Abstract
Somatic embryogenesis (SE) is a model system for understanding the physiological, biochemical, and molecular biological events occurring during plant embryo development. Plant somatic cells have the ability to undergo sustained divisions and give rise to an entire organism. This remarkable feature is called plant cell totipotency. SE is a notable illustration of plant totipotency and involves reprogramming of development in somatic cells toward the embryogenic pathway. Plant growth regularities, especially auxins, are key components as their exogenous application recapitulates the embryogenic potential of the mitotically quiescent somatic cells. It has been observed that there are genetic and also physiological factors that trigger in vitro embryogenesis in various types of plant somatic cells. Analysis of the proteome and transcriptome has led to the identification and characterization of certain genes involved in SE. Most of these genes, however, are upregulated only in the late developmental stages, suggesting that they do not play a direct role in the vegetative-to-embryogenic transition. However, the molecular bases of those triggering factors and the genetic and biochemical mechanisms leading to in vitro embryogenesis are still unknown. Here, we describe the plant factors that participate in the vegetative-to-embryogenic transition and discuss their possible roles in this process.
Keywords: Somatic embryogenesis, Embryogenesis cell, Gene
Introduction
Embryogenesis is a crucial developmental process in the life cycle of plants spanning the transition from the fertilized egg to the generation of a mature embryo. In this process, the embryo acquires a defined apical–basal pattern along the main body axis with shoot and root poles, a hypocotyl and cotyledons. Alternatively, embryogenesis can take place without the involvement of fertilization or gamete fusion. The origins of such asexual embryos are quite diverse; e.g., apomictic embryos are derived from an unfertilized egg cell or from maternal tissue. Somatic embryogenesis (SE) is the developmental restructuring of somatic cells toward the embryogenic pathway and forms the basis of cellular totipotency in higher plants [76, 130]. While carrot was the first plant species in which SE was reported, during the last 50-plus years of culturing experiments, the induction of in vitro SE has been shown to be successful in many plant species, including angiosperms and gymnosperms.
SE provides an attractive model system for studying zygotic embryogenesis, particularly because zygotic embryos are encased by maternal tissues and are difficult to access using biochemical and molecular tools. In contrast to zygotic embryogenesis, SE is a nonsexual propagation process where somatic cells differentiate somatic embryos. Therefore, somatic embryos can be used for studying the regulation of embryo development. However, SE has been viewed as a tool for massive propagation of commercial crops and as a potential model system for the study of the regulation of gene expression required for the earliest developmental events in the life of higher plants, such as the developmental mechanism of embryogenesis [228]. In addition, as the initial basis of cellular and genetic engineering, SE plays an important role in genetic transformation, somatic hybridization, and somaclonal variation.
Although, genetic programs controlling embryo development in zygotic and SE display many similarities [122, 228], the mechanisms determining the induction phase of these two processes are different. Zygotic embryo development begins with the formation of the zygote following fertilization, while somatic cells acquire embryogenic competence as a result of different chemical and physical stimuli. Thus, plant SE is a developmental process involving the reprogramming of gene expression patterns involving cascades of genetic triggers turning on and off the expression of specific genes [42, 127].
Historically, it has been observed that there are genetic and also physiological factors that trigger in vitro embryogenesis in various types of plant somatic cells. However, the molecular bases of those triggering factors and the genetic and biochemical mechanisms leading to in vitro embryogenesis are still unknown [127, 211]. Understanding the key factors promoting vegetative-to-embryogenic transition and identification of genes involved in the induction of competence for embryogenesis and subsequent embryo development presents a challenge for modern molecular biology. There are now many new molecular techniques, which will enable the dissection of these early events in the stages of commitment and differentiation of the plant. We can expect that over the next decade, there will be many basic advances in stem cell biology in both plant and animal systems. Furthermore, these advances will benefit the lives of humans, animals, plants, agriculture, and the environment.
Some reviews have been published dealing specifically with the molecular basis of vegetative-to-embryogenic transition [26, 42, 127, 150]. Although recent analysis of the proteome and transcriptome have led to the identification and characterization of new genes induced SE, there is no review on recent information of prime events SE induction. In this review, recent information on molecular basis of vegetative-to-embryogenic transition is described. Possible molecular basis by which different factors induce or modify embryogenic competence in cultured cells are discussed.
Genes and proteins
There are two different ways of induction of somatic embryogenesis: direct somatic embryogenesis (DSE) and indirect somatic embryogenesis (ISE). DSE is when a minimal proliferation of unorganized tissue precedes embryo formation; while in ISE, callus proliferates profusely before embryo formation. It has been suggested that in DSE, proembryogenic competent cells are already present and the expression of the embryogenetic program merely depends on favorable conditions and a minimal gene reprogramming is required for DSE, whereas in ISE, a major cell gene reprogramming is necessary for de-differentiation to acquire the embryogenic status [218].
During the past two decades, considerable efforts have been made to identify genes with altered expression patterns during SE. Various systems have been exploited to understand the mechanisms of gene regulation during SE and carrot has served as the model system [26, 42, 73, 74, 150, 152, 228]. Analysis of the proteome and transcriptome has led to the identification and characterization of certain genes involved in SE [7, 21, 22, 26, 57, 77, 107, 154, 158, 167, 187, 207, 220, 226]. Most of these genes, however, are upregulated only in the late developmental stages, suggesting that they do not play a direct role in the vegetative-to-embryogenic transition.
SERK genes
The search for genes that mark single somatic cells in transit to become embryogenic resulted in the discovery of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK). SERK is transiently expressed in a subpopulation of enlarged vacuolated cells in an embryogenic culture derived from cultured hypocotyl explants [173]. This cell type was the same as described earlier by Guzzo et al. [62] as being the embryogenic cell type. SERK-marked single cells detached from the proliferating hypocotyl provascular tissue only after prolonged exposure to 2,4-dichlorophenoxyacetic acid (2,4-d) [173], with an exposure time similar to that found by Guzzo et al. [62] to generate embryogenic cells [173]. Cell tracking experiments showed that the SERK-expressing cells could initiate SE. SERK was expressed in cells with varying morphology, but only cells of the enlarged vacuolated type formed somatic embryos. Both in somatic and zygotic carrot embryos SERK expression ceased in most cells after the globular stage, emphasizing the molecular similarity between somatic and zygotic embryogenesis. In addition, SERK was detected during flower development and was highest 10 days after pollination [173]. Homologs of SERK have been discovered in several plant species, which includes Dactylis glomerata [184], Zea mays [5], Medicago truncatula [130], Helianthus annuus [198], Oryza sativa [79], Citrus unshiu [178], Theobroma cacao [169, 170], and Solanum tuberosum [177]. Analysis of the sequenced Arabidopsis genome revealed that a family of five homologs of the carrot SERK was present. Also in maize and Medicago multiple SERK homologs were found three and five, respectively [5, 130]. The Arabidopsis SERKI protein contains all the main protein motifs found in other species [66, 72, 130, 198]. Postembryonically, Arabidopsis SERKI is expressed in vascular bundles [66, 100] and in developing lateral roots [100]. Mutants with enlarged SHOOT MERISTEMLESS (SAM) retain embryogenic competence after germination in Arabidopsis [124]. The SE system described by Mordhorst et al. [124] was used to follow the expression of SERKI in Arabidopsis during the initiation of an embryogenic culture in an altered meristem programl (ampl) mutant background. In response to the presence of 2,4-d in the induction medium, SERKI expression increased in the SAM and the vascular bundles [62, 124, 151, 173, 184]. Embryogenic structures originating from the SAM area showed SERKI expression. No expression was seen in non-embryogenic calli. SERKI expression was enhanced in the highly embryogenic amp1 cultures. Overexpression of SERKI does not result in any obvious plant phenotypes but gives a 3- to 4-fold increase in embryogenic competence, which indicates that SERKI not only marks embryogenic competence (EC) but also promotes the transition of somatic cells to an embryogenic state [66]. Arabidopsis SERKI is expressed before fertilization during both male and female sporophytic and gametophytic development [2, 66, 100] and after fertilization in the developing embryo until heart stage [66, 100]. In the monocot D. glomerata, SERKI was also found to closely follow the development of cells competent to form somatic embryos [184]. In Helianthus, SERKI expression increased in the morphogenetic zone of immature zygotic embryos under embryogenic culture conditions until 2 days of incubation, after which SERKI levels decreased. The increase and subsequent decrease in SERKI expression correlated with the acquisition and loss of EC. Similar to Daucus, Dactylis, and Arabidopsis, Helianthus SERKI continued to be expressed in developing embryonic structures, and expression ceased after 7 days of development. In addition, SERKI expression was detected in the provascular tissue and leaf primordia of the embryo [198]. The suppression of Oryza SERKI by RNA interference resulted in an inhibition and SERKI overexpression resulted in induction of shoot regeneration from callus. Whether this also reflects the embryogenic capacity of the tissue is not known. Interestingly, overexpression of Oryza SERKI also resulted in an increased resistance to blast fungus [72]. In both maize [5] and Medicago [130], SERKI expression was not tightly correlated with SE, because SERKI expression was found in both embryogenic and non-embryogenic callus.
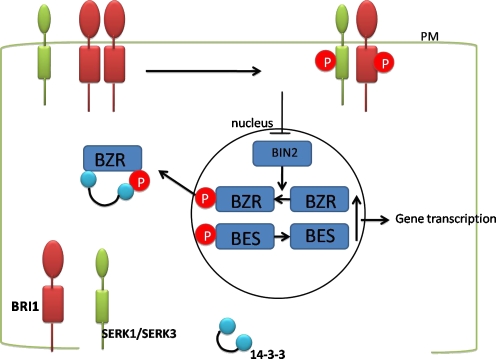
Membrane-located Leucine-Rich Repeat Receptor-Like Kinases (LRR-RLKs) play important roles in plant signaling pathways [204]. The SERK gene encodes a LRR-RLK. The predicted protein contains an N-terminal Leucine zipper domain followed by five LRRs, a serine and proline rich SPP domain, a transmembrane domain, and an intracellular serine/threonine kinase domain. The SPP domain is a unique feature of the SERK family of receptor kinases [66, 173]. Recently, SERK1 were shown in protein complexes that include components of the brassinosteroid signaling pathway such as BRASSINOSTEROID-INSENSITIVE 1 (BRI1) and its co-receptor BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1)/SERK3 [86]. Receptors such as BRI1 and SERK1 are synthesized in the endoplasmic reticulum, from where they pass through the Golgi network to be inserted into the plasma membrane (PM). Indeed, the use of fluorescently tagged BRI1 and SERK proteins has shown clearly both plasma and internal membrane localization [100]. Upon binding of brassinosteroid (BR) to the extracellular domain of BRI1, a negative regulator of BRI1 activity is released from the PM and increases the affinity of BRI1 for SERK3 [216]. Oligomerization of the BRI1/SERK3 receptors and transphosphorylation of the kinase domains takes place [216]. This inhibits, via an unknown pathway, the phosphorylation of BZR1 (for brassinazole resistant 1) by the BIN2 (for BR insensitive 2) kinase that, when phosphorylated, is translocated to [164] and retained in the cytoplasm via 14-3-3 proteins [52]. Subsequently, accumulation of dephosphorylated nuclear-localized BES1 and dephosphorylated BZR1 transcription factors induces the genetic response to brassinolide in Arabidopsis (Fig. 1). BAK1/SERK3 is not the only coreceptor of BRI1; SERK1 also interacts with BRI1 [86], and evidence has been presented that BKK1, identical to SERK4, also participates in BR signaling [65]. Thus, the classical model for ligand-induced heterooligomerization followed by auto- and transphosphorylation as well as translocation of target proteins appears to apply to receptors of the BRI1/SERK class while complexes of different composition may have different specificity. These results suggest that SERK1 may be involved in brassinolide signaling as well as in the acquisition of EC.
Fig. 1.
Interacting partners in brassinosteroid (BR) signaling. Upon binding of brassinolide (BL) to BRI1, SERK3 dissociates from the receptor and transphosphorylation take place with SERK1/SERK3. Subsequently phosphorylation of BZR1 by BIN2 is inhibited. This leads to accumulation of dephosphorylated BZR1 and BES1 in the nucleus, which induces gene transcription. Furthermore, phosphorylated BZR1 translocated to the cytoplasm is retained there by 14-3-3 proteins, and only dephosphorylated BZR1 translocates back to the nucleus
To summarize, it appears that in most plant species investigated, SERKI expression marks the acquisition of EC in tissue culture, but the gene is also expressed in non-embryogenic cells. In addition, there may be a correlation with organogenic competence in some tissue culture systems.
LEC genes
Arabidopsis leafy cotyledon (LEC) genes, LEC1 and LEC2, were identified originally as loss-of-function mutations resulting in defects in both embryo identity and seed maturation processes [63]. LEC genes play a central role in controlling many aspects of plant embryogenesis and their identification and analysis provided insight into their functions.
The LEC1 gene of Arabidopsis thaliana encodes a protein related to the Heme-Activated Proteins 3 (HAP3) subunit of the CCAAT box-Binding F LRR-RLKs actor (CBF) [102, 110]. Kwong et al. [101] identified genes encoding Arabidopsis HAP3 subunits in Phaseolus coccineus. One of them, closely related to LEC1 and named LEC1-LIKE (L1L), is required for normal embryo development [101]. When ectopically expressed, L1L can complement LEC1 functions [101]. A homolog of the L1L gene that encodes the HAP3 subunit of the CBF was found in the cocoa genome (TcL1L) [3]. The TcL1L conceptual translation product showed sequence similarity with the putative L1L protein isolated from P. coccineus [101] and H. annuus [40]. High TcL1L mRNA levels were detected in young somatic embryos and non-embryogenic explants did not show TcL1L expression. Its expression was restricted to young and immature embryos, and no expression was detected in mature embryos. The observation that TcL1L expression was detected before inside/outside patterning was initiated and was then strongly maintained in the protoderm confirmed the role of LEC genes in coordinating primary events leading to embryonic competence [63, 101]. The cells of this special cell layer are precisely the place of origin of secondary somatic embryos. The carrot homolog of Arabidopsis LEC1 was expressed in embryogenic carrot cells and in somatic embryos developing from them, but not in cells from a non-embryogenic culture [223].
LEC2 regulates many distinct aspects of embryogenesis [191]. For example, during the early morphogenesis phase of embryogenesis in which the basic body plan of the embryo is established, loss-of-function mutations in LEC2 affect the maintenance of embryonic cell fate and specification of cotyledon identity. Later in embryogenesis, lec2 mutants have cotyledon tips that do not accumulate storage reserves nor acquire desiccation tolerance, indicating defects in the initiation and/or maintenance of the maturation phase. Consistent with the pleiotropic effects of the lec2 mutation, LEC2 encodes a transcription factor with a B3 domain, a DNA binding region found thus far only in plant proteins [169, 170, 191].
Stone et al. [191] have shown that LEC1 and LEC2 genes may be involved in establishing EC in zygotic embryogenesis where both genes were detected in the earliest embryogenic stages tested. LEC1 and LEC2 were found to be sufficient to induce embryo development in vegetative cells when expressed ectopically [12, 50, 110, 191].
In Arabidopsis, substantial loss of embryogenic potential in cultured somatic cells caused by mutations in LEC genes suggests that LEC transcription factors activate genes essential for SE initiation. The complete inability of LEC mutants for direct SE and infrequent formation of single somatic embryos from callus tissue suggests that two genetically different pathways lead to somatic embryo formation: (1) a rapid and efficient direct embryogenesis which requires active LEC genes and (2) a much less efficient, slower pathway of indirect embryogenesis, in which LEC genes may not be necessary and which is preceded by cell dedifferentiation [50]. These assumptions are consistent with the observation that in wild-type Arabidopsis cultures, both direct and indirect developmental pathways can be induced [48, 75, 124].
Stone et al. [190] have shown that gene-encoding enzymes involved in auxin biosynthesis in SE of Arabidopsis are activated within 1 h after induction of LEC2 activity, and LEC2 may induce somatic embryogenesis in vegetative tissues, in part, through its enhancement of auxin activity. Consistent with a role for LEC2 in the induction of auxin-related processes, LEC2 promoter activity colocalizes with auxin maxima in the cells displaying embryogenic patterns of cell division in tissues undergoing somatic embryogenesis [98]. Auxin-induced somatic embryogenesis in Arabidopsis requires LEC1 and LEC2 expression [50]. Braybrook et al. [12] showed that ectopic expression of a 35S::LEC2:GR transgene activates IAA30 gene expression, indicating a potential link between auxin signaling and LEC2-induced somatic embryogenesis. We suggested that LEC gene may act by stimulating the production of plant hormones and/or increasing the sensitivity of the cell to these substances.
To summarize, the LEC genes are essential for in vitro SE induction. The strongly impaired in vitro embryogenic responses of lec mutant explants, manifested by the frequent formation of non-embryogenic callus, suggests that LEC genes are likely essential for changing cell fate from somatic to EC.
BBM genes
The Baby Boom (BBM) gene, which was isolated from microspore embryo cultures of Brassica napus [11], encodes a transcriptional factor belonging to the AP2/ERF family. The AP2/ERF transcription factor family is one of the largest in Arabidopsis, comprising of almost 150 genes that are differentially expressed (database of Arabidopsis transcription factors: http://datf.cbi.pku.edu.cn; [90, 91, 126, 168]. The AP2/ERF family has been organized into five phylogenetically distinct subfamilies that differ in the number of AP2/ERF domains, as well as the amino acid similarity between these domains [168]. Genes belonging to two of these subfamilies have been shown to enhance in vitro regeneration [4, 11], while others play a role in related processes controlling meristem cell fate and organ development [25, 38, 205]. One of these genes, BBM also bypasses the requirement for plant growth regulators to induce regeneration [186]. Passarinho et al. [142] used DNA microarray analysis in combination with a post-translationally regulated BBM:GR protein and cycloheximide to identify target genes that are directly activated by BBM expression in Arabidopsis seedlings. They suggest that the BBM transcription factor activates a complex network of developmental pathways associated with cell proliferation and growth. BBM AP2/ERF domain protein is a seed and root-meristem expressed transcription factor that was identified as a marker for embryo development in B. napus microspore-derived embryo cultures [11], as a gene showing preferential expression in the basal region of the Arabidopsis embryo [17] and as an auxin-inducible root expressed gene in M. truncatula [78]. Ectopic expression of BBM in Arabidopsis primarily induces spontaneous somatic embryo formation from seedlings, although ectopic shoots and callus also develop at a lower frequency [11]. In tobacco, heterologous BBM expression induces spontaneous shoot and callus formation, while a cytokinin pulse is required for somatic embryo formation [186]. The ability of BBM to promote organogenesis and embryogenesis in the absence of exogenously applied growth regulators suggested that BBM may act by stimulating the production of plant hormones and/or increasing the sensitivity of the cell to these substances. Klucher et al. [95] speculated that AP2/ERF domain proteins, being unique to plants, might have coevolved with plant-specific pathways such as hormone signal transduction.
AGL15 gene
Harding et al. [64] demonstrated that ectopic expression of AGAMOUS-Like 15 (AGL15) could enhance production of somatic embryos from cultured zygotic embryos and from the SAMs of seeds that complete germination in liquid media that contains 2,4-d and enhances production of secondary embryonic tissue from cultured zygotic embryos in Arabidopsis. Recently, Thakare et al. [197] reported that loss-of-function mutants of AGL15, alone or when combined with a loss-of-function mutant of a closely related family member, AGL18, show decreased ability to produce somatic embryos. AGL15 was initially identified using differential display of mRNA as an embryo expressed gene as well as during characterization of MADS-box genes in Arabidopsis [67]. Although the gene is expressed and the protein accumulates to its highest level in developing embryos, AGL15 is expressed in subsets of cells, generally at lower levels after the completion of germination [43, 67, 146]. MADS-domain proteins are a family of transcriptional regulatory factors found in eukaryotic organisms. In plants, MADS-domain proteins are central players in many developmental processes, including control of flowering time, homeotic regulation of floral organogenesis, fruit development, and seed pigmentation [139]. Interestingly and perhaps relevant for SE, AGL15 has been identified as a component of a SERK1 protein complex [86], and both SERK1 and AGL15, are expressed in response to auxin treatment [53, 130, 227]. Also intriguing are recent results that indicate that LEC2 may directly induce expression of AGL15 [12]. Like LEC2 and AGL15 impacts upon bioactive GA accumulation, but AGL15 mediates its effect at least in part by directly inducing expression of GA 2-oxidase6 that encodes a GA 2-oxidase that catabolizes biologically active GA [215]. Expression of this GA 2-oxidase affects somatic embryo development from the SAM of liquid culture-grown seedlings in the presence of 2,4-d [215].
MtSERF1 gene
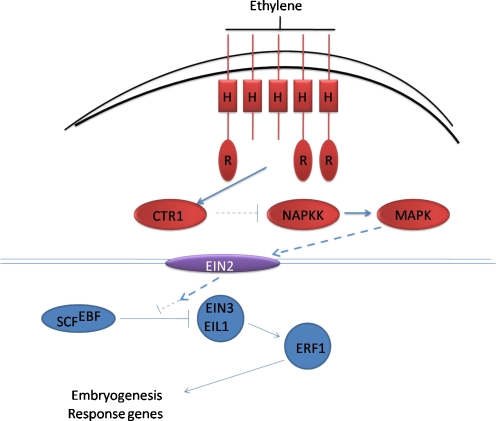
Mantiri et al. [114] reported that in inhibitors of ethylene biosynthesis and perception, it was shown that ethylene was necessary for SE in M. truncatula. They demonstrate that SOMATIC EMBRYO RELATED FACTOR1 (MtSERF1) is induced by ethylene and is expressed in embryogenic calli. RNA interference knockdown of this gene causes strong inhibition of SE. They have shown that MtSERF1 expression is inhibited by AVG and Ag+, indicating its ethylene dependence. This gene is a member of the ERF subfamily based on the classification of Nakano et al. [126]. ERF play an important role in hormone signal transduction, and they interconnect different hormone pathways [212]. Ethylene is perceived by a family of five receptors: ETR1, ERS1, ETR2, ERS2, and EIN4. Genetic and molecular studies have positioned these receptors upstream of the Raf-like MAP kinase kinase kinase, CTR1, which interacts with the receptors and also acts as a negative regulator (Fig. 2). The integral membrane protein, EIN2, and the transcription factors EIN3 and EIL1 are positive regulators of ethylene signaling downstream of CTR1. Current models propose that hormone binding inactivates the receptors, thus resulting in downregulation of CTR1 activity. Since the identification of CTR1, biologists have speculated that a MAP kinase cascade may be involved. Only recently, however, have putative MAP kinase and MAP kinase components of the ethylene pathway been identified [18]. Interestingly, these kinases appear to positively regulate ethylene response, suggesting that CTR1 must inhibit their function. If so, this would represent a novel twist on the traditional MAP kinase signaling paradigm. Precisely how the ethylene signal is transduced to the EIN3 and EIL1 transcription factors remains unclear. However, the recent finding that ethylene stabilizes these transcription factors, which are targeted for degradation by an SCF complex in the absence of ethylene, clearly indicates a role for the ubiquitin pathway [61, 147]. One of the known targets for EIN3 is the ERF1 transcription factor, which activates several genes involved in a subset of ethylene responses.
Fig. 2.
A model for ERF1 play role in ethylene signal transduction. Ethylene is perceived by a family of two-component receptors containing a consensus or degenerate HK domain (H). Three of the receptors also contain a C-terminal receiver domain (R). The receptors negatively regulate ethylene response together with CTR1 in a complex on the endoplasmic reticulum membrane. Perception results in reduced receptor and CTR1 activities and activation of a MAP kinase kinase, which transmits the signal through the EIN2 membrane protein, ultimately resulting in the activation of a transcriptional cascade in the nucleus
The finding of a relationship between an ERF subfamily gene and the formation of somatic embryos in vitro is consistent with an emerging picture of the involvement of ERF transcription factors in developmental processes studied in vitro.
MtSK1gene
The stress response induced by explant wounding and culture is increasingly recognized as an important component of somatic embryo induction [24]. Nolan et al. [131] have cloned and investigated the M. truncatula Stress Kinase1 (MtSK1) gene in relation to SE in M. truncatula. The induction of expression of MtSK1 in culture is not dependent on hormones in the culture medium with elevated expression on both hormone-containing and hormone-free media. Excising the tissue has upregulated MtSK1 expression. The likely role of MtSK1 in stress-induced signaling provides a way forward in relating the stress-response pathway to the auxin and cytokinin-induced pathways involved in the induction of SE in the M. truncatula culture system. MtSK1 is a member of the class of plant kinases called the SNF1-Related Kinase (SnRK) group. Members of the SnRK group of kinases are thought to play a role in stress responses of plants, and some of them are induced by Abscisic Acid (ABA) [71]. The connection between stress and SE has received increasing attention over recent years [42].
GST gene
The transcripts of Glutathione-S-Transferase (GST) genes were detected in abundance during auxin induction and in somatic embryos. GST transcripts have been shown to accumulate in Chicorium intybus [51], Medicago sativa (Thibaud-Nissen et al. 2003), Cyclamen persicum [220], and Gossypium hirsutum [226] somatic embryos and GST appears to be a major regulator of the interacting genes sequenced in the present case in response to auxin. Some GSTs are induced by auxin [125] and H2O2 [106], and might target transcription factors like WRKY [29] and transport certain gene products produced during oxidative stress to the vacuole [36]. Reactive oxygen species have been shown to act as second messenger during auxin and stress-induced embryogenesis [115]. On the other hand, GST are not only responsive to auxin but are also induced by other hormones, e.g., ABA and ethyl jasmonate and under various biotic and abiotic stresses and may have a possible role in detoxifying excessive amounts of auxin, thus regulating the intracellular concentration or its inactive analogs [60]. These results suggest that the roles of GST genes during acquisition of embryogenic potential are likely to be associated with protecting the cell against the harmful effects of reactive oxygen species.
WUS gene
WUSCHEL (WUS) is a homeobox gene which encodes a transcription factor that regulates the pool of stem cells in the shoot meristem and is regulated by a feedback loop involving the CLAVATA (CLV) genes [9]. Mutations in the CLVl, CLV2, and CLV3 genes result in a larger expression domain of WUS, which in turn results in an increase in SAM size [174]. Mutants with an enlarged SAM such as clv genes retain EC after germination [124, 213]. Zuo et al. [229] reported WUS gene promotes vegetative-to-embryonic transition in Arabidopsis in all tissues and organs tested, without any external plant hormones. This is because appropriate auxin transport and distribution are needed for embryo development and pattern formation. WUS transient overexpression causes highly embryogenic callus formation in the presence of auxin, whereas it directly induces somatic embryo formation from different plant organs in the absence of any exogenous auxin. Therefore, it appears that WUS can reprogram cell fate, bypassing the auxin requirement, or simply taking advantage of the endogenous auxin flux.
PKL gene
PICKLE (PKL) is necessary to ensure that traits expressed during embryogenesis and seed formation are not expressed after germination [133]. pkl seedlings are capable of expressing embryo-associated traits throughout the plant. In particular, the primary roots of pkl seedlings have been demonstrated to express many embryo specific traits after germination, including the accumulation of seed storage reserves and the ability to undergo SE [69, 133, 157]. During post-embryonic growth, PKL inhibits embryonic traits via transcriptional repression of seed storage proteins [133] and LEC genes [132, 160], and, therefore, is a master regulator of embryogenesis. The PKL gene encodes a CHD3 protein, a chromosome remodeling factor which is ubiquitously expressed in Arabidopsis [39, 133]. This suggests a possible role for chromatin remodeling in the coordination of transcription during the context of a stress-induced developmental switch, especially in the derepression of gene expression programs associated with somatic embryo induction.
GLPs
Germin-like proteins (GLPs) are a group of proteins sharing homology to cereal germins. Germins/GLPs are apoplastic proteins and are part of the cupin superfamily [34] that includes various proteins identified in many eukaryotes. The common feature of all these proteins is a conserved 3D structure that forms a six stranded beta barrel [35]. Some GLPs such as tobacco Nectarin I [175, 193] are reported to have SOD activity. It is interesting in terms of the physiological role of GLPs that both SOD and OXO generate hydrogen peroxide (H2O2) and that germin and GLPs are located on the apoplast. OXO can catalyze oxalic acid produced by several plant pathogens such as Sclerotinia sclerotiorum, and SOD can dismute superoxide radicals produced by the oxidative burst. In several cases, GLPs also seem to have non-enzymatic biochemical activities. They can act as auxin-binding proteins in peach [136] or serine protease inhibitors in wheat [175]. Thus, germins/GLPs gene expression is induced during biotic or abiotic stresses but can also be related with developmental regulation. In a developmental context, germin/GLP genes are often expressed during the early growth stages in wheat embryos [199], callus [14], pine [128], Arabidopsis (Member et al. 1997), and cotton [116] and during organ formation in Arabidopsis [119], barley [33], and potato [15]. Germins/GLPs could prevent cell expansion by increasing the number of links between polysaccharides and/or proteins within the cell wall as well as favoring lignification. Caliskan and Cuming [13] emphasize that wheat germins accumulate not only in cells that have ceased to expand within the plant but also in auxin-treated callus cells [14]. The role of apoplastic proteins in cell differentiation and organogenesis has been extensively studied in the conifer SE field. Variations in morphology between different embryonic cell lines have been correlated with differential protein secretion [37]. Such extracellular proteins can even restore embryogenic capacity to developmentally blocked conifer cell lines as observed in carrot [108]. In Pinus caribaea, comparison between the profiles of extracellular proteins of non-embryogenic and embryogenic cell lines [30] lead to the characterization of the first GLP identified in gymnosperms [30]. The cDNA corresponding to this protein was later isolated in a library and expression analysis confirmed the embryogenic specificity of this GLP [128].
ECPs
The induction of carrot SE, by treatment with various stresses, has been exploited to isolate those proteins and genes that are thought related to the acquisition of EC. These proteins, i.e., embryogenic cell proteins (ECPs), belong to the LEA protein groups [92, 93, 194]. The expression of the ECP genes is positively regulated by ABA, a phytohormone that is involved in abscission, dormancy, and drought tolerance [89]. The ABI3 gene was isolated based on studies of ABA-insensitive Arabidopsis mutants [97]. This gene is believed to be related to the seed-specific signal transduction of ABA [138]. A homolog of this gene in carrot was isolated and named C-ABI3 [180]. This gene is mainly expressed in embryonic tissue and positively regulates the expression of the ECP genes [89, 179, 180]. The endogenous levels of ABA also increase in response to stress treatments in various plants including Z. mays [165], Pisum sativum [41], and Brassica napus [171]. It has been reported that carrot embryogenic cell contain about 2.5 times more endogenous ABA than somatic embryos at the torpedo stage and about 67.5 times more than non-embryogenic cells that have lost the ability to form somatic embryos [92]. Furthermore, treatment with 10−4 M ABA induces embryo formation in carrot apical tip explants [129], and ABA also plays an important role in the induction of secondary SE in carrot [134]. Kikuchi et al. [89] reported that Somatic embryo formation was inhibited by the application of fluridone, a potent inhibitor of ABA biosynthesis, during stress treatment. These results suggest that the stress-induced accumulation of endogenous ABA is involved in the induction of carrot SE.
Trx H protein
One of the most interesting proteins identified in studies of proteomic analysis of SE of M. truncatula using mesophyll protoplasts [76] and explant cultures [77] was Trx H. This MtTrx H protein is highly homologous (71% identical) to the A. thaliana ah1 protein, which belongs to the subgroup 1 of the plant Trx H family. The members of the Trx H group are encoded by a multigenic family of eight genes in Arabidopsis [156]. The Trx H group is ubiquitous proteins, which regulate a myriad of posttranscriptional biological functions in eukaryotic cells and are involved in reserve breakdown that sustains early seedling growth [56]. The reduction of the first subgroup of Trx H are mediated by NADPH-thioredoxin reductase, and many of these reactions take place in specific cells and play a role in the redox regulation of components of the vascular tissues [56]. The Trx H group of proteins is involved in a wide scope of biological functions, acting as cofactors, transcription regulators, protein binding regulators, protein folding catalysts, growth factors, and antioxidants. In somatic embryo formation from explant cultures, MtTrx H was reduced in expression at 5 weeks and could not be detected in the 8-week-old cultures. These results suggest that MtTrx H plays an important role during early stages of commitment from the vegetative stage to a pathway of cellular differentiation and proliferation.
PGRs
Among different external stimuli that induce an embryogenic pathway of development plant growth regulators (PGRs) such as auxins and cytokinins used for in vitro media have been the most frequently considered, as they regulate the cell cycle and trigger cell divisions [45]. The level of endogenous phytohormones is considered as one of the crucial factors influencing embryogenic potential of explants [49, 80, 81].
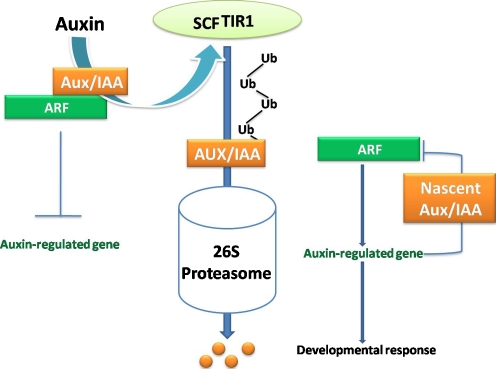
Auxin is a PGR that elicits diverse plant responses ranging from cell division, differentiation, cell elongation, root initiation, and apical dominance to tropic responses [6]. Auxin is considered to be a positional and pattering signal molecule that plays a major role in zygotic embryogenesis [185] and SE [42, 143, 26]. There have been numerous studies concerning the hormonal induction of SE in a wide range of species. A significant amount of literature on auxin biosynthesis, metabolism, and transport in embryos has grown out of extensive analysis which shows that auxin plays important roles both in induction of embryo formation in culture and in the subsequent elaboration of proper morphogenesis during embryo development [42, 149]. One of the well-established functions of the plant ubiquitin/proteasome pathway is in auxin signaling as illustrated in Fig. 3 [68]. In this pathway, auxin promotes the breakdown of certain auxin/indole-3-acetic acid (Aux/IAA) repressor proteins through the action of the ubiquitin protein ligase SCFTIR1, which are believed to block the auxin-response factors. Mutations in the Transport Inhibitor Responsor1 (TIR1) gene confer reduced auxin response [163]. TIR1 encodes a nuclear protein belonging to the F-box protein family of Arabidopsis, which has approximately 700 members. TIR1 interacts with the core SCF subunits [58], the SCFTIR1 complex as a positive regulator of auxin response and suggested a model invoking the SCFTIR1- mediated ubiquitinylation of a repressor of auxin signaling. Additional Auxin Signaling F-Box protein is highly related to TIR1 and were recently shown to exhibit auxin-dependent binding to Aux/IAA proteins [29]. The Early known candidate for an auxin receptor mediating auxin-regulated cell expansion is Axin-Bnding Potein 1 (ABP1), discovered 31 years ago [155]. The knockout mutant of the ABP1 in Arabidopsis was characterized by an embryo-lethal phenotype [23], thus indicating its important role during embryogenesis. In carrot, auxin depletion leads to the inactivation of some genes, thus enabling the embryogenic program to proceed [28].
Fig. 3.
The Ubiquitin-mediated proteolysis of Aux/IAA proteins regulates auxin response. In the absence of an auxin stimulus, Aux/ IAA proteins inhibit ARF transcriptional activity by forming heterodimers. Auxin perception (by an unknown receptor) targets the Aux/IAA proteins to the SCFTIR1 complex, resulting in their ubiquitination and degradation, thereby derepressing the ARF transcription factors. Among the ARF targets are the Aux/IAA genes themselves, which produce nascent Aux/IAA proteins that restore repression upon the pathway in a negative feedback loop
The abolition and reestablishment of polarity in the cultured explant/cells is the foremost event enabling SE to proceed. One possible polarity-controlling mechanism is the orientation of auxin movement in the surrounding tissues, which in seed bearing plants is in the reverse direction (endoscopic embryogenesis) to that of the developing embryo [219]. Treatments such as explant wounding, or exposure to medium containing 2,4-d, may result in the rapid proliferation of cells that become undifferentiated (or de-differentiated) and lose their original polarity [219] or attain a hyperpolarized state [59]. This interference with polar auxin transport abolishes auxin gradients, which subsequently halts the programmed organogenesis and tropic responses [195, 196]. This involves the erasure of existing transcriptional and translational patterns in order to redirect the developmental program of cells [42] and is marked by an increase in endogenous auxin levels [143], which along with exogenous auxin application, is a critical factor during the induction and expression of SE [81]. At the molecular level, Auxin influx carrier protein1/ polar auxin transport inhibitor resistant protein1 (AUX1), PINFORMED (PIN), and P-GLYCOPROTEIN (PGP) classes of auxin influx and efflux facilitators are responsible for maintaining the characteristic polar auxin transport or gradient [55, 137, 141] by their asymmetric localization across the plasma membrane [10, 55, 192]. This coordinated transporter-dependent differential auxin distribution is crucial for embryo development and other patterning and developmental events in plants. AUX1 and PIN regulate proton gradient-driven movement of auxin across the plasma membrane, while PGP mainly operates through an energized auxin transport [120]. The Arabidopsis PIN gene family consists of eight members and their polarity rearrangements define one of the earliest events in the regulation of different patterning and organogenesis processes [120]. During the earliest developmental stages, PIN1 is first expressed in pro-embryogenic cells in a nonpolar manner and then becomes polarized to basal side of provascular cells by the attainment of early globular stage [46, 189].
Among different auxins, 2,4-d was the most commonly applied for somatic embrogenesis induction. In more than 65% of the recent protocols, 2,4-d as applied alone or in combination with other PGRs and many in vitro SE systems rely on the use of exogenous 2,4-d as an inducer [49]. In general, competent cells arise from explants cultured in media supplemented with strong synthetic auxins such 2,4-d. 2,4-d may have several roles in this process, acting as an auxin directly or modifying intracellular indole acetic acid (IAA) metabolism and/or as a “stressor” [42, 151]. Genes which were studied for auxin induction were also found to be responsive to various abiotic stresses. 2,4-d is known to induce many stress-related genes [27, 131, 140, 143, 148]. In soybean, somatic embryos is induced by 2,4-d in cotyledons and is associated with upregulation of oxidative stress and defense genes (Thibaud-Nissen et al. 2003). However, 2,4-d is also a strong herbicide and the concentration of IAA required for the induction of SE is over 103 times the endogenous free IAA level [159]. Therefore, 2,4-d is thought to function as a stress substance rather than as a phytohormone, triggering the acquisition of EC by plant cells. However, it is not known how and why 2,4-d is so effective in the induction of EC.
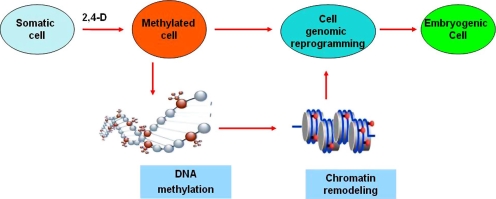
In many reports have been shown that the formation of an embryogenic cell is related to nuclear DNA hypermethylation in the presence of 2,4-d [104, 109, 222]. In Cucurbita pepo, the highest rate of DNA methylation occurred in the early embryo stages, predominantly on medium containing 2,4-d and DNA methylation decreased during embryo maturation on auxin-free medium [104]. A carrot DNA methyltransferase gene, Met1–5 was expressed transiently after the induction of SE by 2,4-d, before the formation of embryogenic cell clumps and 5-azacytidine, an inhibitor of DNA methylation suppressed the formation of embryogenic cell clumps from epidermal carrot cells [222]. Arabidopsis plants with an antisense MET1 transgene, partial-loss-of-function met1 mutations, or cmt3 drm1 drm2 mutations revealed that reduced DNA methylation results in abnormal postembryonic plant development [16, 44, 83–85, 87, 162, 172, 221]. How does DNA methylation affect in acquiring the EC? DNA methylation is a unique and noteworthy process because it involves the covalent modification of a cell’s genetic material [54, 94] and plays an important role to modify the information content of the underlying genetic sequence and gene expression [8]. Xiao et al. [221] show that DNA methylation performed by MET1 influences gene expression during embryogenesis in Arabidopsis. Therefore, dynamic changes in chromatin structure by DNA methylation at presence of 2,4-d leads to genomic reprogramming in somatic cells and hundreds of genes specifically required for acquiring the embryogenic competence are expressed (Fig. 4).
Fig. 4.
A model of the acquiring the embryogenic competence by DNA methylation at present of 2,4-d. DNA methylation afterward chromatin remolding take place in somatic cell. At last, somatic cell was undergoing genomic reprogramming and acquiring the embryogenic competence
Cytokinins are plant hormones that influence diverse processes of growth and development, such as cell proliferation and differentiation, vascular morphogenesis, shoot development, chloroplast morphogenesis, leaf senescence, and axillary bud dormancy [90, 91, 112, 113, 195, 196]. There is support for the idea that cytokinin, in general, are essential during the initial cell division phase of SE, but not for later stages of embryo development and maturation in carrot [200]. It has been previously observed that incorporation of zeatin into the medium during days 3 and 4 of culture, promotes the formation of carrot embryos to a great extent, probably due to enhancement of the cell division that occurs during this period in the cell clusters that is considered to be one of the most important events during the embryogenetic process [47]. In general, auxins are considered to have a major impact on the induction of somatic embryos in most regeneration systems [153]. However, initiation of somatic embryos on medium containing cytokinin as the sole PGR has also been reported in few species such as, H. annuus [20], Spinacia oleracea [225], Corydalis yanhusuo [166], Leptadenia reticulate [117], Dianthus caryophyllus [86], and Elaeagnus angustifolia (Karami O, Piri K, under review). In carrot, purine riboside, an anticytokinin, inhibited direct SE, and this effect was nullified by the application of cytokinin [200]. This observation does not rule out auxins as a major link in the signal transduction chain because the internal auxin concentration, either globally or on a local scale, can be indirectly influenced by the media composition and general growth conditions. Such hypothetical local variation of auxin could be the result of de novo synthesis, catabolism, conjugation, or a relocation of preexisting, auxin forms, within the explant. If such changes in internal auxin concentrations indeed contributed to the induction of somatic embryos, they could be expected to be mimicked, in part, by external addition of auxin. It is clear that external supply can only be a gross approximation of the natural situation because local differences are likely to be swamped.
Cell–cell communication
Cell–cell communication and signaling are controlled by the intercellular trafficking of signal molecules through the cytoplasmic channels called plasmodesmata (PD) and plasma membrane receptors (PMRs) [1, 161]. PD and PMR play an important role in development, coordination of the physiology between different tissues, and disease defenses [161]. Signals can be transmitted via receptor–ligand interactions in both plant and animal cells. However, PD and PMR provide plants with a unique means of intercellular communication, where each plant cell can form direct conduits to its neighbors, forming domains of cells sharing common components.
The acquisition of an embryogenic state is not a cell-autonomous process, but requires cell–cell communication [123]. It was found that higher cell densities in a culture containing both embryogenic and non-embryogenic cells increases the amount of embryogenic cells [28, 96, 201]. Next to that, adding preconditioned medium obtained from an embryogenic culture [28] or co-cultivation with zygotic embryos [70] also increases the embryogenic potential of embryogenic cultures. Labeling of cells with the JIM8 antibody, which recognizes a certain arabinogalactan protein (AGP) epitope present in embryogenic cultures, is indicative for the EC of a cell line [144, 202]. Cells labeled with JIM8 were believed to be in a transition towards an embryogenic state because a subpopulation of small cytoplasm rich cells in an embryogenic culture was recognized by JIM8 [144]. Cell tracking of the JIM8-labeled cells, however, did not reveal a relationship between embryogenesis and JIM-8 labeling, suggesting that the JIM8-labeled cells perform an accessory function in SE [202]. The finding that in the absence of the JIM8-labeled cell population, no somatic embryos develop, but callus is formed, demonstrates the importance of cell–cell communication in the maintenance of EC in culture [145].
The carrot EP3 endochitinase gene is expressed in an embryogenic culture, but its expression is not correlated with embryogenic cells and is absent from somatic embryos [208]. EP3 is secreted into the medium and addition of purified EP3 to a suspension culture of the temperature sensitive carrot mutant tsII rescued somatic embryo development. So, the expression of EP3 in non-embryogenic cells and its importance for SE again suggested that cell–cell communication is essential for SE in culture [82]. AGPs are a family of glycosylated hydroxyproline-rich glycoproteins analogous to animal proteoglycans [181]. AGPs are widely distributed in the plant kingdom, mainly attached to the plasma membrane or in cell walls. However, AGPs are also present in plant secretions. AGPs are implicated in three fundamental cellular processes: cell proliferation, cell expansion, cell differentiation, programmed cell death, and cell–cell communication [181, 188]. Furthermore, various AGPs play an important role in plant embryogenesis [19, 105, 188, 203, 206, 209, 214]. The presence of AGPs stimulating somatic embryogenesis was reported in carrot [206] and Caribbean pine [31]. van Hengel et al. [209] presented evidence that AGP side chains with intact arabinogalactan carbohydrate moieties are essential for the effect on somatic embryogenesis, whereas hydrolytic activation with endochitinases appears essential for full embryo-forming activity of the AGPs. It was found that non-embryogenic carrot lines can become embryogenic again after the addition of certain AGPs [99]. In addition, AGPs restore the EC after cell wall removal, and this restoration was more efficient when using chitinase cleaved forms of the AGPs [209]. These findings combined indicate that complex interactions between cells and substances secreted in the medium of embryogenic cultures are essential to establish and maintain EC in culture. So, in other words, it seems that as observed in intact tissues the unorganized embryogenic culture "niche" maintains its own population of totipotent stem cells.
Conclusions and future perspectives
Somatic embryogenesis is a unique system to investigate the mechanisms that operate during the transition of a single somatic cell into an embryogenic entity with the potential of developing into a complete plant. Early phases of SE are characterized by the induction of many genes. Despite the progress achieved during the last few years in understanding the muscular mechanisms involved in SE, there are still many aspects that are not fully understood and need to be studied in more detail. It is also not known the key molecular steps in common in all cases and why so many different conditions can be used to initiate somatic embryogenesis. Future research in this area must center not only on isolating and characterizing large numbers of genes expressed in early phases of SE but also on deciphering the significance of these genes by demonstrating what happens when their function is disrupted. This is being attempted either by creating transgenic plants that express an antisense construct or by working with genes that have already been disrupted through loss-of-function mutations.
References
- 1.Aker J, Vries SC. Plant Physiol. 2008;147:1560. doi: 10.1104/pp.108.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht C, Russinova E, Hecht V, Baaijens E, Vries S. Plant Cell. 2005;17:3337. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alemanno L, Devic M, Niemenak N, Sanier C. Planta. 2008;227:853. doi: 10.1007/s00425-007-0662-4. [DOI] [PubMed] [Google Scholar]
- 4.Banno H, Ikeda Y, Niu Q-W, Chua N-H. Plant Cell. 2001;13:2609. doi: 10.1105/tpc.13.12.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudino S, Hansen S, Brettschneider R, Hecht VRG, Dresselhaus T, Lorz H, Dumas C, Rogowsky PM. Planta. 2001;213:1. doi: 10.1007/s004250000471. [DOI] [PubMed] [Google Scholar]
- 6.Becker D, Hedrich R. Plant Mol Biol. 2002;49:349. doi: 10.1023/A:1015211231864. [DOI] [PubMed] [Google Scholar]
- 7.Ben C, Hewezi T, Jardinaud MF, Bena F, Ladouce N, Moretti S, Tamborindeguy C, Liboz T, Petitprez M, Gentzbittel L. Plant Molecular Biology. 2005;57:255. doi: 10.1007/s11103-004-7532-2. [DOI] [PubMed] [Google Scholar]
- 8.Bender J. Annul Review Plant Biology. 2004;55:41. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 9.Bhalla PL, Singh MB. Plant Cell Rep. 2006;25:249. doi: 10.1007/s00299-005-0071-8. [DOI] [PubMed] [Google Scholar]
- 10.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J. Nature. 2005;433:39. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 11.Boutilier K, OVringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, Lammeren AA, Miki BL, Custers JB, Lookeren Campagne MM. Plant Cell. 2002;14:1737. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braybrook SA, Stone SL, Park S, Bui AQ, Lee BH, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2006;103:3468. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caliskan M, Cuming AC. Plant J. 1998;15:165. doi: 10.1046/j.1365-313X.1998.00191.x. [DOI] [PubMed] [Google Scholar]
- 14.Caliskan M, Turet M, Cuming AC. Planta. 2004;219:132. doi: 10.1007/s00425-003-1199-9. [DOI] [PubMed] [Google Scholar]
- 15.Campbell MA, Lara MH, Jennifer C. Plant Physiol. 1998;118:711. doi: 10.1104/pp.118.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao X. Curr Biol. 2003;13:2212. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 17.Casson S, Spencer M, Walker K, Lindsey K. Plant J. 2005;42:111. doi: 10.1111/j.1365-313X.2005.02355.x. [DOI] [PubMed] [Google Scholar]
- 18.Chang C. Trends Plant Sci. 2003;8:365. doi: 10.1016/S1360-1385(03)00156-0. [DOI] [PubMed] [Google Scholar]
- 19.Chapman A, Blervacq AS, Vasseur J, Hilbert JL. Planta. 2000;211:305. doi: 10.1007/s004250000299. [DOI] [PubMed] [Google Scholar]
- 20.Charriere F, Hahne G. Plant Sci. 1998;137:63. doi: 10.1016/S0168-9452(98)00128-9. [DOI] [Google Scholar]
- 21.Che P, Lall S, Nettleton D, Howell SH. Plant Physiol. 2006;141:620. doi: 10.1104/pp.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Che P, Love TM, Frame BR, Wang K, Carriquiry AL, Howell SH. Plant Mol Biol. 2006;62:1. doi: 10.1007/s11103-006-9013-2. [DOI] [PubMed] [Google Scholar]
- 23.Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. Genes Dev. 2001;15:902. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. Plant Physiology. 2002;129:661. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuck G, Meeley RB, Hake S. Genes Dev. 1998;12:1145. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chugh A, Khurana PJ. Curr Sci. 2002;83:715. [Google Scholar]
- 27.Davletova S, Meszaros T, Miskolczi P, Oberschall A, Torok K, Magyar Z, Dudits D, Deak M. J Exp Bot. 2001;52:215. doi: 10.1093/jexbot/52.355.215. [DOI] [PubMed] [Google Scholar]
- 28.Vries SC, Booij H, Meyerink P, Huisman G, Wilde HD, Thomas TL. Planta. 1988;176:196. doi: 10.1007/BF00392445. [DOI] [PubMed] [Google Scholar]
- 29.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. Dev Cell. 2005;9:109. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Domon JM, Dumas B, Laine E, Meyer Y, David A, David H. Plant Physiology. 1995;108:141. doi: 10.1104/pp.108.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domon JM, Neutelings G, Roger D, David A, David H. J Plant Physiol. 2000;156:33. [Google Scholar]
- 32.Du L, Chen Z. Plant J. 2000;24:837–47. doi: 10.1046/j.1365-313x.2000.00923.x. [DOI] [PubMed] [Google Scholar]
- 33.Dumas B, Freyssinet G, Pallett KE. Tissue-specific expression of germin-like oxalate oxidase during development and fungal infection of barley seedlings. Plant Physiol. 1995;107:1091. doi: 10.1104/pp.107.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunwell JM. Biotechnol Genet Eng Rev. 1998;15:1. doi: 10.1080/02648725.1998.10647950. [DOI] [PubMed] [Google Scholar]
- 35.Dunwell JM, Purvis A, Khuri S. Phytochemistry. 2004;65:7. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Edwards R, Dixon DP, Walbot V. Trends Plant Sci. 2000;5:193. doi: 10.1016/S1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- 37.Egertsdotter U, Arnold S. Physiology Plant. 1995;93:334. doi: 10.1111/j.1399-3054.1995.tb02237.x. [DOI] [Google Scholar]
- 38.Elliot RC, Betzner AS, Huttner E, Oakes M, Tucker WQJ, Gerentes D, Parez P, Smith DR. Plant Cell. 1996;8:155. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eshed Y, Baum SF, Bowman JL. Cell. 1999;99:199. doi: 10.1016/S0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- 40.Fambrini M, Durante C, Cionini G, Geri C, Giorgetti L, Michelotti V, Salvini M, Pugliesi C. Development Genes Evaluation. 2006;1216:253. doi: 10.1007/s00427-005-0050-7. [DOI] [PubMed] [Google Scholar]
- 41.Fedina IS, Tsonev TD, Guleva EI. J Plant Physiol. 1994;143:245. [Google Scholar]
- 42.Feher A, Pasternak TP, Dudits D. Plant Cell Tissue Organ Cult. 2003;74:201. doi: 10.1023/A:1024033216561. [DOI] [Google Scholar]
- 43.Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC. Plant Cell. 2000;12:183. doi: 10.1105/tpc.12.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finnegan EJ, Peacock WJ, Dennis ES. Development. 1996;93:8449. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Francis D, Sorrell DA. Plant Growth Regul. 2001;33:1. doi: 10.1023/A:1010762111585. [DOI] [Google Scholar]
- 46.Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T. Nature. 2003;426:147. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 47.Fujimura T, Komamine A. Plant Physiol. 1980;64:162. doi: 10.1104/pp.64.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaj MD. Plant Cell Tissue Organ Cult. 2001;64:39. doi: 10.1023/A:1010679614721. [DOI] [Google Scholar]
- 49.Gaj MD. Plant Growth Regul. 2004;43:27. doi: 10.1023/B:GROW.0000038275.29262.fb. [DOI] [PubMed] [Google Scholar]
- 50.Gaj MD, Zhang S, Harada JJ, Lemaux PG. Planta. 2005;222:977. doi: 10.1007/s00425-005-0041-y. [DOI] [PubMed] [Google Scholar]
- 51.Galland R, Randoux B, Vasseur J, Hilbert JL. Biochemistry Biophysics Acta. 2001;1522:212. doi: 10.1016/s0167-4781(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 52.Gampala S, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron J. Dev Cell. 2007;13:177. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. Dev Cell. 2004;7:373. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 54.Gehring M, Henikoff S. Biochemistry Biophysics Acta. 2007;1769:276. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Geisler M, Murphy A. FEBS Lett. 2006;580:1094. doi: 10.1016/j.febslet.2005.11.054. [DOI] [PubMed] [Google Scholar]
- 56.Gelhaye E, Rouhier N, Jacquot JP. Plant Physiol Biochem. 2004;42:265. doi: 10.1016/j.plaphy.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Giroux RW, Pauls KP. Plant Mol Biol. 1997;33:393. doi: 10.1023/A:1005786826672. [DOI] [PubMed] [Google Scholar]
- 58.Gray WM, Hellmann H, Dharmasiri S, Estelle M. Plant Cell. 2002;14:213744. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grebe M, Friml J, Swarup R, Ljung K, Sandberg G, Terlou M. Curr Biol. 2002;12:329. doi: 10.1016/S0960-9822(02)00654-1. [DOI] [PubMed] [Google Scholar]
- 60.Guilfoyle TJ. Auxin-regulated genes and promotors. In: Hooykaas PJJ, Hall MA, Libbenga KR, editors. Biochemistry and molecular biology of plant hormones. Amsterdam: Elsevier; 1999. p. 423. [Google Scholar]
- 61.Guo H, Ecker JR. Cell. 2003;115:667. doi: 10.1016/S0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 62.Guzzo F, Baldan B, Levi M, Sparvoli E, Schiavo FL, Terzi M, Mariani P. Protoplasma. 1995;185:1. doi: 10.1007/BF01272751. [DOI] [Google Scholar]
- 63.Harada JJ. J Plant Physiol. 2001;158:405. doi: 10.1078/0176-1617-00351. [DOI] [Google Scholar]
- 64.Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE. Plant Physiol. 2003;133:653. doi: 10.1104/pp.103.023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, Li J. Curr Biol. 2007;17:1109. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 66.Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, Vries SC. Plant Physiol. 2001;127:803. doi: 10.1104/pp.010324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heck GR, Perry SE, Nichols KW, Fernandez DE. Plant Cell. 1995;7:1271. doi: 10.1105/tpc.7.8.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hellmann H, Estelle M. Science. 2002;297:793. doi: 10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- 69.Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, Vries SC, Ogas J. Plant Physiology. 2004;134:995. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holm PB, Knudsen S, Mouritzen P, Negri D, Olsen FL, Roue C. Plant Cell. 1994;6:531. doi: 10.1105/tpc.6.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halfordm N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu J-K, Harmon AC. Plant Physiol. 2003;132:666. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu H, Xiong L, Yang Y. Planta. 2005;222:107–117. doi: 10.1007/s00425-005-1534-4. [DOI] [PubMed] [Google Scholar]
- 73.Ikeda M, Umehara M, Kamada H. Plant Biotechnol. 2006;23:153. [Google Scholar]
- 74.Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH. Plant Cell Physiology. 2006;47:1443. doi: 10.1093/pcp/pcl023. [DOI] [PubMed] [Google Scholar]
- 75.Ikeda-Iwai M, Satoh S, Kamada H. J Exp Bot. 2002;53:1575. doi: 10.1093/jxb/erf006. [DOI] [PubMed] [Google Scholar]
- 76.Imin N, Jong F, Mathesius U, Noorden G, Saeed NA, Wang XD, Rose RJ, Rolfe BG. Proteomics. 2004;4:1883. doi: 10.1002/pmic.200300803. [DOI] [PubMed] [Google Scholar]
- 77.Imin N, Nizamidin M, Daniher D, Nolan KE, Rose RJ, Rolfe BG. Plant Physiol. 2005;137:1250. doi: 10.1104/pp.104.055277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imin N, Nizamidin M, Wu T, Rolfe BG. J Exp Bot. 2006;58:439. doi: 10.1093/jxb/erl224. [DOI] [PubMed] [Google Scholar]
- 79.Ito Y, Takaya K, Kurata N. Biochemistry Biophysics Acta. 2005;1730:253. doi: 10.1016/j.bbaexp.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Jimenez VM. Plant Growth Regul. 2005;47:91. doi: 10.1007/s10725-005-3478-x. [DOI] [Google Scholar]
- 81.Jimenez VM. Revista Brasileira de Fisiologia Vegetal. 2001;13:196. doi: 10.1590/S0103-31312001000200008. [DOI] [Google Scholar]
- 82.Jong AJ, Hendriks T, Meijer EA, Penning M, LoSchiavo F, Terzi M, Kammen A, Vries SC. Development Genetic. 1995;16:332. doi: 10.1002/dvg.1020160406. [DOI] [Google Scholar]
- 83.Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Proc Natl Acad Sci USA. 1996;93:12406. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kakutani T, Kato M, Kinoshita T, Miura A. Cold Spring Herb Symptom Quant Biology. 2004;69:139. doi: 10.1101/sqb.2004.69.139. [DOI] [PubMed] [Google Scholar]
- 85.Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Genetics. 2003;163:1109. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karami O, Deljou A, Karimi Kordestani G. Plant Cell Tissue Organ Cult. 2008;92:273. doi: 10.1007/s11240-007-9332-2. [DOI] [Google Scholar]
- 87.Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, Vries S. Plant Cell. 2006;18:626. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. Curr Biol. 2003;13:421. doi: 10.1016/S0960-9822(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 89.Kidner CA, Martienssen RA. Development Biology. 2005;280:504. doi: 10.1016/j.ydbio.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 90.Kikuchi A, Sanuki N, Higashi K, Koshiba T, Kamada H. Planta. 2006;223:637. doi: 10.1007/s00425-005-0114-y. [DOI] [PubMed] [Google Scholar]
- 91.Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. Proc Natl Acad Sci USA. 2006;103:814. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim S, Soltis PS, Wall K, Soltis DE. Mol Biol Evol. 2006;23:107. doi: 10.1093/molbev/msj014. [DOI] [PubMed] [Google Scholar]
- 93.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K, Higashi K, Satoh S, Kamada H, Harada H. Planta. 1992;186:337. doi: 10.1007/BF00195313. [DOI] [PubMed] [Google Scholar]
- 94.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K, Kamada H, Harada H. Plant Mol Biol. 1993;21:1053. doi: 10.1007/BF00023602. [DOI] [PubMed] [Google Scholar]
- 95.Klose RJ, Bird AP. Trends Biochem Sci. 2006;31:89. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 96.Klucher KM, Chow H, Reiser L, Fischer RL. Plant Cell. 1996;8:137. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Komamine A, Matsumoto M, Tsukahara M, Fujiwara A, Kawahara R, Ito M, Smith J, Nomura K, Fujimura T. Mechanism of somatic embryogenesis in cell cultures-physiology, biochemistry and molecular biology. In: Nijkamp HJJ, Plas LHW, Aartrijk J, editors. Progress in plant cellular and molecular biology, current plant science and biotechnology in agriculture. Dordrecht: Kluwer; 1990. p. 307. [Google Scholar]
- 98.Koornneef M, Reuling G, Karessen CM. Plant Physiol. 1984;61:377. doi: 10.1111/j.1399-3054.1984.tb06343.x. [DOI] [Google Scholar]
- 99.Kreuger M, Holst GJ. Planta. 1993;189:243. doi: 10.1007/BF00195083. [DOI] [Google Scholar]
- 100.Kurczynska EU, Gaj MD, Ujczak A, Mazur E. Planta. 2007;226:619. doi: 10.1007/s00425-007-0510-6. [DOI] [PubMed] [Google Scholar]
- 101.Kwaaitaal MA, Vries SC, Russinova E. Protoplasma. 2005;226:55. doi: 10.1007/s00709-005-0111-9. [DOI] [PubMed] [Google Scholar]
- 102.Kwong RM, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. Plant Cell. 2003;15:5. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee H, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2003;100:2152. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Legrand S, Hendriks T, Hilbert Je-L, Quillet M-C. BMC Plant Biology. 2007;7:27. doi: 10.1186/1471-2229-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leljak-Levanic D, Naana B, Jelaska MS. Plant Cell Report. 2004;23:120. doi: 10.1007/s00299-004-0819-6. [DOI] [PubMed] [Google Scholar]
- 106.Letarte J, Simion E, Miner M, Kasha-Ken J. Plant Cell Report. 2006;24:691. doi: 10.1007/s00299-005-0013-5. [DOI] [PubMed] [Google Scholar]
- 107.Levine A, Tenhaken R, Dixon R, Lamb C. Cell. 1994;79:583. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 108.Lin X, Hwang GJ, Zimmerman JL. Plant Physiol. 1996;112:1365. doi: 10.1104/pp.112.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lo Schiavo F, Giuliano G, Vries S, Genga A, Bollini R, Pitto L, Cozzani F, Nuti-Ronchi V, Terzi M. Molecular Gene Genetic. 1990;223:385. doi: 10.1007/BF00264444. [DOI] [PubMed] [Google Scholar]
- 110.Loschiavo F, Pitto L, Giuliano G, Torti G, Nuit-Ronchi V, Marazziti D, Vergara R, Orselli S, Terzi M. Theory Apply Genetic. 1989;77:325. doi: 10.1007/BF00305823. [DOI] [PubMed] [Google Scholar]
- 111.Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Tamagishi K, Fisher RL, Goldberg RB, Harada JJ. Cell. 1998;93:1195. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 112.Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. Development. 1999;126:469. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- 113.Mahonen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Tormakangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y. Science. 2006;311:94. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 114.Mahonen AP, Higuchi M, Tormakangas K, Miyawaki K, Pischke MS, Sussman MR, Helariutta Y, Kakimoto T. Curr Biol. 2006;16:1116. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 115.Mantiri FR, Kurdyukov S, Lohar DP, Sharopova N, Saeed NA, Wang X-D, VandenBosch KA, Rose RJ. Plant Physiol. 2008;146:1622. doi: 10.1104/pp.107.110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maraschin SF, Priester W, Spaink HP, Wang M. J Exp Bot. 2005;56:1711. doi: 10.1093/jxb/eri190. [DOI] [PubMed] [Google Scholar]
- 117.Suarez MF, Bozhkov PV. Methods Mol Biol. 2008;427:1–14. doi: 10.1007/978-1-59745-273-1_14. [DOI] [PubMed] [Google Scholar]
- 118.Martin KP. BIOLOGIA, PLANTARUM. 2004;48:285. doi: 10.1023/B:BIOP.0000033457.09115.f3. [DOI] [Google Scholar]
- 119.Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Cell. 1998;95:805. doi: 10.1016/S0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 120.Membre N, Berna A, Neutelings G, David A, David H, Staiger D, Saez Vasquez J, Raynal M, Delseny M, Bernier F. Plant Mol Biol. 1997;35:459. doi: 10.1023/A:1005833028582. [DOI] [PubMed] [Google Scholar]
- 121.Michniewicz M, Brewer PB, Friml J. Polar auxin transport and asymmetric auxin distribution. Somerville CR, Meyerowitz EM (eds) Arabidopsis book. Rockville: American Society of Plant Biologists; 2007. p. 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kalo P, Prabhu R, Marsh JF, Mitra RM, Kereszt A. Plant Cell. 2007;19:1221. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mordhorst AP, Hartog MV, El Talmer MK, Laux T, Vries SC. Planta. 2002;214:829. doi: 10.1007/s00425-001-0700-6. [DOI] [PubMed] [Google Scholar]
- 124.Mordhorst AP, Toonen MAJ, Vries SC. Critical Review Plant Science. 1997;16:535. doi: 10.1080/713608156. [DOI] [Google Scholar]
- 125.Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, VanWent J, Koornneef M, deVries SC. Genetics. 1998;149:549. doi: 10.1093/genetics/149.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nagata T, Ishida S, Hasezawa S, Takahashi Y. Int J Dev Biology. 1994;38:321. [PubMed] [Google Scholar]
- 127.Nakano T, Suzuki K, Fujimura T, Shinshi H. Plant Physiol. 2006;140:411. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Namasivayam P. Plant Cell Tissue Organ Cult. 2007;90:1. doi: 10.1007/s11240-007-9249-9. [DOI] [Google Scholar]
- 129.Neutelings G, Domon JM, Membre N, Bernier F, Meyer Y, David A, David H. Plant Mol Biol. 1998;38:1179. doi: 10.1023/A:1006033622928. [DOI] [PubMed] [Google Scholar]
- 130.Nishiwaki M, Fujino K, Koda Y, Masuda K, Kikuta Y. Planta. 2000;211:756. doi: 10.1007/s004250000387. [DOI] [PubMed] [Google Scholar]
- 131.Nolan KE, Irwanto RR, Rose RJ. Plant Physiol. 2003;133:218. doi: 10.1104/pp.103.020917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nolan KE, Saeed NA, Rose RJ. Plant Cell Report. 2006;25:711. doi: 10.1007/s00299-006-0135-4. [DOI] [PubMed] [Google Scholar]
- 133.Ogas J, Kaufmann S, Henderson J, Somerville C. Proc Natl Acad Sci USA. 1999;96:13839. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ogas J, Cheng JC, Sung ZR, Somerville C. Science. 1997;277:91. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- 135.Ogata Y, Iizuka M, Nakayama D, Ikeda M, Kamada H, Koshiba T. Planta. 2005;221:417. doi: 10.1007/s00425-004-1449-5. [DOI] [PubMed] [Google Scholar]
- 136.Ohmiya A, Tanaka Y, Kadowaki K, Hayashi T. Plant Cell Physiology. 1998;39:492. doi: 10.1093/oxfordjournals.pcp.a029396. [DOI] [PubMed] [Google Scholar]
- 137.Okamura JK, Caster B, Villaroel R, Montagu M, Jofuku KD. Proc Natl Acad Sci USA. 1997;94:7076. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Paponov IA, Teale WD, Trebar M, Blilou K, Palme K. Trends Plant Sci. 2005;10:170. doi: 10.1016/j.tplants.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 139.Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Plant Cell. 1994;6:1567. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Parenicova L, Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B. Plant Cell. 2003;15:1538. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. J Biology Chemistry. 2007;282:10036. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 142.Parry G, Marchant A, May S, Swarup R, Swarup K, James N. Plant Growth Regul. 2001;20:217. doi: 10.1007/s003440010030. [DOI] [Google Scholar]
- 143.Passarinho P, Ketelaar T, Xing M, Arkel J, Maliepaard C, Hendriks MW, Joosen R, Lammers M, Herdies L, Boer B, Geest Lo, Boutilier K. Plant Mol Biol. 2008;68:225. doi: 10.1007/s11103-008-9364-y. [DOI] [PubMed] [Google Scholar]
- 144.Pasternak TP, Prinsen E, Ayaydin F, Miskolczi P, Potters G, Asard H, Onckelen HA, Dudits D, Feher A. Plant Physiol. 2002;129:1807. doi: 10.1104/pp.000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pennell RI, Graham NLJ, Hilbert S, Vries SC, Robertsw K. J Cell Biol. 1992;119:1371. doi: 10.1083/jcb.119.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pennell RI, Quentin CB, Cronk L, Scott F, Christine S, Lars S, Per K, Paul FM. Biol Sci. 1995;350:87. doi: 10.1098/rstb.1995.0142. [DOI] [Google Scholar]
- 147.Perry SE, Nichols KW, Fernandez DE. Plant Cell. 1996;8:1977. doi: 10.1105/tpc.8.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S. Cell. 2003;115:679. doi: 10.1016/S0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 149.Puigderrajols P, Jofre A, Mir G, Pla M, Verdaguer D, Huguet G, Molinas M. J Exp Bot. 2002;53:1445. doi: 10.1093/jexbot/53.373.1445. [DOI] [PubMed] [Google Scholar]
- 150.Quint M, Gary WM. Curr Opin Plant Biol. 2006;9:448. doi: 10.1016/j.pbi.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Quiroz-Figueroa FR, Rojas-Herrera R, Galaz-Avalos RM, Loyola-Vargas VM. Plant Cell Tissue Org Culture. 2006;86:285. doi: 10.1007/s11240-006-9139-6. [DOI] [Google Scholar]
- 152.Raghavan V. Am J Bot. 2004;9:1743. doi: 10.3732/ajb.91.11.1743. [DOI] [PubMed] [Google Scholar]
- 153.Raghavan V. Curr Sci. 2006;90:1336. [Google Scholar]
- 154.Raghavan V. Somatic embryogenesis. In: Raghavan V, editor. Embryogenesis in angiosperms. Cambridge: Cambridge University Press; 1986. p. 115. [Google Scholar]
- 155.Rani AR, Reddy VD, Prakash Babu P, Padmaja G. Biology Plant. 2005;49:347. doi: 10.1007/s10535-005-0006-9. [DOI] [Google Scholar]
- 156.Ray PM, Dohrmann U, Hertel R. Plant Physiol. 1977;60:585. doi: 10.1104/pp.60.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reichheld JP, Mestres-Ortega D, Laloi C, Meyer Y. Plant Physiol Biochem. 2002;40:685. doi: 10.1016/S0981-9428(02)01406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rensing PB, Daniel L, Schumann, Reski R, Hohe A. Journal Plant Growth Regulators. 2005;24:102. doi: 10.1007/s00344-005-0033-y. [DOI] [Google Scholar]
- 159.Ribnicky DM, Ilic N, Cohen JD, Cooke TJ. Plant Physiology. 1996;112:549. doi: 10.1104/pp.112.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rider SD, Hemm MR, Hostetler HA, Li HC, Chapple C, Ogas J. Planta. 2004;219:489. doi: 10.1007/s00425-004-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rider SD, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Plant J. 2003;35:33. doi: 10.1046/j.1365-313X.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Roberts AG, Oparka K. Plant Cell Environmental. 2003;26:103. doi: 10.1046/j.1365-3040.2003.00950.x. [DOI] [Google Scholar]
- 163.Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Science. 1996;273:654. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 164.Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. Genes Dev. 1998;12:198. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ryu H, Kim K, Hwang I. Plant Signal Behavior. 2008;3:278. doi: 10.4161/psb.3.4.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Saab IN, Sharp RE, Pritchard J. Plant Physiol. 1992;99:26. doi: 10.1104/pp.99.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sagare AP, Lee YL, Lin TC, Chen CC, Tsay HS. Plant Sci. 2000;160:139. doi: 10.1016/S0168-9452(00)00377-0. [DOI] [PubMed] [Google Scholar]
- 168.Sakuma Y, Liu Q, Dubouzet JG, Abes H, Shinozak K, Yamaguchi- Shinozaki K. Biochemical and Biophysical Research Communications. 2002;290:998. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 169.Sallandrouze A, Faurobert M, Maataoui M, Espagnac H. Electrophoresis. 1999;20:1109. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1109::AID-ELPS1109>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 170.Santos A, Oliveira MD, Romano E, Clemente YKS, Penha TML, Andrade BB. Plant Sci. 2005;168:723. doi: 10.1016/j.plantsci.2004.10.004. [DOI] [Google Scholar]
- 171.Santos MM, Dubreucq B, Miquel M, Caboche M, Lepiniec L. FEBS Lett. 2005;579:4666. doi: 10.1016/j.febslet.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 172.Sauser C, Kwiatkowski J, Jung J, Grossamann K. J Plant Physiol. 1992;140:747. [Google Scholar]
- 173.Saze H, Scheid OM, Paszkowski J. Nat Genet. 2003;34:65. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 174.Schmidt EDL, Guzzo F, Toonen MAJ, Vries SC. Development. 1997;124:2049. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- 175.Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T. Cell. 2000;100:635. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 176.Segarra CI, Casalongue CA, Pinedo ML, Ronchi VP, Conde RD. J Exp Bot. 2003;54:1335. doi: 10.1093/jxb/erg139. [DOI] [PubMed] [Google Scholar]
- 177.Shah K, Schmidt EDL, Vlak JM, Vries SC. Biochimie. 2001;83:415. doi: 10.1016/S0300-9084(01)01257-3. [DOI] [PubMed] [Google Scholar]
- 178.Sharma SK, Millam S, Hein I, Glenn BJ. Planta. 2008;228:319. doi: 10.1007/s00425-008-0739-8. [DOI] [PubMed] [Google Scholar]
- 179.Shimada T, Hirabayashi T, Endo T, Fujii H, Kita M, Omura M. Scientia Horticulture. 2005;103:233. doi: 10.1016/j.scienta.2004.07.005. [DOI] [Google Scholar]
- 180.Shiota H, Kamada H. J Plant Physiol. 2000;156:510. [Google Scholar]
- 181.Shiota H, Satoh R, Watabe K, Harada H, Kamada H. Plant Cell Physiology. 1998;39:1184. doi: 10.1093/oxfordjournals.pcp.a029319. [DOI] [PubMed] [Google Scholar]
- 182.Showalter AM. Cell Molecular Life Science. 2001;58:1399. doi: 10.1007/PL00000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Singla B, Tyag AK, Khurana JP, Khurana P. Plant Mol Biol. 2007;65:677. doi: 10.1007/s11103-007-9234-z. [DOI] [PubMed] [Google Scholar]
- 184.Skriver K, Mundy J. Plant Cell. 1990;2:503. doi: 10.1105/tpc.2.6.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Somleva MN, Schmidt EDL, Vries SC. Plant Cell Report. 2000;19:718. doi: 10.1007/s002999900169. [DOI] [PubMed] [Google Scholar]
- 186.Souter M, Lindsey K. J Exp Bot. 2000;51:971. doi: 10.1093/jexbot/51.347.971. [DOI] [PubMed] [Google Scholar]
- 187.Srinivasan C, Liu ZR, Heidmann I, Supena EDJ, Fukuoka H, Joosen R, Lambalk J, Angenent G, Scorza R, Custers JBM. Planta. 2007;225:341. doi: 10.1007/s00425-006-0358-1. [DOI] [PubMed] [Google Scholar]
- 188.Stasolla C, Bozhkov PV, Chu TM, Zyl L, Egertsdotter U, Suarez MF, Craig D, Wolfinger RD, Arnold S, Sederoff RR. Tree Physiology. 2004;24:1073. doi: 10.1093/treephys/24.10.1073. [DOI] [PubMed] [Google Scholar]
- 189.Steele-King CG, Willats WGT, Knox JP. Arabinogalactan-proteins and cell development in roots and somatic embryos. In: Nothnagel EA, Bacic A, Clarke AE, editors. Cell and developmental biology of Arabinogalactan-proteins. New York: Kluwer; 2000. pp. 95–107. [Google Scholar]
- 190.Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S. Science. 1999;286:316. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- 191.Stone SL, Braybrook SA, Paula S, Kwon LW, Meuser J, Pelletier J, Hsieh T-F, Fischer RL, Goldberg B, Harada JJ. Proc Natl Acad Sci USA. 2008;105:3151. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Stone SL, Kwong LW, Yee KM, Pelletier J, Loic L, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2001;98:11806. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K. Genes Dev. 2001;15:2648. doi: 10.1101/gad.210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tabuchi T, Kumon T, Azuma T, Nanmori T, Yasuda T. Physiology Plant. 2003;118:523. doi: 10.1034/j.1399-3054.2003.00133.x. [DOI] [Google Scholar]
- 195.Tachikawa Y, Saitou T, Kamada H, Harada H. Plant Biotechnology. 1998;15:17. [Google Scholar]
- 196.Tanaka H, Dhonukshe P, Brewer PB, Friml J. Cell Molecular Life Science. 2006;63:2738. doi: 10.1007/s00018-006-6116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. Plant J. 2006;45:1028. doi: 10.1111/j.1365-313X.2006.02656.x. [DOI] [PubMed] [Google Scholar]
- 198.Thakare D, Weining T, Hill K, Perry SE. Plant Physiol. 2008;146:1663. doi: 10.1104/pp.108.115832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Thomas C, Meyer D, Himber C, Steinmetz A. Plant Physiol Biochem. 2004;42:35. doi: 10.1016/j.plaphy.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 200.Thompson EW, Lane BG. J Biology Chemistry. 1980;255:5965. [PubMed] [Google Scholar]
- 201.Tokuji Y, Kuriyama K. Journal of Plant Physiology. 2003;160:133. doi: 10.1078/0176-1617-00892. [DOI] [PubMed] [Google Scholar]
- 202.Toonen MAJ, Hendriks T, Schmidt Ed DL, Verhoeven HA, Kammen Ab, Vries SC. Planta. 1994;194:565. doi: 10.1007/BF00714471. [DOI] [Google Scholar]
- 203.Toonen MAJ, Schmidt EDL, Heo TH, Verhoeven HA, Kammen A, Vries SC. Planta. 1996;200:167. doi: 10.1007/BF00208305. [DOI] [Google Scholar]
- 204.Toonen MAJ, Schmidt EDL, Kammen A, Vries SC. Planta. 1997;203:188. doi: 10.1007/s004250050181. [DOI] [Google Scholar]
- 205.Torii KU. Int Rev Cytol. 2004;234:1. doi: 10.1016/S0074-7696(04)34001-5. [DOI] [PubMed] [Google Scholar]
- 206.Graa VE, Dulk-Ras A, Hooykaas PJJ, Keller B. Development. 2000;127:4971. doi: 10.1242/dev.127.22.4971. [DOI] [PubMed] [Google Scholar]
- 207.Thibaud-Nissen FO, Shealy RT, Khanna A, Vodkin LO. Plant Physiology. 2003;132:118. doi: 10.1104/pp.103.019968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hengel AJ, Kammen A, Vries SC. Physiology Plant. 2002;114:637. doi: 10.1034/j.1399-3054.2002.1140418.x. [DOI] [Google Scholar]
- 209.Hengel AJ, Guzzo F, Kammen A, Vries SC. Plant Physiol. 1998;117:43. doi: 10.1104/pp.117.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hengel AJ, Tadesse Z, Immerzeel P, Schols H, Kammen A, Vries SC. Plant Physiol. 2001;125:1880. doi: 10.1104/pp.125.4.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vergne P, Dumas C. Plant Mol Biol. 2000;44:559. doi: 10.1023/A:1026521506952. [DOI] [PubMed] [Google Scholar]
- 212.Vogel G. Science. 2005;300:86. doi: 10.1126/science.309.5731.86. [DOI] [PubMed] [Google Scholar]
- 213.Vogler H, Kuhlemeier C. Curr Opin Plant Biol. 2003;6:51. doi: 10.1016/S1369-5266(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 214.Recklinghausen IR, Iwanowska A, Kieft H, Mordhorst AP, Schel JHN, Lammeren AAM. Protoplasma. 2000;211:217. doi: 10.1007/BF01304489. [DOI] [Google Scholar]
- 215.Vroemen C, deVries S, Quatrano R. Cell Development Biology. 1999;10:157. doi: 10.1006/scdb.1999.0291. [DOI] [PubMed] [Google Scholar]
- 216.Wang H, Caruso LV, Downie AB, Perry SE. Plant Cell. 2004;16:1206. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wang X, Chory J. Science. 2006;313:1118. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 218.Willemsen V, Scheres B. Annul Review Genetic. 2004;38:587. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- 219.Williams EG, Maheswaran G. Annals Botany. 1986;57:443. [Google Scholar]
- 220.Wilson JW, Wilson PMW. J Plant Physiology. 1993;20:555. [Google Scholar]
- 221.Winkelmann T, Heintz D, Dorsselaer A, Serek M, Braun HP. Planta. 2006;224:508. doi: 10.1007/s00425-006-0238-8. [DOI] [PubMed] [Google Scholar]
- 222.Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL. Plant Cell. 2006;18:805. doi: 10.1105/tpc.105.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yamamoto N, Kobayashi H, Togashi T, Mori Y, Kikuchi K, Kuriyama K, Tokuji Y. J Plant Physiol. 2005;162:47. doi: 10.1016/j.jplph.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 224.Yazawa K, Takahata K, Kamada H. Plant Physiol Biochem. 2004;42:215. doi: 10.1016/j.plaphy.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 225.Zambryski P, Crawford K. Annul Review Cell Development Biology. 2000;16:393. doi: 10.1146/annurev.cellbio.16.1.393. [DOI] [PubMed] [Google Scholar]
- 226.Zdravkovic-Korac S, Neskovic M. Plant Cell Tissue Organ Cult. 1999;55:109. doi: 10.1023/A:1006258426969. [DOI] [Google Scholar]
- 227.Zeng F, Zhang X, Zhu L, Tu L, Guo X, Nie Y. Plant Mol Biol. 2006;60:167. doi: 10.1007/s11103-005-3381-x. [DOI] [PubMed] [Google Scholar]
- 228.Zhu C, Perry SE. Plant J. 2005;41:583. doi: 10.1111/j.1365-313X.2004.02320.x. [DOI] [PubMed] [Google Scholar]
- 229.Zimmerman JL. Plant Cell. 1993;5:1411. doi: 10.1105/tpc.5.10.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zuo Jianru Q-W, Giovanna F, Nam-Hai C. Plant J. 2002;30:349. doi: 10.1046/j.1365-313X.2002.01289.x. [DOI] [PubMed] [Google Scholar]