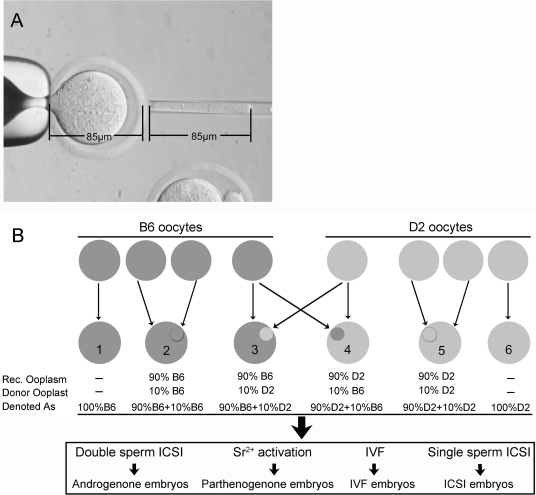
Cytoplasm transfer was performed with MII oocytes in M2 medium containing 5 µg/ml cytochalasin B and 0.2 µg/ml demicolcine. Approximately 10% of the total oocyte volume was removed from each recipient oocyte. This permitted an equivalent volume of donor ooplasm to be transferred. A 10% volume was chosen to mimic procedures employed in the human (Cohen et al., 1998; Barritt et al., 2001a). The inner diameter of the pipette was 10 µm. Calculating the cylindrical volume of the transfer pipette [πr2h = (4/3)πR3/10, ‘R’ and ‘r’ refer to the radius of the oocyte and pipette, respectively], the length of space in the pipette (h) to approximate 10% oocyte volume should be 85 µm, or approximately one oocyte diameter, which was used to estimate the ooplasm volume transferred (Fig. 1A). Ooplasts approximating 10% ooplasm volume were transferred from a donor MII oocyte to the perivitelline space of the recipient oocyte. After recovery in CZB medium (Han et al., 2008) for 1 h, the donor ooplast and recipient oocyte were fused by electrofusion using an electro cell manipulator (BTX, San Diego, CA, USA), with a single DC pulse of 0.9 kV/cm DC for 10 µs in 0.27 M mannitol medium supplemented with 0.1 mM MgSO4, 0.05 mM CaCl2 and 0.3% BSA. One hour after fusion, the fused oocytes were selected and cultured for further use.
Figure 1.
Illustration of ooplast transfers.
(A) Volume of 10% ooplasm of total oocyte. The volume of donated ooplasm can be controlled. The inner radius of pipette used for ooplasm transfer is 5 µm (r), the average radius of a mouse oocyte is 42.5 µm (R). Then the formula is: πr2h = (4/3)πR3/10, thus h = 85 µm, which equals the oocyte diameter. (B) C57BL/6 (B6) components are dark-gray and DBA/2 (D2) components are light-gray. Resulting combinations from cytoplasm transfers and denotations are below. Six kinds of cytoplasm combinations were produced with B6 and D2 oocytes. The oocytes without ooplasmic transfer [denoted as 100% B6 (number 1) or 100% D2 (number 6)] and those with intra-strain ooplasmic transfer [denoted as 90% B6+10% B6 (number 2) or 90% D2+10% D2 (number 5)] were used as controls. The oocytes with inter-strain ooplasmic transfer [denoted as 90% B6+10% D2 (number 3) or 90% D2+10% B6 (number 4)] were used to investigate the effects of ‘foreign’ ooplast on early embryo development. Each kind of construct was subjected to make androgenone, parthenogenone, IVF or ICSI embryos.
Six kinds of oocytes were produced with B6 and D2 oocytes in this study: oocytes without ooplasm transfer (denoted as 100% B6 or 100% D2), oocytes with intra-strain ooplasm transfer (denoted as 90% B6+10% B6 or 90% D2+10% D2) served as controls and oocytes with inter-strain ooplasm transfer (denoted as 90% B6+10% D2 or 90% D2+10% B6) used to investigate the effects of ‘foreign’ ooplasm on early embryo development. Each kind of oocyte was used to make androgenones, parthenogenones, and control embryos by In vitro fertilization (IVF) or ICSI techniques (Fig. 1B).
ICSI to obtain fertilized control embryos was performed as described (Yoshida and Perry, 2007) using a piezopipet micromanipulator (PMM Primer Tech, Ltd, Ibaraki-ke, Japan). ICSI was also performed to construct diploid androgenones with or without additional ooplast fusion. The spindle–chromosome complex was first removed, and then two sperm heads were injected. Sperm were injected into the oocyte near the point of ooplasm transfer, as marked by the notch in the zona remaining after ooplast fusion. IVF embryos were constructed as described (Janssenswillen et al., 1997). Oocyte activation for parthenogenetic control embryos was achieved by 6 h incubation in calcium-free CZB-Glutamine supplemented with 10 mM SrCl2 and 5 µg/ml cytochalasin B (Janssenswillen et al., 1997; Gao et al., 2004).
Embryos were examined for pronucleus formation at 6 h after sperm injection or oocyte activation, and those with PN were cultured in the CZB medium for 2 days followed by 3 days of culture in Whitten's medium supplemented with 100 µM EDTA as described (Latham and Solter, 1991; Latham, 1994) and as in previous studies (Abramczuk et al., 1977; Chatot et al., 1989). Embryos were checked at 48, 72, 96 and 120 h post-hCG injection to record the 2-cell, 4-cell, morula and blastocyst development, respectively.
Embryo staining, cell number counting and diameter measurement
After 120 h post-hCG injection, the cultured embryos were stained with Hoechst 33342 (25 µg/ml) for 15 min, and mounted on a glass slide for image record under the microscope equipped with UV light. The images were analyzed with Image-Pro Plus software (Media Cybernetics, Inc., Bethesda, MD, USA) for cell number counting and blastocyst or blastocoel diameter measurement.
Statistical analysis
Each group consisted of at least 30 constructed embryos, with a minimum of three replicates for every experiment. The statistical significance of differences in development was evaluated using a χ2 test. Student's t-test was used for cell number and blastocyst or blastocoel diameter comparison. A probability value P < 0.05 was considered statistically significant.
Results
Experimental design
One could postulate that ooplasm transfer in mammalian oocytes should have little or no effect on the formation, processing and subsequent function of the embryonic genome, because this process should be driven largely by the recipient ooplasm, which is present in great excess over the donor ooplasm. However, because genetic differences exist in genome function between unmanipulated embryos of different strains and because the transferred ooplasm might fail to disperse rapidly after transfer, there exists the potential for unexpected effects of the donor ooplasm on embryonic genome function, particularly with respect to the paternal pronucleus, which is extensively remodeled after fertilization. The overall objective of these studies was to determine whether foreign ooplasm of one genotype could either positively or negatively affect the function of paternal genomes within recipient oocytes of an opposing genotype. In order to detect such effects, it is necessary that the paternal genome function be assayed in the absence of any maternal genetic contribution that could mask such effects. The androgenetic mouse embryo provides an ideal model for this analysis, because ooplasm from genetically distinct mouse strains is known to modify the paternal genome differently. Androgenones can be produced readily with and without inter-genotype ooplasm transfer, and the developmental potentials of manipulated embryos can be compared. Our overall experimental design is summarized in Fig. 1B. Oocytes were collected from females of the two strains, B6 and D2. These oocytes are then either subjected to ICSI, IVF or parthenogenetic activation, with or without prior ooplasm exchange. Comparisons of embryos prepared from intra-strain ooplasm transfer oocytes to embryos produced from non-manipulated oocytes provide a control for possible effects of the ooplasm transfer itself. Comparisons between inter- and intra-strain ooplasm transfer embryos provide the ability to assess effects of foreign ooplasm on genome function. Comparisons between androgenones, parthenogenones and fertilized (IVF or ICSI) embryos provide the ability to determine whether the effects of foreign ooplasm are specific for maternal or paternal genomes. Last, it is important to note that for all androgenone, IVF and ICSI studies, the sperm are derived from (B6D2) F1 males, and hence the results are unaffected by genetic variation in the genotype of sperm donor.
Effect of oocyte genotype on androgenetic, parthenogenetic, IVF and ICSI embryos development
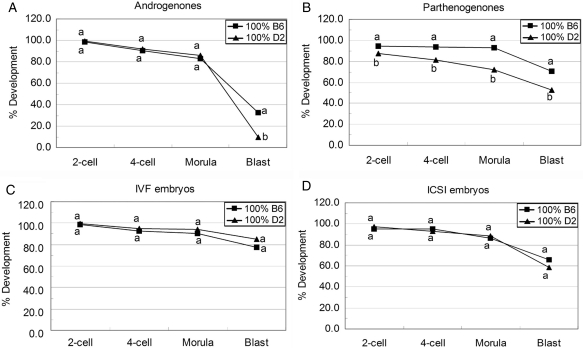
To determine whether the oocytes derived from B6 and D2 mice have the same capacity to support androgenetic, parthenogenetic, IVF and ICSI embryo development, the developmental efficiency of each kind of embryo was evaluated without ooplasm transfer (denoted as 100% B6 and 100% D2) (Fig. 2). We compared the ratio of embryos attaining each stage. In androgenones, there were no significant differences between B6 and D2 at the 2-cell, 4-cell and morula stages. However, androgenones produced with B6 oocytes showed the ‘higher’ rate (high relative to D2 androgenones) of blastocyst formation, an average of 32.9% (23 of 70), with the majority of these being expanded blastocysts of fairly normal morphology. Androgenones prepared with D2 oocytes developed poorly, with only 9.6% (9 of 94) forming abnormal blastocysts of poor morphology and with rudimentary blastocoels (P < 0.0002). Hence, the difference in the blastocyst formation between the two strains was due to differences in progression from the morula to blastocyst stage (Fig. 2A). These results confirmed results from earlier studies of androgenone production by pronucleus transfer (Latham and Solter, 1991).
Figure 2.
Development of different kind of embryos constructed with B6 or D2 oocytes without ooplasm transfer.
Androgenone, parthenogenone, IVF and ICSI embryos were constructed with B6 (square) or D2 (triangle) oocytes without ooplasm donation. Embryos were checked for the ratio of 2-cell, 4-cell, morula and blastocyst stages at 48, 72, 96 and 120 h post-hCG. (A) Development of androgenetic embryos (number of embryos: B6 = 70; D2 = 94); (B) development of parthenogenetic embryos (number of embryos: B6 = 203; D2 = 310); (C) development of IVF embryos (number of embryos: B6 = 71; D2 = 123); (D) development of ICSI embryos (number of embryos: B6 = 96; D2 = 169). Within the same stage, percentages without a common letter are different (P < 0.05).
Parthenogenones constructed with B6 oocytes displayed a higher potential to develop to blastocyst stage compared with D2 oocyte [70.4% (143 of 203) versus 52.9% (164 of 310), P < 10−4]. However, the developmental differences in the parthenogenetic embryos were manifested earlier than with the androgenones, with lesser cleavage beyond the 2-cell stage for D2 parthenotes [94.6% (192 of 203) versus 87.1% (270 of 310), P < 0.01] (Fig. 2B).
Embryos constructed with IVF or ICSI methods were also compared between B6 and D2 oocytes. There was no significant difference between these two strains in the development of these embryos from the 2-cell stage to blastocyst stage (Fig. 2C and D).
Effect of ooplasm donation on androgenetic, parthenogenetic, IVF and ICSI embryo development
In order to study the effect of ‘foreign’ ooplasm donation on the early developmental efficiency of different kinds of embryos, 10% of the ooplasm was transferred from donor oocytes of one strain to recipient oocytes of the other (denoted as 90% B6+10% D2 and 90% D2+10% B6). As controls, 10% ooplasm was exchanged between oocytes of the same strain (denoted as 90% B6+10% B6 and 90% D2+10% D2).
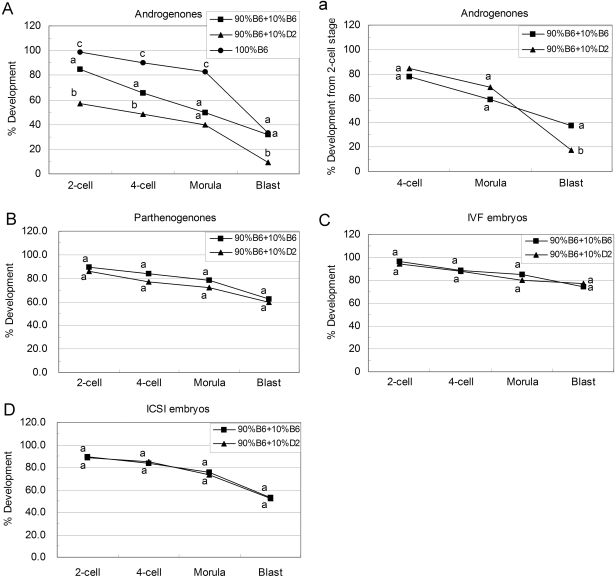
Among androgenones, intra-strain transfer of B6 ooplasm to B6 oocytes or D2 ooplasm to D2 oocytes diminished the development to the morula stage, but had no significant effect on blastocyst formation compared with the non-cytoplasm transferred groups (Figs 3A and 4A, Table I), indicating that the ooplasm transfer procedure itself was not producing any significant deficiency in development for these embryos. The transfer of 10% ooplasm from D2 to B6 oocytes significantly decreased the 2-cell, 4-cell and blastocyst rates for androgenones, when compared with the control group, in which 10% ooplasm was transferred from B6 to B6 oocytes [2-cell: 57.1% (52 of 91) versus 85.1% (80 of 94), P < 10−4; 4-cell: 48.4% (44 of 91) versus 65.9% (62 of 94), P < 0.05; blastocyst: 9.8% (9 of 91) versus 31.9% (30 of 94), P < 0.0002; Fig. 3A]. Because the rates of development to both 2-cell and blastocyst stages were lower in androgenones constructed with 90% B6+10% D2 than that of the controls, we tested whether this decrease was the result of a deficiency in progression beyond the 2-cell stage (i.e. ‘2-cell block’), or a deficiency in development thereafter. Normalizing developmental data to the number of 2-cell stage embryos in each group (Fig. 3B) revealed no significant difference at the 4-cell and morula stages, but a significant difference in blastocyst formation as a fraction of 2-cell stage embryos, when comparing inter- and intra-strain cytoplasm transfer androgenones [17.3% (9 of 52) versus 37.5% (30 of 80) P < 0.05], indicating that the deficiency in development was attributable primarily to deficient transition from morula to blastocyst stage.
Figure 3.
Development of different kind of embryos constructed with B6 oocytes coupled with ooplasm donation.
Androgenone, parthenogenone, IVF and ICSI embryos were constructed with B6 oocytes coupled with intra- (square) or inter-strain (triangle) ooplasm donation. Embryos were checked for the ratio of 2-cell, 4-cell, morula and blastocyst stages at 48, 72, 96 and 120 h post-hCG. (A) Development of androgenetic embryos (based on pronuclear) (number of embryos: 90% B6+10% B6 = 94; 90% B6+10% D2 = 91; 100% B6 = 70); (a) development of androgenetic embryos (based on 2-cell); (B) development of parthenogenetic embryos (number of embryos: 90% B6+10% B6 = 177; 90% B6+10% D2 = 131); (C) development of IVF embryos (number of embryos: 90% B6+10% B6 = 121; 90% B6+10% D2 = 91); (D) development of ICSI embryos (number of embryos: 90% B6+10% B6 = 103; 90% B6+10% D2 = 80). Within the same stage, percentages without a common letter are different (P < 0.05). To compare non-cytoplasm transferred androgenones with intra-strain cytoplasm transferred androgenones, the data of 100% B6 (circle) were reproduced from Fig. 2.
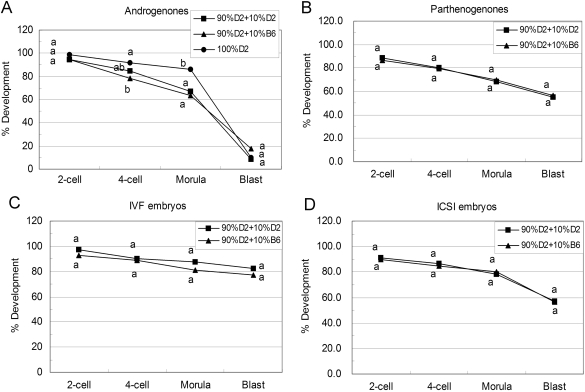
Figure 4.
Development of different kind of embryos constructed with D2 oocytes coupled with ooplasm donation.
Androgenone, parthenogenone, IVF and ICSI embryos were constructed with D2 oocytes coupled with intra- (square) or inter-strain (triangle) ooplasm donation. Embryos were checked for the ratio of 2-cell, 4-cell, morula and blastocyst stages at 48, 72, 96 and 120 h post-hCG. (A) Development of androgenetic embryos (number of embryos: 90% D2+10% D2 = 106; 90% D2+10% B6 = 79; 100% D2 = 94); (B) development of parthenogenetic embryos (number of embryos: 90% D2+10% D2 = 204; 90% D2+10% B6 = 96); (C) development of IVF embryos (number of embryos: 90% D2+10% D2 = 136; 90% D2+10% B6 = 70); (D) development of ICSI embryos (number of embryos: 90% D2+10% D2 = 105; 90% D2+10% B6 = 60). Within the same stage, percentages without a common letter are different (P < 0.05). To compare non-cytoplasm transferred androgenones with intra-strain cytoplasm transferred androgenones, the data of 100% D2 (circle) were reproduced from Fig. 2.
Table I.
Comparison of blastocysts formation between intra-strain cytoplasm transferred and non-cytoplasm transferred embryos
| Type of embryo | Blastocyst formation* |
P-value | ||
|---|---|---|---|---|
| Non-cytoplasm transfer | 10% intra-strain cytoplasm transfer | |||
| B6 | Androgenone | 32.9% (23) | 31.9% (30) | 0.8985 |
| Parthenogenone | 70.4% (143) | 62.1% (110) | 0.0872 | |
| IVF | 77.5% (55) | 74.4% (90) | 0.6313 | |
| ICSI | 65.6% (63) | 53.4% (55) | 0.0794 | |
| D2 | Androgenone | 9.6% (9) | 8.5% (9) | 0.7892 |
| Parthenogenone | 52.9% (164) | 54.9% (112) | 0.6566 | |
| IVF | 85.4% (105) | 82.4% (112) | 0.5112 | |
| ICSI | 58.6% (99) | 57.1% (60) | 0.8147 | |
Numbers in parentheses show the numbers of blastocysts scored.
*Blastocysts defined by the presence of a fluid-filled cavity. Embryos were scored at 120 h post-hCG injection.
Interestingly, the reciprocal inter-strain transfers, 10% ooplasm from B6 transferred to D2 oocytes, did not enhance D2 androgenone development (Fig. 4A). Thus, whereas D2 ooplasm can exert a dominant negative effect on development in embryos prepared with B6 recipient oocytes, B6 ooplasm was not able to overcome the negative effects of a recipient D2 ooplasm on androgenone development.
The effects of inter-strain ooplasm donation on parthenogenetic, IVF and ICSI embryos were also investigated. There were no significant differences between inter- and intra-strain ooplasm donation groups amongst parthenogenetic, IVF and ICSI embryos (Figs 3B–D and 4B–D). The ooplasm transfer protocol itself had no effect on blastocyst formation since we did not find any significant difference between the intra-strain cytoplasm transferred and non-cytoplasm transferred parthenogenetic, IVF and ICSI embryos (Table I).
Effect of ooplasm donation on embryo quality
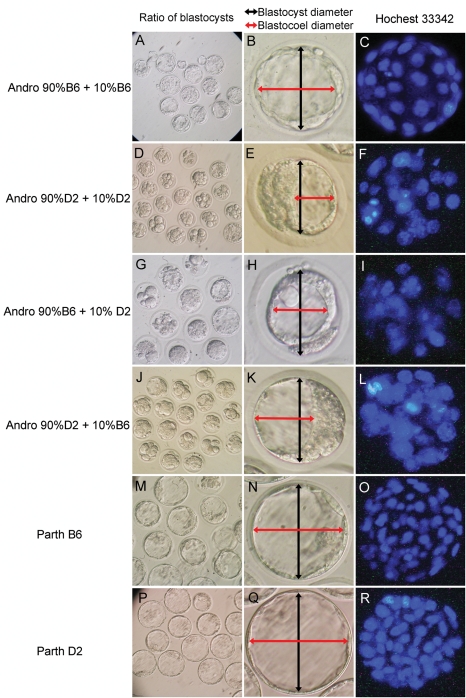
Embryo quality was compared between groups with respect to total embryo cell number, number of cells in embryos attaining the blastocyst stage, average blastocyst diameter and average blastocoel diameter (Table II). Diploid androgenones developed to the blastocyst stage at a high frequency when intra-strain ooplasm transferred B6 oocytes were used (Fig. 5A). These blastocysts were relatively normal in morphology, with larger cell numbers (30.73 ± 2.47 versus 19.36 ± 3.66, P < 10−4) (Fig. 5C), larger blastocyst diameters (102.67 ± 7.74 versus 83.94 ± 5.72, P < 0.005) and better-expanded blastocoel cavities (69.69 ± 8.94 versus 43.70 ± 6.39, P < 10−4) than their D2 counterparts (Fig. 5B). Diploid androgenetic embryos formed blastocysts poorly when made with intra-strain ooplasm transferred D2 oocytes (Fig. 5D). The few androgenetic blastocysts that developed from D2 oocytes had very abnormal morphologies, with small blastocoel cavities (Fig. 5E), reduced cell numbers and many excluded cells compared with their B6 counterparts (Fig. 5F). However, for inter-strain ooplasm donated androgenones, 10% ooplasm donation from D2 to B6 oocytes significantly decreased the cell number (Fig. 5I) compared with intra-strain controls (16.57 ± 1.70 versus 26.09 ± 4.59, P < 10−4), as well as the blastocyst (90.31 ± 3.50 versus 102.67 ± 7.74, P < 0.01) and blastocoel (42.36 ± 6.95 versus 69.69 ± 8.94, P < 0.001) diameters (Fig. 5H) of the androgenones. In contrast, 10% ooplasm donation from B6 to D2 oocytes did not increase the quality of the androgenones (Fig. 5J–L). A previous study reported cell numbers for non-ooplasm transfer androgenones prepared with C57BL/6 and DBA/2 oocytes as 25 and 15, respectively (Latham and Solter, 1991), similar to our current study. The combined data indicate that inter-strain ooplasm transfer of DBA/2 ooplasm to C57BL/6 oocytes diminished embryo quality for androgenones, whereas the reciprocal inter-strain transfer had no significant effect, and the intra-strain ooplasm transfer had no significant effect. For other kinds of embryos, including parthenogenetic, IVF and ICSI embryos, ooplasm donation within inter- or intra-strain oocytes did not affect embryo quality (Fig. 5M–R, Table II).
Table II.
Cell number and diameter of embryos cultured for 120 h post-hCG
| Type of embryo | Total embryo cell number* Mean ± SD (n) | Total cell number in blastocyst Mean ± SD (n) | Blastocyst diameter Mean ± SD (n) (µm) | Blastocoel diameter Mean ± SD (n) (µm) | |
|---|---|---|---|---|---|
| Andro | 100% B6 | 28.11 ± 5.31 (18)a | 32.00 ± 2.40 (9)a | 105.31 ± 6.28 (9)a | 72.78 ± 7.44 (9)a |
| 100% D2 | 18.95 ± 3.15 (20)bc | 21.70 ± 1.94 (9)b | 86.85 ± 4.94 (9)b | 46.51 ± 5.45 (9)b | |
| 90% B6+10% B6 | 26.09 ± 4.59 (15)a | 30.73 ± 2.47 (8)a | 102.67 ± 7.74 (8)a | 69.69 ± 8.94 (7)a | |
| 90% D2+10% D2 | 17.43 ± 2.17 (16)bc | 19.36 ± 3.66 (8)b | 83.94 ± 5.72 (9)b | 43.70 ± 6,39 (8)b | |
| 90% B6+10% D2 | 16.57 ± 1.70 (14)b | 17.83 ± 1.72 (6)c | 90.31 ± 3.50 (6)b | 42.36 ± 6.95 (6)b | |
| 90% D2+10% B6 | 20.12 ± 3.20 (17)c | 19.40 ± 2.40 (5)bc | 88.49 ± 5.17 (5)b | 40.50 ± 3.28 (5)b | |
| Parth | 100% B6 | 44.48 ± 7.32 (31)d | 47.12 ± 5.03 (25)d | 116.47 ± 3.61 (25)a | 98.38 ± 5.30 (25)c |
| 100% D2 | 44.03 ± 4.59 (29)d | 44.30 ± 3.90 (15)d | 112.52 ± 4.95 (15)a | 96.33 ± 4.29 (15)c | |
| 90% B6+10% B6 | 42.75 ± 5.96 (28)d | 43.31 ± 6.72 (21)d | 111.83 ± 8.93 (20)a | 92.37 ± 6.90 (21)c | |
| 90% D2+10% D2 | 41.99 ± 6.47 (25)d | 40.06 ± 5.62 (17)d | 104.48 ± 7.36 (15)a | 90.11 ± 5.74 (16)c | |
| 90% B6+10% D2 | 42.75 ± 5.40 (21)d | 43.71 ± 4.44 (17)d | 109.49 ± 5.39 (17)a | 89.80 ± 3.31 (17)c | |
| 90% D2+10% B6 | 43.66 ± 4.54 (19)d | 45.89 ± 3.91 (15)d | 110.86 ± 4.75 (15)a | 89.58 ± 3.89 (15)c | |
| IVF | 100% B6 | 46.40 ± 7.60 (10)d | 50.59 ± 3.54 (8)d | 118.68 ± 4.91 (8)a | 96.15 ± 5.29 (8)c |
| 100% D2 | 41.78 ± 6.55 (9)d | 46.39 ± 3.75 (7)d | 114.39 ± 5.10 (7)a | 94.34 ± 4.48 (7)c | |
| 90% B6+10% B6 | 43.39 ± 7.52 (8)d | 48.34 ± 5.52 (8)d | 110.53 ± 6.87 (7)a | 93.41 ± 6.25 (8)c | |
| 90% D2+10% D2 | 40.73 ± 7.35 (7)d | 44.66 ± 4.78 (6)d | 106.90 ± 7.79 (8)a | 90.01 ± 4.99 (7)c | |
| 90% B6+10% D2 | 43.59 ± 4.90 (9)d | 45.82 ± 3.89 (6)d | 108.41 ± 6.22 (6)a | 90.50 ± 5.39 (6)c | |
| 90% D2+10% B6 | 44.47 ± 3.83 (9)d | 46.79 ± 4.40 (6)d | 103.63 ± 5.54 (6)a | 89.45 ± 4.40 (6)c | |
| ICSI | 100% B6 | 45.52 ± 4.39 (27)d | 47.38 ± 3.69 (14)d | 111.39 ± 6.29 (14)a | 92.96 ± 5.76 (14)c |
| 100% D2 | 45.56 ± 5.62 (16)d | 48.52 ± 2.48 (10)d | 109.71 ± 5.87 (10)a | 95.82 ± 3.71 (10)c | |
| 90% B6+10% B6 | 42.62 ± 6.33 (10)d | 44.51 ± 5.98 (10)d | 107.33 ± 7.38 (11)a | 89.64 ± 6.53 (11)c | |
| 90% D2+10% D2 | 41.69 ± 7.21 (12)d | 46.70 ± 4.62 (10)d | 104.34 ± 7.47 (10)a | 92.83 ± 5.39 (10)c | |
| 90% B6+10% D2 | 44.53 ± 5.50 (10)d | 45.30 ± 3.60 (7)d | 104.48 ± 4.37 (7)a | 91.44 ± 6.23 (7)c | |
| 90% D2+10% B6 | 43.62 ± 4.86 (10)d | 45.96 ± 2.71 (7)d | 106.37 ± 4.65 (7)a | 91.70 ± 4.92 (7)c |
Within column, different letters indicate significantly different values (P < 0.05).
*Average cell numbers for all embryos. The number attaining blastocyst stage is indicated in column 4.
Figure 5.
Morphology analysis of embryos developed to 120 h post-hCG.
Androgenetic embryos coupled with ooplasm donation were constructed. After culture for 120 h post-hCG, the morphology of each embryo was analyzed with the criterion of ratio of blastocysts formation (A, D, G and J), diameter of blastocyst/blastocoel (B, E, H and K) and cell number (C, F, I and L). The blastocysts derived from parthenogenetic embryos were also analyzed as controls (M–R). Diploid androgenones constructed with 90% B6+10% B6 develop to the blastocyst stage at a high frequency (A). These blastocysts are relatively normal in morphology, with well-expanded blastocoel cavities (B) and larger cell numbers (C). Diploid androgenetic embryos form blastocysts poorly when made with 90% D2+10% D2 ooplasm (D). The few androgenetic blastocysts that develop from D2 oocytes have very abnormal morphologies, with small blastocoel cavities (E), reduced cell numbers and many excluded cells (F). For inter-strain ooplasm donated androgenones, 10% ooplasm donation from D2 oocytes to B6 oocytes significantly decreased the cell number (I), blastocyst and blastocoel diameter (H) of the androgenones. In contrast, 10% ooplasm donation from B6 oocytes to D2 oocytes did not increase the androgenones quality (J–L). For other kinds of constructions such as parthenogenetic embryos, the blastocysts quality is much better than the androgenones (M–R).
Discussion
Abundant data exist to indicate significant epigenetic effects of the ooplasm on the paternal genome during the early post-fertilization period, as well as genetic diversity in these effects (Latham, 1999). Unexpected and undesired epigenetic modifications could thus arise via effects of ‘foreign’ ooplasm of one genotype on the formation of the paternal pronucleus in oocytes of a different genotype, and the subsequent function of those paternal genomes may also be affected. Investigating this possibility is crucial for evaluating the safety of ooplasm donation procedures in humans. We report here the differential effects of foreign ooplasm on paternal genome function following ooplasm transfer combined with ICSI. Our data reveal that ooplasm from D2 oocytes can negatively affect the efficiency and quality of blastocyst development from androgenetic mouse embryos prepared by dual sperm injection into B6 oocytes. These embryos provide a sensitive indicator of paternal genome function, with no maternal genome present to obscure phenotype. The observed effect on paternal genome function is quite specific. No effect of inter-strain ooplasm transfer is observed in parthenogenetic, IVF or ICSI controls, and no negative effect is associated with intra-strain ooplasm transfer. Additionally, the inter-genotype effect in androgenones is restricted to a negative effect of D2 ooplasm in B6 oocytes, with no reciprocal positive effect of B6 ooplasm observed with D2 oocytes.
Earlier pronuclear transfer studies revealed that the reduced developmental capacity of androgenones made with D2 oocytes was the result of aberrant epigenetic processing, rather than a metabolic insufficiency of the D2 cytoplasm; i.e. paternal genomes that form in D2 oocytes and are then transferred to B6 cytoplasm still cannot support the higher rate of blastocyst formation (Latham and Solter, 1991; Latham and Sapienza, 1998). Those earlier studies, however, could not distinguish whether the D2 ooplasmic effect was a negative effect upon the paternal PN, or the absence of a positive effect otherwise imposed by the B6 strain of ooplasm. The studies performed here are distinct from these earlier studies because they test for an effect of a small fraction (10%) of foreign ooplasm, rather than testing an effect of total ooplasm replacement, as with nuclear (including GV or pronuclear) transfer, on paternal genome function. Moreover, the data presented here reveal that the inter-strain difference in androgenone development is the result of a negative or unfavorable modification of the paternal genome by the D2 ooplasm, rather than a lack of positive effects of B6 ooplasm. The simplest explanation for these observations is that strain-dependent differences in male pronucleus modifications are mediated by ooplasmic factors and that these modifications affect the expression of imprinted genes that are required for blastocyst formation and must be expressed from the paternal genome.
Clinical procedures have combined foreign ooplasm injection with ICSI, so that the paternal pronucleus forms in the same region as that occupied by the injected ooplasm. The studies here approximated this procedure by injecting sperm nuclei into the region of the transferred ooplasm, as marked by the position of the slit in the zona pellucida. The negative effect of transferred D2 ooplasm indicates that the injected sperm nuclei are negatively affected by the surrounding D2 ooplasm even within a larger volume of recipient B6 ooplasm. Moreover, it appears that this negative effect is long-lasting, as the inter-strain ooplasm effect persists even after normalization to the number of 2-cell stage embryos obtained. As with earlier pronuclear transfer studies (Latham and Solter, 1991), these results support a stable epigenetic effect on paternal genome function. The lack of any beneficial effect of B6 ooplasm on D2 androgenones indicates that negative attributes of the native ooplasm cannot always be overcome by transferring a small amount of positive acting foreign ooplasm. This observation indicates that clinical procedures seeking to ‘cure’ poor oocyte quality may not be effective in certain instances. Knowledge of the specific molecular basis for reduced oocyte quality, therefore, would be needed in order to judge a priori whether ooplasm donation could be beneficial.
We did not observe a significant effect of inter-strain versus intra-strain ooplasm transfer upon parthenogenetic development. Thus, the foreign ooplasm itself does not perturb development, and neither does the foreign ooplasm compromise maternal genome function. The specificity of the foreign ooplasm effect for the paternal genome could reflect an effect upon target genes that are regulated by gamete genomic imprinting, or genes that are differentially modified between the maternal and paternal genomes post-fertilization, which would mimic the effects of gamete imprinting (Latham, 1999).
We also failed to observe a significant effect of inter-strain ooplasm transfer on the development of IVF or ICSI embryos. Other studies from this laboratory have revealed that a similar ooplasm transfer procedure can yield subtle effects on embryo survival and post-term growth, but not yield pronounced effects in inter-strain versus intra-strain transfers (Cheng et al., 2009). Collectively, the results from parthenotes, ICSI and IVF embryos indicate that the paternal genome may be more highly affected by inter-strain ooplasm transfer than the maternal genome and that the presence of a maternal genome may mitigate the deleterious effects during later stages of development. However, this does not negate the fact that the inter-strain ooplasm transfer can alter paternal genome function and that this alteration could contribute to an altered phenotype after birth. It is becoming increasingly apparent that imprinted genes regulate a wide range of phenotypic characteristics other than growth and morphogenesis (Massa et al., 1992; Brenton et al., 1995; Polychronakos et al., 1995; Davies et al., 2005; Reiner, 2005), any of which could lead to long-term health consequences and possible behavioral or cognitive effects not readily assayed in the mouse model. It is also becoming widely appreciated that early epigenetic effects in the embryo can exert long-lasting effects on a range of characteristics such as imprinted gene expression, behavior, cognition and hypertension (Doherty et al., 2000; Khosla et al., 2001; Ecker et al., 2004; Fernandez-Gonzalez et al., 2004; Mann et al., 2004; Lee et al., 2005; Fetita et al., 2006; Kwong et al., 2006; Sinclair et al., 2007; Chelbi and Vaiman, 2008; Woo and Patti, 2008). Hence, our demonstration here that foreign ooplasm transfer can alter paternal genome function reinforces the view that the potential risks of such procedures have not yet been defined, and thus cannot yet be reliably evaluated for clinical application.
Funding
This work was supported by a grant from the National Institutes of Health, National Center for Research Resources (RO1 RR18907 and HD 41440).
Acknowledgements
We thank Bela Patel for her technical assistance. We thank Carmen Sapienza for his helpful comments on the manuscript.
References
- Abramczuk J, Solter D, Koprowski H. The beneficial effect EDTA on development of mouse one-cell embryos in chemically defined medium. Dev Biol. 1977;61:378–383. doi: 10.1016/0012-1606(77)90308-6. [DOI] [PubMed] [Google Scholar]
- Barritt JA, Brenner CA, Malter HE, Cohen J. Mitochondria in human offspring derived from ooplasmic transplantation. Hum Reprod. 2001;a 16:513–516. doi: 10.1093/humrep/16.3.513. [DOI] [PubMed] [Google Scholar]
- Barritt JA, Brenner CA, Malter HE, Cohen J. Rebuttal: interooplasmic transfers in humans. Reprod Biomed Online. 2001;b 3:47–48. doi: 10.1016/s1472-6483(10)61966-9. [DOI] [PubMed] [Google Scholar]
- Bredenoord AL, Pennings G, de Wert G. Ooplasmic and nuclear transfer to prevent mitochondrial DNA disorders: conceptual and normative issues. Hum Reprod Update. 2008;14:669–678. doi: 10.1093/humupd/dmn035. [DOI] [PubMed] [Google Scholar]
- Brenton JD, Viville S, Surani MA. Genomic imprinting and cancer. Cancer Surv. 1995;25:161–171. [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- Chelbi ST, Vaiman D. Genetic and epigenetic factors contribute to the onset of preeclampsia. Mol Cell Endocrinol. 2008;282:120–129. doi: 10.1016/j.mce.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wang K, Kellam LD, Lee YS, Liang CG, Han Z, Mtango NR, Latham KE. Effects of ooplasm manipulation on DNA methylation and growth of progeny in mice. Biol Reprod. 2009;80:464–472. doi: 10.1095/biolreprod.108.073593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Mann MR, Bartolomei MS, Latham KE. Nuclear-cytoplasmic “tug of war” during cloning: effects of somatic cell nuclei on culture medium preferences of preimplantation cloned mouse embryos. Biol Reprod. 2002;66:1178–1184. doi: 10.1095/biolreprod66.4.1178. [DOI] [PubMed] [Google Scholar]
- Cohen J, Scott R, Schimmel T, Levron J, Willadsen S. Birth of infant after transfer of anucleate donor oocyte cytoplasm into recipient eggs. Lancet. 1997;350:186–187. doi: 10.1016/S0140-6736(05)62353-7. [DOI] [PubMed] [Google Scholar]
- Cohen J, Scott R, Alikani M, Schimmel T, Munne S, Levron J, Wu L, Brenner C, Warner C, Willadsen S. Ooplasmic transfer in mature human oocytes. Mol Hum Reprod. 1998;4:269–280. doi: 10.1093/molehr/4.3.269. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–430. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Stein P, Xu Z, Williams CJ, Kopf GS, Bilker WB, Abel T, Schultz RM. Long-term effects of culture of preimplantation mouse embryos on behavior. Proc Natl Acad Sci USA. 2004;101:1595–1600. doi: 10.1073/pnas.0306846101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Moreira P, Bilbao A, Jimenez A, Perez-Crespo M, Ramirez MA, Rodriguez De Fonseca F, Pintado B, Gutierrez-Adan A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc Natl Acad Sci USA. 2004;101:5880–5885. doi: 10.1073/pnas.0308560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetita LS, Sobngwi E, Serradas P, Calvo F, Gautier JF. Consequences of fetal exposure to maternal diabetes in offspring. J Clin Endocrinol Metab. 2006;91:3718–3724. doi: 10.1210/jc.2006-0624. [DOI] [PubMed] [Google Scholar]
- Gao S, Czirr E, Chung YG, Han Z, Latham KE. Genetic variation in oocyte phenotype revealed through parthenogenesis and cloning: correlation with differences in pronuclear epigenetic modification. Biol Reprod. 2004;70:1162–1170. doi: 10.1095/biolreprod.103.024216. [DOI] [PubMed] [Google Scholar]
- Han Z, Mtango NR, Patel BG, Sapienza C, Latham KE. Hybrid vigor and transgenerational epigenetic effects on early mouse embryo phenotype. Biol Reprod. 2008;79:638–648. doi: 10.1095/biolreprod.108.069096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes SM, Sapienza C, Latham KE. Ooplasmic donation in humans: the potential for epigenic modifications. Hum Reprod. 2002;17:850–852. doi: 10.1093/humrep/17.4.850. [DOI] [PubMed] [Google Scholar]
- Janssenswillen C, Christiaens F, Camu F, Van Steirteghem A. The effect of propofol on parthenogenetic activation, in vitro fertilization and early development of mouse oocytes. Fertil Steril. 1997;67:769–774. doi: 10.1016/s0015-0282(97)81381-7. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Kwong WY, Miller DJ, Ursell E, Wild AE, Wilkins AP, Osmond C, Anthony FW, Fleming TP. Imprinted gene expression in the rat embryo-fetal axis is altered in response to periconceptional maternal low protein diet. Reproduction. 2006;132:265–277. doi: 10.1530/rep.1.01038. [DOI] [PubMed] [Google Scholar]
- Latham KE. Strain-specific differences in mouse oocytes and their contributions to epigenetic inheritance. Development. 1994;120:3419–3426. doi: 10.1242/dev.120.12.3419. [DOI] [PubMed] [Google Scholar]
- Latham KE. Epigenetic modification and imprinting of the mammalian genome during development. Curr Top Dev Biol. 1999;43:1–49. doi: 10.1016/s0070-2153(08)60377-4. [DOI] [PubMed] [Google Scholar]
- Latham KE, McGrath J, Solter D. Mechanistic and developmental aspects of genetic imprinting in mammals. Int Rev Cytol. 1995;160:53–98. doi: 10.1016/s0074-7696(08)61553-3. [DOI] [PubMed] [Google Scholar]
- Latham KE, Sapienza C. Localization of genes encoding egg modifiers of paternal genome function to mouse chromosomes one and two. Development. 1998;125:929–935. doi: 10.1242/dev.125.5.929. [DOI] [PubMed] [Google Scholar]
- Latham KE, Solter D. Effect of egg composition on the developmental capacity of androgenetic mouse embryos. Development. 1991;113:561–568. doi: 10.1242/dev.113.2.561. [DOI] [PubMed] [Google Scholar]
- Lee HK, Park KS, Cho YM, Lee YY, Pak YK. Mitochondria-based model for fetal origin of adult disease and insulin resistance. Ann N Y Acad Sci. 2005;1042:1–18. doi: 10.1196/annals.1338.001. [DOI] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- Massa S, Gardini F, Sinigaglia M, Guerzoni ME. Klebsiella pneumoniae as a spoilage organism in mozzarella cheese. J Dairy Sci. 1992;75:1411–1414. doi: 10.3168/jds.S0022-0302(92)77894-1. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel De Villena F, de La Casa-Esperon E, Williams JW, Malette JM, Rosa M, Sapienza C. Heritability of the maternal meiotic drive system linked to Om and high-resolution mapping of the Responder locus in mouse. Genetics. 2000;155:283–289. doi: 10.1093/genetics/155.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard B, Dean W, Engemann S, Bergmann K, Fuermann M, Jung M, Reis A, Allen N, Reik W, Walter J. Epigenetic targeting in the mouse zygote marks DNA for later methylation: a mechanism for maternal effects in development. Mech Dev. 2001;103:35–47. doi: 10.1016/s0925-4773(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Polychronakos C, Giannoukakis N, Deal CL. Imprinting of IGF 2, insulin-dependent diabetes, immune function, and apoptosis: a hypothesis. Dev Genet. 1995;17:253–262. doi: 10.1002/dvg.1020170310. [DOI] [PubMed] [Google Scholar]
- Reiner SL. Epigenetic control in the immune response. Hum Mol Genet. 2005;14:R41–R46. doi: 10.1093/hmg/ddi115. [DOI] [PubMed] [Google Scholar]
- Sapienza C, Paquette J, Pannunzio P, Albrechtson S, Morgan K. The polar-lethal Ovum mutant gene maps to the distal portion of mouse chromosome 11. Genetics. 1992;132:241–246. doi: 10.1093/genetics/132.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci USA. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M, Patti ME. Diabetes risk begins in utero. Cell Metab. 2008;8:5–7. doi: 10.1016/j.cmet.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Perry AC. Piezo-actuated mouse intracytoplasmic sperm injection (ICSI) Nat Protoc. 2007;2:296–304. doi: 10.1038/nprot.2007.7. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang CW, Krey L, Liu H, Meng L, Blaszczyk A, Adler A, Grifo J. In vitro maturation of human preovulatory oocytes reconstructed by germinal vesicle transfer. Fertil Steril. 1999;71:726–731. doi: 10.1016/s0015-0282(98)00549-4. [DOI] [PubMed] [Google Scholar]