Summary
Campylobacter jejuni is a gastrointestinal pathogen of humans but can asymptomatically colonize the avian gut. C. jejuni therefore grows at both 37°C and 42°C, the internal temperatures of humans and birds respectively. Microarray and proteomic studies on temperature regulation in C. jejuni strain 81–176 revealed the upregulation at 42°C of two proteins, Cj0414 and Cj0415, orthologous to gluconate dehydrogenase (GADH) from Pectobacterium cypripedii. 81–176 demonstrated GADH activity, converting d-gluconate to 2-keto-d-gluconate, that was higher at 42°C than at 37°C. In contrast, cj0414 and cj0415 mutants lacked GADH activity. Wild-type but not cj0415 mutant bacteria exhibited gluconate-dependent respiration. Neither strain grew in defined media with d-gluconate or 2-keto-d-gluconate as a sole carbon source, revealing that gluconate was used as an electron donor rather than as a carbon source. When administered to chicks individually or in competition with wild-type, the cj0415 mutant was impaired in establishing colonization. In contrast, there were few significant differences in colonization of BALB/c-ByJ mice in single or mixed infections. These results suggest that the ability of C. jejuni to use gluconate as an electron donor via GADH activity is an important metabolic characteristic that is required for full colonization of avian but not mammalian hosts.
Introduction
Campylobacter jejuni is a major cause of gastroenteritis throughout the world (Friedman et al., 2000; Miller and Mandrell, 2005). Campylobacteriosis consists of a 5–7 day course of fever, severe abdominal cramping and watery diarrhoea with or without blood (Miller and Mandrell, 2005). Although the majority of C. jejuni infections are self-limiting, up to 10% of patients may require medical intervention. Sequelae of infection can be severe and include arthropathies and Guillain–Barré syndrome (GBS), an autoimmune disease resulting in acute motor nerve paralysis (Nachamkin et al., 2000).
Campylobacter jejuni is found in numerous environmental niches, including surface waters, cattle, sheep, migratory birds and poultry (Miller and Mandrell, 2005). A number of studies have implicated consumption of undercooked poultry meat or its juices as major sources of C. jejuni infection (Kramer et al., 2000; Newell and Wagenaar, 2000; Allos, 2001; Jorgensen et al., 2002; Nadeau et al., 2002). Because birds (including poultry) have a higher body temperature (42°C) than humans (37°C), it is clear that C. jejuni is exposed to different temperatures during growth in poultry and humans. Gene regulation in response to growth temperature occurs in many pathogenic bacteria including C. jejuni (Rappuoli et al., 1992; Stevenson et al., 1995; Straley and Perry, 1995; Hurme et al., 1997; Konkel et al., 1998; Thies et al., 1999a,b,c; Konkel and Tilly, 2000; Stintzi, 2003; Stintzi and Whitworth, 2003; M. Pajaniappan and S.A. Thompson, unpubl. data, 2008). Temperature regulation can be critical for pathogenesis, and may prevent the bacteria from inappropriate expression of energetically costly proteins until they are in the environment in which the proteins are required (often the host). The few studies undertaken on C. jejuni temperature regulation have primarily identified heat shock proteins (DnaJ, DnaK, GroEL and ClpB) regulated by elevated temperature (48°C) (Konkel et al., 1998; Thies et al., 1999a,b,c; Stintzi, 2003), and mutants lacking either the heat shock protein DnaJ or the temperature-responsive two component regulatory system RacRS are attenuated in chicken colonization (Konkel et al., 1998; Brás et al., 1999). Together, these results suggest that temperature is an important regulatory signal for C. jejuni, and that temperature-regulated proteins are necessary for the optimal colonization of chickens.
Although not traditionally defined as a virulence characteristic, metabolism is critical for the success of bacterial pathogens. Metabolic pathways allow bacteria to acquire nutrients for growth, provide substrates for the assembly of important cellular structures, and generate energy for the function of virulence-related organelles such as flagella. Consequently, understanding aspects of metabolism can shed light on how bacteria grow and persist in their hosts. The ability of a pathogen to alter its metabolism in response to different conditions can also significantly affect its ability to traverse diverse environments related to pathogenesis (i.e. in vivo, intracellular, the ambient atmosphere, etc.). The nutritional requirements of C. jejuni are largely unknown, although several studies have begun to shed light on its metabolism (Smith et al., 2000; Kelly, 2005). C. jejuni lacks a complete glycolytic pathway, which explains the inability of this organism to utilize carbohydrates (Kelly, 2005). Instead, C. jejuni appears to grow primarily on amino acids such as serine (Velayudhan et al., 2004).
One sugar, metabolism of which may be important in bacterial survival and colonization, is gluconate (an oxidation product of glucose). In some bacteria, gluconate can be transported intracellularly and converted by various pathways to a number of related sugars that can enter pathways of central intermediary metabolism such as the Entner-Doudoroff or pentose phosphate pathways (Matsushita et al., 1979; Matsushita et al., 1982; McIntire et al., 1985; Yum et al., 1997; Yum et al., 1998). The importance of gluconate metabolism has been demonstrated by the colonization defect of an Escherichia coli mutant unable to catabolize gluconate, which is abundant (at a concentration of 0.69 mM) in the mouse intestinal tract (Peekhaus and Conway, 1998; Chang et al., 2004; Leatham et al., 2005). Alternatively, some sugars with favourable redox potentials (such as gluconate) can also serve as periplasmic electron donors during electron transport (White, 2007). The C. jejuni NCTC11168, RM1221 and 81–176 genomes (Parkhill et al., 2000; Fouts et al., 2005; Hofreuter et al., 2006) predict orthologues of several membrane-bound oxidoreductases (e.g. hydrogenase, and dehydrogenases for succinate, formate, lactate, sulphite and malate) that may catalyse reactions that donate electrons to the menaquinone pool, initiating electron transport and energy conservation through a highly branched electron transport chain (Kelly, 2005; Myers and Kelly, 2005; Pittman and Kelly, 2005). The current study was undertaken to determine the enzymatic function of a predicted C. jejuni oxidoreductase operon that was upregulated at 42°C and exhibited homology to gluconate dehydrogenases (GADHs) in other organisms, and to determine the importance of this system in survival or persistence of C. jejuni within avian or mammalian hosts.
Results
Expression of cj0414 and cj0415 are higher at 42°C than at 37°C
In the course of studies designed to investigate differential expression of C. jejuni genes in response to steady-state growth temperature (37°C versus 42°C), a number of genes expressed more highly at 42°C than at 37°C were identified (M. Pajaniappan and S.A. Thompson, unpubl. data, 2008). In these experiments, C. jejuni cells were grown overnight at 37°C, then back-diluted into fresh media and two cultures grown in parallel at 37°C and 42°C to mid-log growth phase. RNA samples prepared from 37°C- and 42°C-grown cells were hybridized against microarrays of genes found in the chromosome of C. jejuni NCTC11168 (Leonard et al., 2003). Among the genes whose expression was most increased at 42°C were cj0414 and cj0415, and these were induced a mean of 3.7- and 2.7-fold at 42°C respectively (data not shown).
As an additional assay of temperature regulation, proteomics experiments were performed on cultures of C. jejuni 81–176 grown at the two temperatures. Two proteins that had the approximate pIs and molecular masses of the predicted Cj0414 and Cj0415 proteins were the most highly upregulated at 42°C (Fig. 1). These proteins were 3.4- and 2.9-fold more highly expressed at 42°C respectively. These proteins were excised from the polyacrylamide gel and subjected to MALDI-ToF/ToF mass spectrometry. The proteins were identified with 100% confidence as Cj0414 and Cj0415 (data not shown), supporting the microarray experiments that the genes encoding these proteins were substantially upregulated at 42°C.
Fig. 1.
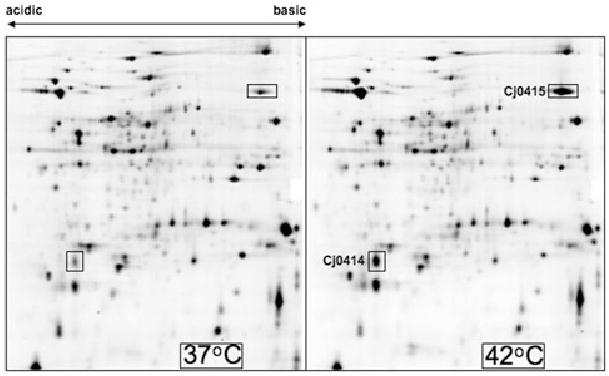
Temperature-dependent changes in the expression levels of the Cj0414 and Cj0415 proteins. Shown is a two-dimensional SDS-PAGE protein gel showing the expression of C. jejuni 81–176 proteins at 37°C and 42°C. The Cj0414 and Cj0415 proteins identified using mass spectrometry are shown.
Although both microarray and proteomics experiments indicated that the cj0414 and cj0415 genes were more highly expressed at 42°C than at 37°C, real-time quantitative reverse transcription (qRT)-PCR was used to confirm this increased transcription. RNA samples prepared from cells grown at 37°C and 42°C were reverse-transcribed and subjected to real-time PCR using primers specific for cj0414 or cj0415. 16S rRNA was used as a non-regulated control. The qRT-PCR primers amplified cj0414- and cj0415-specific cDNA more rapidly from samples grown at 42°C than those at 37°C, confirming that cj0414- and cj0415 were 3.3- and 2.6-fold upregulated at 42°C respectively. Together, three separate approaches indicated the induction of cj0414 and/or cj0415 at 42°C in C. jejuni strain 81–176.
cj0414 and cj0415 are co-transcribed
In completed C. jejuni genome sequences (strains NCTC11168, RM1221 and 81–176) known to contain the cj0414 and cj0415 genes, the genes were arranged in an apparent two-gene operon (Fig. 2A), with the two genes separated by one nucleotide. The similar induction of cj0414 and cj0415 noted above, together with analyses of repository data from numerous other microarray-based gene expression studies (Gaynor et al., 2004; MacKichan et al., 2004; Gaynor et al., 2005) further supported the co-transcription of these genes. However, to confirm that the genes were co-transcribed in strain 81–176, reverse transcription PCR was performed. RNA purified from strain 81–176 was copied to cDNA using reverse transcriptase and random hexamer primers. This cDNA was used in PCR reactions using forward and reverse PCR primers located in the cj0414 and cj0415 genes respectively (Table S1). These primers were designed to amplify a product only if the cj0414 and cj0415 genes were present on the same RNA transcript. The expected 2.4 kb PCR product was obtained (Fig. 2B, lane 1), indicating co-transcription of cj0414 and cj0415, consistent with the prediction from the genome sequences. A control PCR reaction in which the template RNA was not reverse transcribed yielded no product, confirming the absence of contaminating genomic DNA (Fig. 2B, lane 2).
Fig. 2.
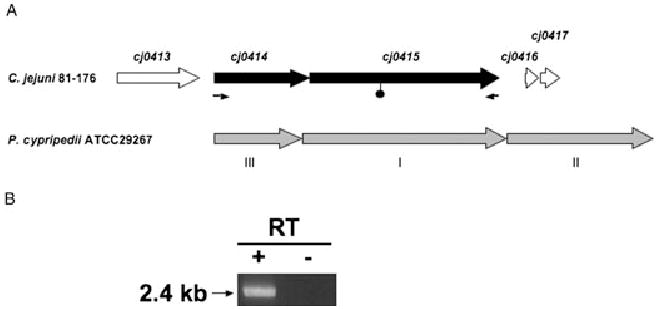
A. Comparison of the C. jejuni 81–176 cj0414/cj0415 locus and the GADH locus of Pectobacterium (Erwinia) cypripedii (adapted from Yum et al., 1997). The filled symbol shows the position at which a kanamycin-resistant transposon was inserted in 81–176cj0415. The small arrows below cj0414 and cj0415 indicate the positions of PCR primers used in the RT-PCR experiments shown in B. B. Studies on cj0414/cj0415 transcript. Reverse transcription-PCR (RT-PCR) of cj0414/cj0415. DNA-free RNA from 81–176 grown at 42°C was treated with (+) or without (−) reverse transcriptase, followed by PCR using primers cj0414-F1 and cj0415-R1 (Table S1).
cj0414 and cj0415 are conserved in Campylobacter jejuni but absent in most other Campylobacter species
Campylobacter jejuni exhibits considerable genomic diversity, both in gene complement and in gene sequence (Dorrell et al., 2001; Leonard et al., 2003). To determine whether the cj0414 and cj0415 genes are present in other Campylobacter strains, we designed PCR primers based on the C. jejuni NCTC11168 genomic sequence (Parkhill et al., 2000). These primers were located in the flanking cj0413 and cj0417 genes, and were used to amplify a 3.2 kb segment encompassing the cj0414/cj0415 locus from five additional C. jejuni strains and one Campylobacter coli strain. PCR products of the expected size were obtained from each strain, and both strands of each PCR product (total of 3191 bp sequence analysed for each strain) were sequenced directly by primer walking. The analogous regions from five additional Campylobacter genome sequences were also included in the analysis (Fouts et al., 2005). In each case, the cj0414 and cj0415 genes are highly conserved. For both the cj0414 and cj0415 genes, the nucleotide sequences between pairs of C. jejuni strains or C. coli D3088 are at least 98.0% identical. With regard to the encoded proteins, Cj0414 and Cj0415 are also highly similar, with a minimum amino acid identity of 99.0%. Interestingly, none of the recently sequenced non-Campylobacter jejuni genomes contain orthologues of cj0414 and cj0415 (Fouts et al., 2005). Together, these results indicate that cj0414 and cj0415 are highly conserved in C. jejuni (100% of 12 strains), present to a lesser extent in a limited set of C. coli (50% of two strains), but absent in one strain each of C. lari, C. upsaliensis, C. concisus, C. curvus and C. fetus. However, when present, the genes and predicted Cj0414 and Cj0415 proteins are highly similar to the other orthologues. For the remainder of this work, we focused on Cj0414/Cj0415 from C. jejuni strain 81–176, although all features noted for the 81–176 proteins are also found in all of the other strains tested.
Features of Cj0414 and Cj0415 proteins in strain 81–176
The proteins predicted from the genome sequence of NCTC11168 were annotated as two components of a putative oxidoreductase most similar to the GADH of Pectobacterium (Erwinia) cypripedii (Yum et al., 1997; Parkhill et al., 2000), and putative orthologues in several genomes (Peterson et al., 2001). Cj0414 has a predicted molecular mass of 26.9 kDa and a pI of 5.5, and is most similar to subunit III of P. cypripedii ATCC29267 GADH (Yum et al., 1997). Cj0415 is related to subunit I (catalytic subunit) of P. cypripedii GADH, with predicted molecular mass of 63.7 kDa and and a pI 8.8 respectively. A gene homologous to that encoding subunit II (cytochrome) of P. cypripedii GADH is not located adjacent to the cj0414 and cj0415 genes (Fig. 2A), nor are orthologues apparent in BLAST searches of the genome sequences of any other C. jejuni strain.
Cj0414 and Cj0415 were also analysed for the presence of protein motifs (Gasteiger et al., 2003). Cj0415 contains a motif found in the GMC oxidoreductase protein family (PFAM00732), of which GADHs are members. Proteins of the GMC family often bind FAD as a cofactor. Consistent with the predicted nucleotide-binding abilities of this protein, a putative NAD/FAD binding site (Prosite entries PS50204 and PS50205) is located within the first 48 amino acids of Cj0415. Cj0415 is also related to another family of flavoproteins (COG2303, including BetA and choline dehydrogenase).
Because GADH proteins from other bacteria are associated with the bacterial cytoplasmic membrane (Matsushita et al., 1979; Matsushita et al., 1982; McIntire et al., 1985) [possibly on the periplasmic face (Yum et al., 1997)], the SignalP and TatP algorithms were used (Bendtsen et al., 2004; Bendtsen et al., 2005) to analyse the Cj0414 and Cj0415 proteins for the presence of N-terminal signal sequences. Cj0414 is predicted to possess a signal sequence, with a probable cleavage site between amino acids 35 and 36 of the preprotein (VLKA↓AE). However, the predicted signal sequence has features most like those of proteins translocated by the twin-arginine translocation (Tat) pathway (Bendtsen et al., 2005; Palmer et al., 2005). Paired arginine residues are found at positions 8–9 as components of a motif (DRRSFFK) similar to the Tat consensus motif SRRxFLK (Palmer et al., 2005). Furthermore, the lysine residue preceding the putative Cj0414 signal peptidase cleavage site is typically not found in proteins transported by the sec-dependent pathway. The presence of a Tat-related signal sequence in NCTC11168 Cj0414 was previously found using bioinformatics approaches (Park, 2005; Dilks et al., 2003). Cj0415 is predicted to not have an N-terminal signal sequence. Secondary structure predictions indicated that both Cj0414 and Cj0415 are devoid of transmembrane segments (other than the hydrophobic core of the Cj0414 signal sequence) and thus are likely to be periplasmic proteins, possibly peripherally associated with the cytoplasmic membrane.
Inactivation of the cj0414 and cj0415 genes
To investigate the phenotypes associated with the predicted Cj0414/Cj0415 enzyme, mutants lacking the predicted GADH subunits Cj0414 and Cj0415 were constructed as follows. A non-polar cj0414 mutation was created by cloning PCR-amplified cj0414 into pCRII-TOPO, then disrupting cj0414 with a chloramphenicol resistance (cat) cassette such that the translation initiation signals of cj0415 were unaffected. While the cj0414 mutant lacked expression of Cj0414, Western blot showed that Cj0415 was still expressed (data not shown), confirming the non-polar nature of the cj0414 mutation. To disrupt cj0415, PCR-amplified cj0415 gene was cloned into pBluescript II, and the resultant plasmid was used for in vitro transposon reactions with a KmR derivative of EZ∷TN (see Experimental procedures). One plasmid (pBS-cj0415-aphA) containing a transposon inserted after nucleotide 649 of the cj0415 open reading frame, with transcription of the aphA gene oriented in the same direction as that of cj0415 (data not shown), was chosen for further use. The mutagenized cj0414 and cj0415 alleles were introduced into C. jejuni 81–176 by electroporation or natural transformation. Several CmR or KmR transformants were verified as cj0414 or cj0415 mutants, respectively, by PCR and DNA sequencing (data not shown), and representative colonies were chosen for further analysis.
C. jejuni 81–176 has cj0415- and cj0414-dependent GADH activity that is higher at 42°C than at 37°C
The wild-type, cj0414 and cj0415 mutant strains were tested for GADH activity using a coupled assay (Yum et al., 1997; Matsushita et al., 1982). Protein extracts from wild-type 81–176 and cj0414 and cj0415 mutant strains grown at 37°C and 42°C were prepared and added to reaction mixtures containing gluconate and the potential electron acceptors 2,6-dichlorophenolindolphenol (DCIP) and phenazine methosulphate (PMS). C. jejuni 81–176 wild-type extracts exhibited GADH activity which was 2.5-fold higher in extracts of cells grown at 42°C than at 37°C (Fig. 3), consistent with the greater expression of Cj0414 and Cj0415 at 42°C. The cj0414 and cj0415 mutants had negligible GADH activity at either temperature (Fig. 3), indicating that activity was dependent on both Cj0414 and Cj0415. Identical results were obtained with independently derived mutants isolated on a different day. These findings strongly support the prediction based on comparative DNA sequence analysis that both Cj0414 and Cj0415 comprise GADH in C. jejuni.
Fig. 3.
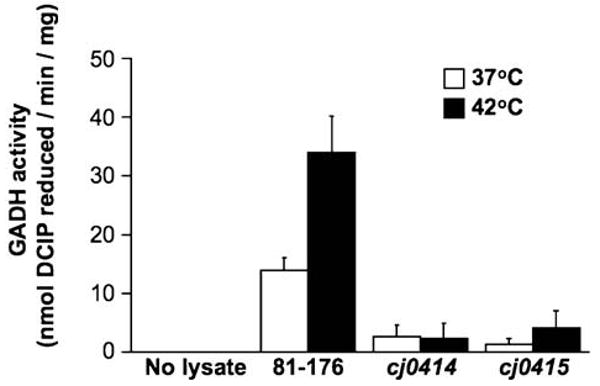
GADH activity of C. jejuni 81–176, 81–176cj0414, and 81–176cj0415. The presence of GADH activity was determined by a coupled enzyme assay, using lysates of wild-type, cj0414, or cj0415 mutant cells grown at 37°C or 42°C, or no cell lysate as a negative control.
We next performed fractionation experiments to localize the GADH activity within the C. jejuni cell (Myers and Kelly, 2005). Cytoplasmic, periplasmic and total membrane fractions of C. jejuni 81–176 cells grown at 42°C were tested for GADH activity. Most of the GADH activity was found in the membrane fraction, although a significant amount was also present in the cytoplasm (Fig. S2). Negligible GADH activity was found in the periplasm. These results indicate that GADH activity is primarily membrane-associated, with additional activity in the cytoplasm probably owing to assembled enzyme activity prior to secretion.
Finally, to explore the impact of loss of GADH function on C. jejuni protein expression, 81–176 and 81–176cj0415 were grown to mid-log phase at 42°C, and protein profiles were compared by 2-D gel electrophoresis and mass spectrometry analyses. Surprisingly, the only observable difference in protein expression was the apparent absence of both Cj0414 and Cj0415 in the cj0415 mutant (Fig. S1). Loss of Cj0415 appeared to result in loss of the entire GADH system. Given this observation, together with the fact that targeted deletion of either cj0414 or cj0415 resulted in near-attenuation of GADH activity, the 81–176cj0415 mutant was used for all subsequent analyses to explore the effects of loss of GADH activity in C. jejuni.
C. jejuni 81–176 does not use gluconate compounds as sole carbon sources
To determine whether C. jejuni 81–176 could use d-gluconate or 2-keto-d-gluconate (2-KG) as sole carbon sources to support growth, we grew cells in Mueller–Hinton broth overnight, then backdiluted them into defined MEMα containing various carbon sources (Velayudhan and Kelly, 2002). Consistent with previous observations (Velayudhan and Kelly, 2002), C. jejuni 81–176 was able to achieve maximal growth in minimal medium when supplied with pyruvate as a sole carbon source (Fig. 4). In contrast, when no additional sugar was added, only a small amount of growth occurred owing to the presence of utilizable amino acids in MEMα (Velayudhan and Kelly, 2002; Fig. 4). When supplied with either d-gluconate or 2-KG, the extent of 81–176 growth was similar to cultures containing no added sugar, indicating that 81–176 was unable to use those sugars as carbon sources. The addition of either d-gluconate or 2-KG did not further stimulate the growth of 81–176 on pyruvate (data not shown). 81–176cj0415 showed an identical growth profile for these sugars (data not shown).
Fig. 4.
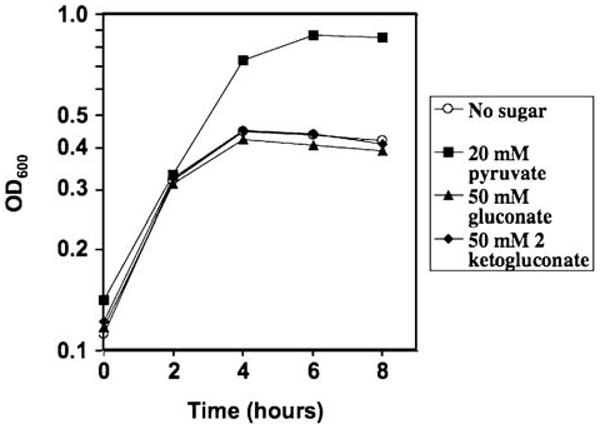
Growth of C. jejuni 81–176 on various carbon sources at 42°C. Growth of C. jejuni in defined αMEM media containing various carbon sources was monitored by optical density at 600 nm. Carbon sources added were: 20 mM pyruvate (filled squares), 20 mM d-gluconate (filled triangles), 20 mM 2-KG (filled diamonds) and no added carbon source (open circles).
C. jejuni respires using d-gluconate as electron donor
To confirm the suspected role of the C. jejuni Cj0414 and Cj0415 proteins as a respiratory GADH, respiration experiments were performed with differing electron donors (Fig. 5). Cell suspensions of 81–176 and 81–176cj0415 grown to late exponential phase at either 37°C or 42°C were provided the known or potential electron donors sodium formate or sodium gluconate, respectively, and respiration-dependent oxygen consumption was then measured. Addition of sodium formate resulted in rapid consumption of oxygen by both wild-type and cj0415 mutant C. jejuni, indicative of electron transport stimulated by formate (Fig. 5A). Growth temperature did not affect the rate of formate-linked oxygen consumption by wild-type or cj0415 mutant strains. Addition of sodium gluconate to 81–176 also resulted in respiratory oxygen consumption; however, 81–176cj0415 was unable to respire on gluconate (Fig. 5B). Oxygen uptake due to gluconate was 3.3-fold higher in wild-type cells grown at 42°C compared with cells grown at 37°C. These results showed that gluconate can be used as an electron donor by C. jejuni, that the dehydrogenase linking gluconate to the electron transport chain required Cj0415, and that gluconate-dependent respiration was significantly higher in cells grown at 42°C than those grown at 37°C.
Fig. 5.

Formate- and gluconate-linked oxygen respiration at 37°C and 42°C. Wild-type (WT) 81–176 and cj0415 mutant strains were grown microaerobically to late exponential phase in Mueller–Hinton broth supplemented with 20 mM serine at both 37°C and 42°C. Intact cells were assayed for formate-linked or gluconate-linked oxygen uptake using a Clark-type oxygen electrode.
A. WT (grey bars) and cj0415 (empty bars) cells exhibited a high rate of formate-linked oxygen consumption after the addition of 5 mM formate at each growth temperature.
B. No rate for gluconate-stimulated oxygen respiration was detected in cj0415 mutant intact cells at either 37°C or 42°C, after the addition of 20 mM gluconate. However, WT whole cells (grey bars) exhibited gluconate-linked oxygen uptake at 37°C and showed a 3.3-fold increase in uptake at 42°C.
To determine the point at which electrons derived from gluconate entered the C. jejuni respiratory chain, we performed experiments using the menaquinol–cytochrome bc1 complex inhibitor HQNO (2-n-heptyl-4-hydroxyquinoline N-oxide; Myers and Kelly, 2005). Wild-type C. jejuni were grown at 42°C, then assayed for formate- or gluconate-dependent oxygen consumption in the presence or absence of HQNO. As expected, treatment with HQNO resulted in a 42% decrease in oxygen consumption using formate as electron donor, as respiration with formate partly involves the cytochrome bc1 complex–cytochrome c oxidase route of electron transfer to oxygen. In contrast, oxygen consumption following gluconate addition was decreased by less than 1%, indicating that the cytochrome bc1 complex is not required for respiration on gluconate. These results suggest that the entry point of electrons from gluconate is after the cytochrome bc1 complex, probably at the level of cytochrome c.
We also tested whether the addition of the electron donors formate and gluconate contributed to the overall growth yield of C. jejuni. Cultures of C. jejuni 81–176 were inoculated at varying concentrations (initial OD600 = 0.02, 0.06 and 0.1), and growth was monitored using OD600 (Fig. S3). The presence of formate or gluconate did not result in any overt changes in the growth of C. jejuni.
81–176cj0415 is defective in chicken colonization
The colonization potential of 81–176cj0415 compared with wild-type 81–176 was tested in 1-day-old chicks (42°C host). First, wild-type and cj0415 mutant strains were each administered orally to groups of 10 chicks, using target doses of either c. 1 × 104 cfu or c. 3 × 103 cfu. The birds were sacrificed after 5 days and the cecal colonization level of each bird was determined. At the 1 × 104 cfu dose, 81–176 and 81–176cj0415 colonized all 10 birds in each group, with statistically similar colonization densities (geometric means of 4.1 × 107 cfu g−1. of cecal contents for wild-type and 6.0 × 107 cfu g−1. of cecal contents for the cj0415 mutant; Fig. 6A, ‘1 × 104 cfu’ dose). At the 3 × 103 cfu dose, however, differences in the abilities of the two strains to colonize chicks were noted. 81–176 colonized all 10 chicks to a mean density of 2.4 × 107 cfu g−1, similar to the higher dose (Fig. 6A, ‘3 × 103 cfu’ dose). In contrast, 81–176cj0415 colonized nine of 10 chicks, with a mean colonization density of only 1.8 × 105 cfu g−1 (Fig. 6A, ‘3 × 103 cfu’ dose). This 130-fold decrease in colonization of the cj0415 mutant relative to wild-type was statistically significant (P = 0.036). At an even lower dose (1 × 103 cfu), 81–176 colonized nine of 10 birds to a mean density of 3.6 × 106 cfu g−1. 81–176cj0415 exhibited a 5000-fold decrease in colonization ability, with only four of nine birds colonized and a mean colonization density of 6.9 × 102 cfu g−1 (P = 0.0068).
Fig. 6.
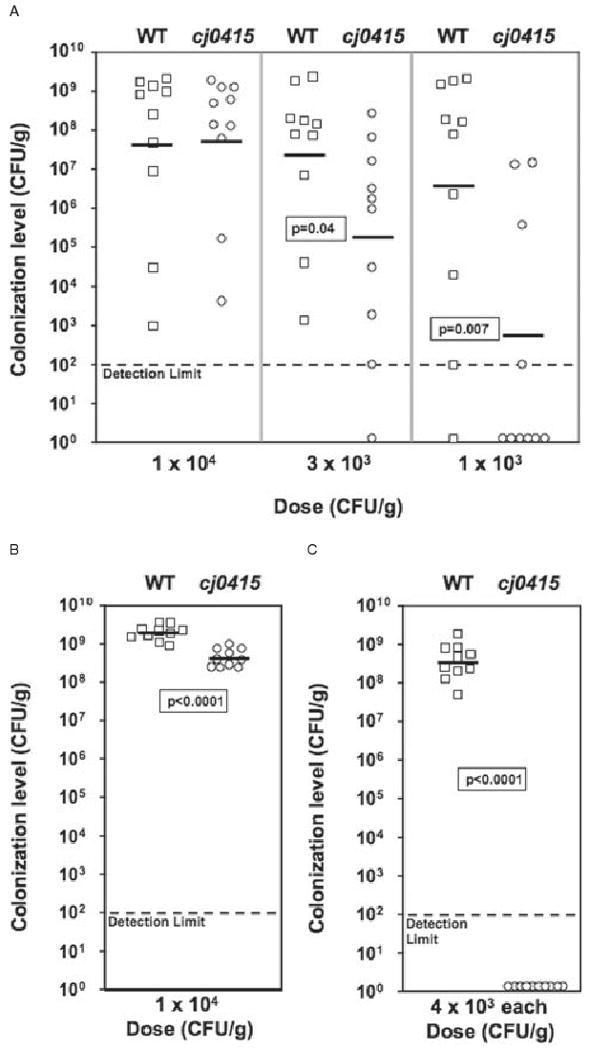
Colonization of 1-day-old chicks by wild-type 81–176 (squares) and 81–176cj0415 (circles). C. jejuni inocula were prepared at various concentrations and administered to groups of 10 1-day-old chicks as described in Experimental procedures. The actual doses administered are shown below each graph, and the resulting colonization level in each bird was determined after a duration of 5 days (A) or 5 weeks (B). In competition experiments, wild-type and mutant bacteria were mixed 1:1 (4 × 103 cfu of each strain) prior to administration to 1-day-old chicks, and the colonization levels of wild-type and mutant bacteria were assessed after 5 days (C). Mean colonization levels are indicated by horizontal bars, and the statistical significance of pairs is indicated. Symbols located on the x-axis represent colonization values below the limit of detection.
To determine whether the duration of colonization was affected by mutation of cj0415, groups of chicks were administered doses of wild-type and cj0415 mutant that yielded similar 5 day colonization levels (c. 1 × 104 cfu, similar to highest dose, Fig. 6A). These birds were sacrificed after 5 weeks and cecal colonization quantified as before. All 10 chicks administered 81–176 showed high-level colonization (mean 2.0 × 109 cfu g−1; Fig. 6B). Although all 10 chicks were colonized with 81–176cj0415 at 5 weeks, the colonization level was significantly lower than wild-type (4.0 × 108 cfu g−1, P < 0.0001).
Finally, the ability of the cj0415 mutant to compete with the wild-type strain for colonization was evaluated. Equal numbers of 81–176 and 81–176cj0415 were mixed (4 × 103 cfu of each strain), then the mixture fed to chicks and colonization again assayed at 5 days. Bacteria were enumerated on media either with or without kanamycin to quantify mutant or total C. jejuni respectively. Control experiments in which equal numbers of cj0415 mutant bacteria were recovered on plates containing or lacking kanamycin showed that the presence of kanamycin in the media did not bias the recovery of the mutant strain (data not shown). 81–176 colonized all 10 chicks to a high colonization density (mean 3.7 × 108 cfu g−1). At 5 days, the cj0415 mutant was not detected in any chick (P < 0.0001), demonstrating an inability to compete with the wild-type strain for colonization (Fig. 6C). To ensure that the colonization defect noted in competition experiments was not due to a general growth deficiency of the cj0415 mutant relative to wild-type, we also performed in vitro growth competition experiments (Fig. S4). In these experiments, wild-type (either unmarked, or using a spontaneous StrR derivative) and cj0415 (KmR) mutant strains were grown in MH broth either individually, or mixed in equal concentration prior to growth (to mimic the in vivo competition in chicks). The growth curves of wild-type, mutant and in vitro competition were indistinguishable (Fig. S4A). Likewise, in the in vitro competition experiment, viable counts revealed nearly identical growth of wild-type and mutant bacteria (Fig. S4B). Therefore, the in vivo competitive disadvantage of the cj0415 mutant was due specifically to conditions encountered during chick colonization and not to a general growth defect of the mutant.
C. jejuni GADH plays a minor role in mouse colonization
To determine whether the colonization defect of the cj0415 mutant also occurred in a mammalian (37°C) host, we used the BALB/c intestinal colonization model previously shown to identify colonization differences between wild-type and colonization-defective mutant strains (Pei et al., 1998; Baqar et al., 1995a). In two separate experiments, mice were inoculated orogastrically with doses of wild-type 81–176 and 81–176cj0415 of 5 × 109, 5 × 107 and 5 × 105 cfu, either individually or in mixtures. Each dose showed similar results, and the results for the lowest dose of 5 × 105 cfu are shown in Fig. 7. In individually inoculated mice, 81–176 achieved a mean colonization density of 2.6 × 106 cfu g−1 of fecal pellet at 7 days, which gradually dropped to a mean of 3.3 × 103 cfu g−1 by 35 days (Fig. 7A). 81–176cj0415 also achieved a high and persistent level of colonization, decreasing from 2.6 × 106 cfu g−1 at 7 days to 1.2 × 104 cfu g−1 at 35 days. The mean colonization densities of wild-type and cj0415 mutant strains were not statistically different at any time point.
Fig. 7.
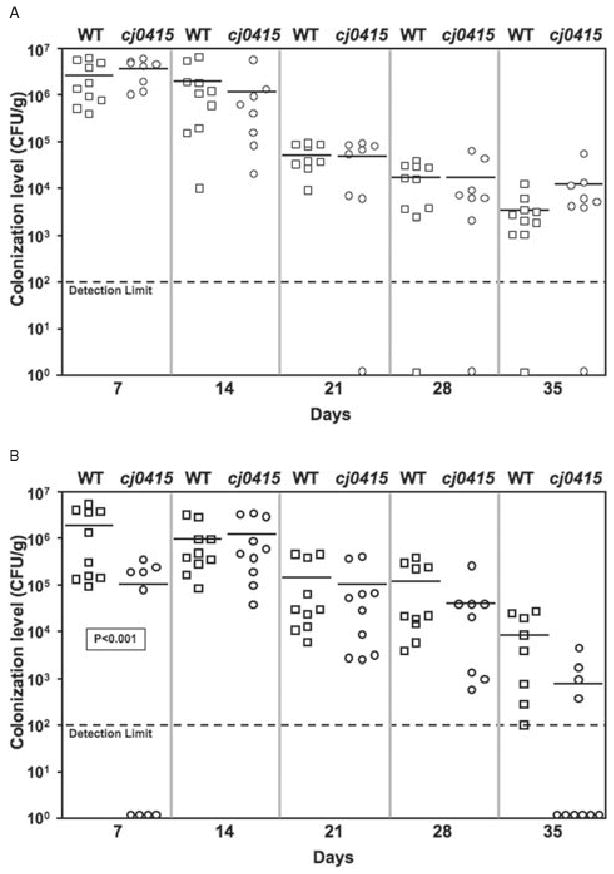
Colonization of BALB/c-ByJ mice by wild-type 81–176 (squares) and 81–176cj0415 (circles).
A. Wild-type or mutant C. jejuni (5 × 105 cfu of each strain) were fed individually to mice by oral gavage and colonization assessed over 35 days.
B. Competition experiment in which mice were fed a 1:1 mixture (2.5 × 105 cfu of each strain) of 81–176 and 81–176cj0415, and the colonization levels of each were determined. Mean colonization levels are indicated by horizontal bars, and the statistical significance of pairs are indicated. Symbols located on the x-axis represent colonization values below the limit of detection.
As an alternative measure of the colonization fitness of each strain in mice, 81–176 and 81–176cj0415 were mixed at a ratio of 1:1 (2.5 × 105 cfu each) prior to inoculation of mice (Fig. 7B). 81–176 demonstrated similar colonization as when it was inoculated individually, with colonization densities ranging from 2 × 106 cfu g−1 fecal pellet at 7 days to 8.8 × 103 cfu g−1 at 35 days. The cj0415 mutant showed a slight (approximately 20-fold), statistically significant delay in the establishment of colonization at 7 days (mutant 1.1 × 105 cfu g−1 versus wild-type 2 × 106 cfu g−1; P < 0.001). The mutant also exhibited a 10-fold reduced level of colonization compared with wild-type after 35 days (mean 8 × 102 cfu g−1 for the mutant, 40% of mice colonized, versus 8 × 103 cfu g−1 for the wild-type, 100% of mice colonized), but this did not reach statistical significance. Through the remainder of the experiment colonization densities were similar to those of wild-type. Together, these results indicate that, in contrast to the chick data, the cj0415 mutant exhibited only a minor colonization defect in mice, even when the two strains were placed in competition for colonization.
Discussion
To establish colonization, and/or infection, bacterial pathogens must be able to acquire nutrients and conserve energy. Among gastrointestinal bacteria, the availability of nutrients within host intestines is thought to determine the niche that specific bacteria can inhabit (Freter, 1983; Freter, 1988; Sweeney et al., 1996a). Sugars are abundant in the intestinal tracts of animals, as are bacterial pathways for utilizing them. While a huge body of literature describes mechanisms by which bacteria can completely oxidize hexoses such as glucose, less is known about the use of the glucose oxidation product gluconate.
In most systems in which gluconate utilization has been studied to date (e.g. E. coli, Salmonella and Pseudomonas), gluconate is used primarily as a carbon source. These bacteria contain extracytoplasmic enzymes that can interconvert glucose, gluconate and gluconate derivatives such as 2-KG, 5-keto-d-gluconate, 2,5-diketo-d-gluconate and l-idonate (Lessie and Phibbs, 1984; Truesdell et al., 1991; Schleissner et al., 1997; Yum et al., 1997; Peekhaus and Conway, 1998; Swanson et al., 2000; Fuhrer et al., 2005). Bacteria that utilize these sugars as carbon sources have both low- and high-affinity membrane transporters for their transport (e.g. GntP, GntS, GntT, GntU or Gat for gluconate; Kgt or KguT for 2-KG; and IdnT for gluconate and idonate) (Peekhaus and Conway, 1998; Sweeney et al., 1996a; Lessie and Phibbs, 1984; Swanson et al., 2000), and transporters for gluconate and related sugars are widely conserved in both Gram-negative and Gram-positive bacterial genomes including the Enterobacteriaceae, Gluconobacter and Pseudomonas (data not shown). Once inside the cell, aldonic acids such as gluconate are not readily metabolized by the glycolytic pathway. Instead, the sugars are converted to 6-phosphogluconate, which can then enter the Entner-Doudoroff and pentose phosphate pathways for carbon assimilation (Peekhaus and Conway, 1998).
Gluconate is an important in vivo carbon source for E. coli (Tsunedomi et al., 2003; Chang et al., 2004). This sugar is extremely abundant in mouse mucus, at a concentration of 0.69 mM (Peekhaus and Conway, 1998), and E. coli uses gluconate present within the mouse cecum to establish and maintain colonization (Sweeney et al., 1996a,b; Peekhaus and Conway, 1998; Tsunedomi et al., 2003; Chang et al., 2004). Chang et al. (2004) showed that gluconate was a major carbon source for E. coli exposed to mouse cecal mucus, that gluconate utilization enzymes were induced in early log phase, and that an E. coli edd mutant (deficient in the Entner-Doudoroff pathway, the primary route for gluconate-derived carbon utilization) had a major defect in the initiation and maintenance of colonization (Chang et al., 2004; Leatham et al., 2005). Similarly, microarray studies showed that gluconate is used as a major carbon source via the Entner-Doudoroff pathway by Salmonella enteritidis during growth within macrophages (Eriksson et al., 2003).
In contrast to bacteria such as E. coli and Salmonella, several lines of evidence suggest that C. jejuni does not use gluconate as a carbon source. First, significant work as well as genome analyses have shown that C. jejuni does not appear to use hexoses such as glucose and gluconate as carbon sources (reviewed in Kelly, 2005). Key enzymes of the main pathways for use of these sugars are missing, such as 6-phosphofructokinase for glycolysis and Edd for the Entner-Doudoroff pathway (Kelly, 2005). Second, although membrane transporters for glucose, gluconate and related sugars are widespread among bacteria, analysis of Campylobacter genome sequences fails to reveal apparent orthologues of these transporters (data not shown), although one cannot rule out the possibility of their transport by divergent sugar transporters. Third, the work presented here shows that C. jejuni 81–176 cannot use either gluconate or 2-KG as a sole carbon source during growth in chemically defined medium (Fig. 4). These data argue that the role of the putative Cj0414/Cj0415 GADH is not sugar conversion for the purpose of carbon utilization. Therefore, gluconate utilization by C. jejuni appears to be quite different from that of E. coli or Salmonella.
Another possible use of gluconate by bacteria is as an electron donor for bacterial respiration. For most bacteria capable of respiration, the transport of electrons via cytoplasmic membrane-bound carrier proteins is a primary means of energy conservation (White, 2007). Bacteria are capable of using a wide variety of electron donors and electron acceptors in electron-transporting oxidation-reduction reactions. However, the role of bacterial electron transport in host colonization and/or pathogenesis is incompletely understood. C. jejuni has a highly branched electron transport chain, and is known or suspected to use a wide variety of inorganic and organic primary electron donors such as hydrogen, sulphite, formate, succinate, malate and lactate (Kelly, 2005; Myers and Kelly, 2005; Hoffman and Goodman, 1982). Homology to membrane-bound GADH enzymes suggests that Cj0414 and Cj0415 might provide C. jejuni with a means for utilizing gluconate as a respiratory electron donor.
To determine the function of the putative Cj0414/Cj0415 GADH, we inactivated the cj0415 and cj0415 genes in C. jejuni strain 81–176 by targeted mutagenesis. GADH enzyme assays on wild-type 81–176, 81–176cj0414 and 81–176cj0415 (Fig. 3) showed that C. jejuni has GADH activity, and that this activity is dependent on both Cj0414 and Cj0415. Wild-type C. jejuni 81–176 GADH activity is also 2.5 times higher at 42°C than at 37°C, consistent with the upregulation of cj0414 and cj0415 at 42°C. To confirm that the role of GADH activity was in respiration rather than for carbon acquisition, oxygen consumption via respiration was measured when cells were provided gluconate as a potential electron donor. Gluconate stimulated C. jejuni respiration, although with a somewhat lower rate of oxygen consumption than that of the control electron donor formate (Fig. 5). The lower oxygen consumption on gluconate could be simply because the enzyme has slower kinetics than formate dehydrogenase. As with GADH activity, gluconate-dependent oxygen consumption was also higher at 42°C than at 37°C.
Many (e.g. P. cypripedii, Gluconobacter, Pseudomonas spp., Bordetella bronchiseptica, Bordetella parapertussis), but not all, known or predicted GADH enzymes are co-transcribed with a cytochrome that links GADH to the electron transport chain. To determine the genomic organization of the cj0414/cj0415 locus in Campylobacter strains, the region encompassing cj0414 and cj0415 was sequenced in a total of six C. jejuni strains and one C. coli strain, and compared with the available sequences from Campylobacter genomes (both C. jejuni and non-jejuni Campylobacter species) as well as other ε-proteobacteria. All 12 C. jejuni and one of two C. coli strains had the cj0414 and cj0415 genes, and in each strain were arranged in a predicted two-gene operon. However, no such cytochrome component is present adjacent to the GADH-encoding genes in any C. jejuni genome for which sequence data are available. Nevertheless, respiration studies using the menaquinol–cytochrome bc1 complex inhibitor HQNO revealed that gluconate-derived electrons most likely enter the respiratory chain via cytochrome c, as HQNO did not inhibit respiration on gluconate.
The present work was undertaken as part of investigations examining the role of temperature-regulated proteins in the colonization potential of C. jejuni. Cj0414 and Cj0415 were identified as two of the proteins that are the most highly upregulated at mid-log phase upon shift in growth temperature from 37°C to 42°C. Interestingly, Cj0414 and Cj0415 are among the most abundant proteins expressed by strain 81–176 at mid-log phase at 42°C (Fig. 1). Microarray and real-time PCR experiments performed in parallel support these proteomics experiments, and indicate that the upregulation of cj0414 and cj0415 occurs at the transcriptional level. The upregulation of cj0414 and cj0415 at 42°C is perhaps surprising, as these genes were identified as being downregulated in chick ceca (Woodall et al., 2005). There are at least two possible explanations for these findings. First, the current work was performed in strain 81–176 rather than with strain NCTC11168 used by Woodall et al. (2005). It is not known whether growth at 42°C regulates the expression of Cj0414 and Cj0415 in strain NCTC11168. Alternatively, it is possible that despite the higher expression of Cj0414 and Cj0415 during in vitro growth at 42°C (when temperature is the only difference), there may be additional signals encountered in chick ceca that override the increased expression of Cj0414/Cj0415 at 42°C. Preliminary evidence suggests that the cj0414 promoter is also slightly activated by gluconate (M. Pajaniappan and S.A. Thompson, unpubl. data, 2008), so it is possible that environmental conditions other than temperature may influence its expression. Stintzi (2003) examined temperature-regulated genes of C. jejuni NCTC11168, using microarrays on cells following upshift from 37°C to 42°C. In that work, cj0414 and cj0415 were not identified as being changed (neither up nor downregulated), although only the first 50 min following temperature upshift were analysed rather than the several hours in the current experiment required to reach mid-log phase growth. Whether this reflects strain heterogeneity (as mentioned above), differences in the kinetics of Cj0414/Cj0415 induction, or involvement of growth phase is not known.
The usage of gluconate as a respiratory substrate by C. jejuni has also been shown to be an important factor during host colonization, although differing effects of GADH on colonization of chickens and mice may reflect the cecal environments (including temperature differences) of the two hosts. In chicks, at the lower individual inoculating doses, clear colonization defects were noted for the cj0415 mutant compared with wild-type (Fig. 6A). When inoculated individually at the higher dose (1 × 104 cfu), there was little difference in the relative abilities of the wild-type and cj0415 mutant strains to initiate colonization measured at 5 days (Fig. 6A). This suggests that the lack of GADH activity was overcome by other metabolic factors present in the cj0415 mutant and that the presence of greater numbers of GADH-deficient bacteria somehow allowed their initial survival and colonization, when not in competition with GADH-expressing C. jejuni. However, when chicks receiving the higher individual 1 × 104 cfu inoculating dose were harvested after 5 weeks (versus 5 days), a statistically significant 10-fold difference in levels of wild-type versus cj0415 mutant C. jejuni were recovered from the ceca, indicating a role for gluconate respiration in long-term maintenance of colonization (Fig. 6B). Finally, the colonization deficiency of GADH mutant C. jejuni was most apparent when they were placed in competition for colonization with wild-type bacteria (Fig. 6C). In this case, GADH-deficient C. jejuni were not recovered at even the earliest time points. It is possible that the competitive deficiency of the cj0415 mutant was due simply to slower growth by the mutant, although the in vitro growth rate of the cj0415 mutant was indistinguishable from wild-type (data not shown). Alternatively, it is possible that 81–176cj0415 is deficient in some other colonization-related phenotype such as in vivo motility or chicken cecal cell adherence. Preliminary experiments revealed no in vitro differences in motility, cell morphology, or adherence to or invasion of INT407 human intestinal epithelial cells (data not shown). These data suggest that utilization of gluconate as a respiratory substrate is important for both the establishment and maintenance of chicken colonization.
The colonization phenotype of GADH-deficient C. jejuni in mice is quite different from that in chicks. In mice, the only statistically significant difference related to GADH activity is in the earliest phases of colonization, and only when the cj0415 mutant was placed in competition with wild-type C. jejuni. At 7 days, the cj0415 mutant exhibited a delay in initiation of colonization, but this was abrogated by 14 days and colonization remained equivalent to wild-type throughout the remainder of the experiment. It is perhaps surprising that loss of GADH activity does not significantly impair the ability of C. jejuni to colonize mice, given the abundance of gluconate (0.69 mM) in the mouse intestinal tract (Peekhaus and Conway, 1998; Chang et al., 2004; Leatham et al., 2005). However, it is possible that there are other electron donors that are of equal or greater abundance in the mammalian gastrointestinal tract that can serve as preferred energy sources during C. jejuni infection. Preliminary data suggests that the genes sdhABC (encoding a putative succinate dehydrogenase) are expressed at higher levels at 37°C than at 42°C, consistent with this hypothesis (S.A. Thompson et al., 2007, unpubl. results).
In several bacteria in which GADH activity has been characterized, this enzymatic activity has been localized to the periplasmic face of the cytoplasmic membrane (Shinagawa et al., 1978; Matsushita et al., 1979; Matsushita et al., 1982; Yum et al., 1997; Deppenmeier et al., 2002). Fractionation experiments show that C. jejuni GADH activity is found in both cytoplasmic and membrane compartments. The association of GADH with the membrane fraction suggests an intimate association of the GADH with a possible membrane-bound cytochrome c. However, the presence of a Tat export signal on Cj0414 strongly suggests an extracytoplasmic localization, probably on the periplasmic face of the cytoplasmic membrane. Using bioinformatics approaches, the Tat signal was previously recognized in Cj0414 (Dilks et al., 2003; Park, 2005). It is likely that Cj0415, which lacks an export signal, is transported using a ‘hitchhiker’ mechanism, similar to many Tat-transported proteins (Rodrigue et al., 1999; Palmer et al., 2005). The cytoplasmic C. jejuni GADH activity is most likely due to GADH enzyme that is assembled in the cytoplasm prior to transport by the Tat apparatus, as occurs with other multisubunit Tat substrates (Palmer et al., 2005).
In summary, we have shown that C. jejuni 81–176 expresses a GADH encoded by the cj0414 and cj0415 genes. This probable membrane-bound GADH enables C. jejuni to utilize an abundant sugar for respiration-dependent energy conservation but not for carbon acquisition. Related proteins are found only in C. jejuni and C. coli but not in any other ε-proteobacteria. GADH activity is induced at the chicken body temperature and is required for high-level colonization of chicks. This method for utilizing gluconate is different from that of most pathogenic bacteria, and is yet another example of the critical importance of bacterial metabolism in establishing host colonization.
Experimental procedures
Bacterial strains and routine culture conditions
Campylobacter jejuni 81–176 (Black et al., 1988; Bacon et al., 2000) was routinely maintained with minimal passage on blood agar plates (Remel; Lenexa, KS) at 37°C in sealed culture boxes [Mitsubishi Gas Chemical (MGC), New York, NY] containing a microaerobic atmosphere generated by Pack-Micro Aero (MGC). Liquid cultures of 81–176 were grown in Brucella broth and cultured in the same environments described above. For the growth of 81–176cj0415, an isogenic mutant derived from 81–176, kanamycin was added to 30 μg ml−1 to select for the mutation.
Escherichia coli JM109 was used as the host strain for cloning experiments, and was cultured in Luria–Bertani broth or agar (Sambrook and Russell, 2001), supplemented with the following antibiotics as appropriate for selection of plasmids: ampicillin, 50 μg ml−1; kanamycin, 30 μg ml−1.
Growth of C. jejuni at two different temperatures
For standardized growth at different temperatures, C. jejuni cells were scraped from blood agar plates grown overnight at 37°C, then re-suspended to an OD600 = 1.0 in a small volume of Brucella broth without antibiotics. A liquid culture was then inoculated with this suspension and grown overnight at 37°C (16 h). The following day, aliquots of this overnight culture were removed and added to two identical, pre-warmed 300 ml sidearm flasks containing 30 ml Brucella broth to an initial OD600 = 0.1. These were quickly placed into two identical incubators (one at 37°C and one at 42°C) with identical microaerophilic atmospheres, and shaken at 100 r.p.m. on identical rotary shaking platforms. Great care was taken to ensure that the only difference between the two cultures was incubation temperature. At the target OD600 = 0.5 (approximately mid-log), cells were harvested rapidly for either microarray or proteomics experiments as follows. To stop RNA synthesis, one tenth volume of a 5% phenol/95% ethanol solution was added and the cells were immediately placed on ice (Gaynor et al., 2005). Chloramphenicol was then immediately added to a final concentration of 187 μg ml−1 to stop protein synthesis (Rivera-Amill et al., 2001). The resulting samples were then split in aliquots for microarrays or proteomics and the cells pelleted by centrifugation at 10 000 r.p.m. for 20 min at 4°C.
Microarray experiments
RNA was prepared for microarray experiments by previously described methods (Gaynor et al., 2005). Briefly, RNA samples from C. jejuni cells grown at 37°C or 42°C were labelled with Cy3 and Cy5 dyes and hybridized against a microarray of C. jejuni genes (Leonard et al., 2003; Gaynor et al., 2005).
Two-dimensional protein gels
Proteomics experiments were performed using differential in-gel electrophoresis technology and a semi-automated workstation supplied by GE Biosystems (Piscataway, NJ), with C. jejuni cells grown to mid-log phase at 37°C and 42°C as described above. Protein lysates were prepared by washing cells three times in wash buffer (10 mM Tris, pH 8.0, 5 mM magnesium acetate) at 4°C for 4 min at 12 000 g. The pellet was re-suspended in 1 ml of lysis buffer [8 M urea, 30 mM Tris, pH 8.5, 5 mM magnesium acetate, 4% (w/v) CHAPS], incubated for 30 min on ice, then sonicated for six ten second bursts (Model 100 Sonic Dismembrator, Fisher Scientific). The lysate was then centrifuged at 4°C for 10 min at 12 000 g and the soluble component used for further studies. Protein concentrations were determined using the BCA assay kit (Pierce, Rockford, IL). Protein samples from cells grown at 37°C and 42°C were labelled individually with Cy3 and Cy5 dyes according to the protocol supplied by the manufacturer (GE Biosystems). Briefly, 25 μg of each protein sample (37°C and 42°C) was labelled in the dark with 1 μl Cy3 and Cy5 dye conjugates, respectively, which label proteins at lysine residues. After 10 min, the reactions were stopped by addition of lysine to a final concentration of 1 mM. The Cy3- and Cy5-labelled proteins were then mixed with an equal amount of unlabelled protein, and finally all proteins mixed together (total of 50 μg of proteins from 37°C and 42°C in a single mixture). The proteins were subjected to isoelectric focusing (IEF) using IPGPhor IEF strips (range of 3–10, nonlinear). Following IEF, the strip was washed with equilibration buffer (6 M urea, 10 mM Tris, pH 6.8, 30% glycerol, 1% SDS), and then placed onto a 12% SDS polyacrylamide gel for second dimension separation.
Following separation, the gel was scanned on a Typhoon fluorescent flatbed scanner (GE Biosystems), at the following wavelengths: Cy3, 540 nm excitation, 590 nm emission; Cy5, 625 nm excitation, 680 nm emission. Images were overlaid and analysed with Decyder Differential In-Gel Analysis (DIA) software (version 4.0, GE Biosystems) for identification of proteins with higher expression under either of the growth conditions.
Proteins of interest were excised by a robotic spot picker and digested with trypsin (Invitrogen). Tryptic peptides were extracted and subjected to mass spectrometry using a matrix-associated laser desorption ionization – time-of-flight/time-of-flight (MALDI-ToF/ToF) spectrometer (Applied Biosystems, Foster City, CA). Protein identifications resulted from parsing protein databases with both tryptic fingerprint data as well as primary amino acid sequence of peptides following fragmentation and MS/MS.
Experiments comparing the proteins expressed at 42°C by 81–176 and its isogenic mutant 81–176cj0415 were performed in an identical manner (data not shown).
Analysis of cj0414 and cj0415 in additional Campylobacter strains
The DNA sequences of the cj0414/cj0415 locus were determined from the seven Campylobacter strains listed in Table S1. Genomic DNA was prepared from these strains using a MasterPure DNA purification kit (Epicentre, Madison, WI). Primers cj0413-F1 and cj0417-R1 (Table S1) were designed based on the published NCTC11168 genome sequence and used in PCR reactions with template DNA to amplify the region encompassing cj0414 and cj0415. Thermocycler parameters were 35 cycles of: 94°C for 30 s, 48°C for 30 s and 72°C for 4 min. PCR products were purified using QIAquick PCR purification kits (Qiagen, Valencia, CA), which were then sequenced directly (both strands) by primer walking, using an ABI 3730xl sequencer (Applied Biosystems). DNA sequences were analysed and assembled using VectorNTI (version 7, Invitrogen, Carlsbad, CA). Protein motifs were determined using the ExPASy Prosite server (http://au.expasy.org/prosite/) (Hulo et al., 2004; Gasteiger et al., 2003), and signal peptides were analysed using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (Bendtsen et al., 2004) and TatP 1.0 (http://www.cbs.dtu.dk/services/TatP/) (Bendtsen et al., 2005).
Studies of cj0414/cj0415 transcription
To determine whether the cj0414 and cj0415 genes were co-transcribed, RNA samples were purified from C. jejuni 81–176 cells grown at 37°C and 42°C using a MasterPure RNA purification kit (Epicentre). The RNA samples were treated with RNase-free DNase to eliminate all traces of genomic DNA. RNA was converted to cDNA with random hexamer primers and 2.5 U Moloney Murine Leukemia Virus (MuLV) reverse transcriptase (Applied Biosystems). A negative control reaction was prepared in which reverse transcriptase was omitted. These samples, primers cj0414-F1 and cj0415-R1 (Table S1), and AmpliTaq polymerase (Qiagen) were used in PCR reactions with 30 amplification cycles of the following parameters: 1 min at 94°C (denaturation), 1 min at 55°C (annealing), 2 min at 72°C (extension). PCR reactions were analysed by electrophoresis in 0.7% agarose gels.
Quantitative real-time reverse transcription PCR
Quantitative real-time reverse transcription (qRT-) PCR was performed according to manufacturer's instructions (Applied Biosystems). Briefly, cDNA was prepared from RNA samples of C. jejuni grown at 37°C and 42°C using a GeneAmp RNA PCR kit (Applied Biosystems). qRT-PCR was run using an ICycler IQ real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) with IQ Sybr Green Super Mix, and primers cj0414-RT-FOR/cj0414-RT-REV and cj0415-RT-FOR/cj0415-RT-REV (Table S1). Data were analysed using on-board Bio-Rad ICycler data analysis software. Primers specific for 16S rDNA (16S-RT-For and 16S-RT-Rev, Table S1) were used in control reactions, and allow amplification of an RNA whose abundance is not regulated (Woodall et al., 2005). The differences in amplification profiles among different samples were calculated using the comparative threshold cycle (ΔΔCT) method to yield fold differences in transcript levels, as previously described (Palyada et al., 2004).
Isolation of C. jejuni cj0414 and cj0415 mutants
For cloning and mutagenesis purposes, we amplified the cj0414 and cj0415 genes from C. jejuni 81–176 as follows. For cj0414 mutagenesis, we performed PCR on C. jejuni 81–176 chromosomal DNA with primers cj0414-F1 and cj0415-R1 (Table S1) designed from the NCTC11168 genome sequence to amplify the entire cj0414 and cj0415 genes. Thermocycler parameters for cj0414-cj0415 amplification were 30 cycles of: 94°C for 1 min, 55°C for 1 min and 72°C for 3 min. The 2.4 kb cj0414-cj0415 PCR product was purified and subcloned in pCRII-TOPO (Invitrogen) to give pTOPO-cj0414/5. To create cloning sites for insertion of the cat cassette, inverse PCR was performed on pTOPO-cj0414/5 using the primers cj0414-inv1 and cj0414-inv2 (Table 1) and the following thermocycler conditions: 94°C for 5 min; then 30 cycles of 94°C for 30 s, 48°C for 30 s and 68°C for 6 min; followed by a final extension at 68°C for 6 min. The cat gene was amplified from pRY111 (Yao et al., 1993) using primers cat-Nhe and cat-Age (Table 1) and the following PCR conditions: 94°C for 2 min; then 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 1 min; followed by a final extension at 72°C for 2 min. The cat and pTOPO-cj0414/5 inverse PCR products were digested with Age I and Nhe I and ligated to each other to create the cj0414 mutation donor construct pTOPO-cj0414-cat. This final construct deleted the 3′ half of the cj0414 gene but maintained the RBS and start codon of cj0415 to avoid transcriptional polarity.
Table 1.
Strains used in this study.
| Strain | HS serotype | Reference |
|---|---|---|
| C. jejuni 81–176 | HS:23; 36 | Black et al. (1988) |
| C. jejuni NCTC11168 | HS:2 | Parkhill et al. (2000) |
| C. jejuni 81116 | HS:6 | Manning et al. (2001) |
| C. jejuni HB95-29 | HS:19 | Nachamkin et al. (2001) |
| C. jejuni INP44 | HS:4 | Nachamkin et al. (2001) |
| C. jejuni INP59 | HS:41 | Nachamkin et al. (2001) |
| C. coli D3088 | HS:19 | Nachamkin et al. (2001) |
For mutating cj0415, we performed PCR on 81–176 DNA with primers cj0415-F1 and cj0415-R1 (Table S1), using the thermocycler parameters: 35 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 2 min. The 1.6 kb cj0415 PCR product was purified, digested with PstI and SalI, and cloned into PstI/SalI-digested pBluescript II to yield pBS-cj0415. We next performed in vitro transposition to interrupt the cj0415 gene, using the EZ∷TN kit (Epicentre). The transposition vector pMOD-2 was digested with BamHI, then ligated with the Tn903 aphA gene (Accession No. X06404) released by BamHI digestion from pUC4K (Vieira and Messing, 1982). The resulting plasmid, pMOD-Kan was used for in vitro transposition as recommended by the manufacturer, with pBS-cj0415 as the target. Plasmids into which transposons had inserted were identified by resistances to ampicillin and kanamycin, and the locations of the transposons within the plasmid were determined by restriction mapping and DNA sequencing.
The cj0414 gene mutated with the cat gene or the cj0415 gene interrupted by the aphA gene were introduced into C. jejuni 81–176 using natural transformation-mediated allelic replacement (Wang and Taylor, 1990). Chloramphenicol- or kanamycin-resistant colonies were screened by colony PCR with primers cj0414-F1 and cj0415-R1 (for the cj0414 mutant) or cj0415-F1 and cj0415-R1 (for the cj0415 mutant). One mutant colony from each donor (designated 81–176cj0414 and 81–176cj0415) was chosen for further study, although cj0414 and cj0415 mutants derived from independent experiments performed on different days were tested in GADH assays (described in the text) and gave identical results. The absence of the Cj0414 and Cj0415 proteins in 81–176cj0415 was verified by proteomics as described above.
GADH assays
Cell lysates were prepared by harvesting C. jejuni cells grown at 37°C and 42°C to mid-log phase as described above, then washing the cells twice with distilled water with centrifugation at 10 000 g at 4°C. The cells were disrupted by sonication in 50 mM acetate buffer (pH 5.0) and cell debris removed by centrifugation at 12 000 g for 20 min at 4°C. GADH enzyme assays were performed as previously described (Yum et al., 1997; Matsushita et al., 1982). Each reaction contained 0.1 ml of 1 M d-gluconate, 1 ml of potassium phosphate buffer (pH 6.0), 0.1 ml of 10 mM DCIP and 0.1 ml of 3 mM PMS made up to 3 ml with water along with 0.2 mg of protein from 81–176, 81–176cj0414, or 81–176cj0415 cell lysates. Enzyme activity was measured by continuously monitoring the reduction rate of DCIP for 15 min at A600 at 25°C. A negative control reaction was included in which the cell lysates were omitted. Subcellular fractions of C. jejuni cells were prepared by the method of Myers and Kelly (Myers and Kelly, 2005), and were assayed for GADH activity as described above.
In vitro growth using different carbon sources and electron donors
The abilities of wild-type 81–176 and mutant 81–176cj0415 to grow on different carbon sources were assessed in defined MEMα medium (Velayudhan and Kelly, 2002). Cells were grown overnight in Brucella Broth, then backdiluted into MEMα containing sodium pyruvate, d-glucose, d-gluconate or 2-KG, each at a final concentration of 20 mM. Growth was monitored by measuring OD600. The potential role of electron donors in stimulating C. jejuni growth was tested by supplementing cultures of 81–176 backdiluted at various densities into MEMα containing 20 mM d-gluconate or 5 mM sodium formate and monitoring growth using OD600.
Measurement of respiration rates by oxygen uptake
Substrate oxidation was determined as described previously (Myers and Kelly, 2005) by measuring the change in dissolved oxygen concentration of cell suspensions in a Clark-type oxygen electrode linked to a chart recorder and calibrated using air saturated 25 mM phosphate buffer (pH 7.5) [220 (nmol dissolved O2) ml−1 at 37°C]. A zero oxygen baseline was determined by the addition of sodium dithionite. Suspensions of wild-type and cj0415 mutant C. jejuni (grown at 37°C and 42°C) were maintained at 37°C and stirred at a constant rate. Substrates were added by injection through a fine central pore in the airtight plug. Rates were expressed in (nmol O2 utilized) min−1 (mg cell protein)−1. For inhibitor experiments, wild-type C. jejuni cells were grown at 42°C and oxygen consumption experiments performed at 37°C as above, in the presence or absence of the menaquinol–cytochrome bc1 complex inhibitor HQNO (2-n-heptyl-4-hydroxyquinoline N-oxide) at a concentration of 20 μM.
Chicken colonization experiments
Colonization of 1-day-old chicks was assayed as previously described (Wassenaar et al., 1993). Briefly, doses of 1 × 104, 3 × 103 and 1 × 103 cfu of wild-type 81–176 and mutant 81–176cj0415 (with similar laboratory passage histories) were administered by oral gavage to 1-day-old chicks hatched from eggs derived from specific-pathogen-free hens (Lohmann's). In different experiments, chickens were sacrificed after either 5 days or 5 weeks, the ceca removed, and the bacteria contained therein were enumerated after growth on sheep's blood agar plates containing cephoperazone (20 μg ml−1). Media were supplemented with kanamycin (30 μg ml−1) for selecting mutant bacteria, although control experiments showed that identical numbers of cj0415 mutant bacteria were obtained whether kanamycin was present in the media or not. For competition experiments, wild-type and mutant bacteria were mixed in equal amounts (4 × 103 cfu of each strain) immediately prior to inoculation. The geometrical mean of colonization densities for each experimental group was calculated and the statistical significance of the means between groups was calculated using INSTAT 2.0 (GraphPad Software, San Diego, CA).
Mouse colonization experiments
Wild-type 81–176 or mutant 81–176cj0415 (at doses of c. 5 × 109, 5 × 107 and 5 × 105 cfu) were administered by oral gavage to BALB/cByJ mice (The Jackson Laboratory, Bar Harbor, ME) as described (Lee et al., 1999; Baqar et al., 1995b). Competition experiments were performed by mixing equal numbers of wild-type and mutant bacteria (c. 2.5 × 109 cfu of each) prior to inoculation. C. jejuni shed in fecal pellets were enumerated on Brucella agar containing 5% (v/v) sheep's blood, cephaperazone (20 μg ml−1), vancomycin (10 μg ml−1) and amphotericin B (2 μg ml−1) without or with kanamycin (30 μg ml−1) to determine the numbers of total or mutant bacteria respectively. All experiments were performed twice, except for the 5 × 107 cfu dose, which was performed once. Statistical significance was calculated as described above for chick colonization experiments.
Supplementary Material
Figure S1. Proteome analysis comparing protein profiles of wild-type 81-176 and mutant 81-176cj0415. Although the downstream cj0415 gene was insertionally inactivated, both the Cj0415 and Cj0414 proteins are missing in the cj0415 mutant.
Figure S2. GADH activity of C. jejuni 81-176 cell fractions. C. jejuni 81-176 cells grown at 42°C were fractionated as described (Myers & Kelly, 2005), and the presence of GADH activity was determined by a coupled enzyme assay. The samples were total membranes (M), cytoplasm (C), periplasm (P), whole cells (WC), or no cell lysate (N) as a negative control.
Figure S3. Growth at 42°C of C. jejuni 81-176 supplemented with electron donors. Growth of C. jejuni in defined αMEM media containing the electron donors gluconate and formate was monitored by optical density at 600 nm. Electron donors added were: 5 mM formate (open circles), 20 mM d-gluconate (open squares), or no added electron donor (open diamonds). Cultures were inoculated at initial OD600 of approximately 0.02 (left panel), 0.06 (center panel), or 0.1 (right panel). Addition of electron donors did not stimulate C. jejuni growth.
Figure S4. In vitro growth profiles of 81-176 and 81-176cj0415. Wild-type and cj0415 mutant strains were grown individually or mixed in equal concentration prior to inoculation; growth of the three cultures was monitored by OD600 (Panel A) and by viable count of the in vitro competition (Panel B). Results showed no differences in the growth of the strains either individually or during in vitro competition.
TABLE S1. Primers used in this study
Acknowledgments
We thank Irv Nachamkin (University of Pennsylvania) for providing some of the strains used in this study, Dawn Israel (Vanderbilt University) for plasmid pUC4K, and David White (Indiana University) and members of the Thompson lab for helpful comments. This work was supported in part by National Institutes of Health grants AI055715 and AI058284 (to S.A.T.) and by grant BB/D008395/1 from the UK Bioctechnology and Biological Sciences Research Council (to D.J.K.). The research of E.C.G. is supported in part by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and also by a Canada Research Chair award, the Michael Smith Foundation for Health Research, and a Canadian Institutes of Health Research Operating Grant (MOP-68981).
Footnotes
Supplementary material: This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2008.06161.x
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baqar S, Applebee LA, Bourgeois AL. Immunogenicity and protective efficacy of a prototype Campylobacter killed whole-cell vaccine in mice. Infect Immun. 1995a;63:3731–3735. doi: 10.1128/iai.63.9.3731-3735.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baqar S, Bourgeois AL, Schultheiss PJ, Walker RI, Rollins DM, Haberberger RL, Pavlovskis OR. Safety and immunogenicity of a prototype oral whole-cell killed Campylobacter vaccine administered with a mucosal adjuvant in non-human primates. Vaccine. 1995b;13:22–28. doi: 10.1016/0264-410x(95)80006-y. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- Brás AM, Chatterjee S, Wren BW, Newell DG, Ketley JM. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J Bacteriol. 1999;181:3298–3302. doi: 10.1128/jb.181.10.3298-3302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppenmeier U, Hoffmeister M, Prust C. Biochemistry and biotechnological applications of Gluconobacter strains. Appl Microbiol Biotechnol. 2002;60:233–242. doi: 10.1007/s00253-002-1114-5. [DOI] [PubMed] [Google Scholar]
- Dilks K, Rose RW, Hartmann E, Pohlschroder M. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol. 2003;185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R. Mechanisms that control the microflora in the large intestine. In: Hentges DJ, editor. Human Intestinal Microflora in Health and Disease. New York: Academic Press; 1983. pp. 33–54. [Google Scholar]
- Freter R. Mechanisms of bacterial colonization of the mucosal surface of the gut. In: Roth JA, editor. Virulence Mechanisms of Bacterial Pathogens. Washington, DC: American Society for Microbiology; 1988. pp. 45–60. [Google Scholar]
- Friedman CR, Neimann J, Wegener HC, Tauxe RV. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser MJ, editors. Campylobacter. Washington, DC: American Society for Microbiology Press; 2000. pp. 121–138. [Google Scholar]
- Fuhrer T, Fischer E, Sauer U. Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol. 2005;187:1581–1590. doi: 10.1128/JB.187.5.1581-1590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucl Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Cawthraw S, Manning G, MacKichan JK, Falkow S, Newell DG. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J Bacteriol. 2004;186:503–517. doi: 10.1128/JB.186.2.503-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol. 2005;56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- Hoffman PS, Goodman TG. Respiratory physiology and energy conservation efficiency of Campylobacter jejuni. J Bacteriol. 1982;150:319–326. doi: 10.1128/jb.150.1.319-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Sigrist CJ, Le Saux V, Langendijk-Genevaux PS, Bordoli L, Gattiker A, et al. Recent improvements to the PROSITE database. Nucl Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurme R, Berndt KD, Normark SJ, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90:55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- Jorgensen F, Bailey R, Williams S, Henderson P, Wareing DR, Bolton FJ, et al. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int J Food Microbiol. 2002;76:151–164. doi: 10.1016/s0168-1605(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Kelly DJ. Metabolism, electron transport and bioenergetics of Campylobacter jejuni: implications for understanding life in the gut and survival in the environment. In: Ketley JM, Konkel ME, editors. Campylobacter: Molecular and Cellular Biology. Norfolk: Horizon Bioscience; 2005. pp. 275–292. [Google Scholar]
- Konkel ME, Kim BJ, Klena JD, Young CR, Ziprin R. Characterization of the thermal stress response of Campylobacter jejuni. Infect Immun. 1998;66:3666–3672. doi: 10.1128/iai.66.8.3666-3672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2000;2:157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- Kramer JM, Frost JA, Bolton FJ, Wareing DR. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J Food Prot. 2000;63:1654–1659. doi: 10.4315/0362-028x-63.12.1654. [DOI] [PubMed] [Google Scholar]
- Leatham MP, Stevenson SJ, Gauger EJ, Krogfelt KA, Lins JJ, Haddock TL, et al. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect Immun. 2005;73:8039–8049. doi: 10.1128/IAI.73.12.8039-8049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LH, Burg E, 3rd, Baqar S, Bourgeois AL, Burr DH, Ewing CP, et al. Evaluation of a truncated recombinant flagellin subunit vaccine against Campylobacter jejuni. Infect Immun. 1999;67:5799–5805. doi: 10.1128/iai.67.11.5799-5805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard EEN, Takata T, Blaser MJ, Falkow S, Tompkins LS, Gaynor EC. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J Infect Dis. 2003;187:691–694. doi: 10.1086/368268. [DOI] [PubMed] [Google Scholar]
- Lessie TG, Phibbs PV., Jr Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–388. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- MacKichan JK, Gaynor EC, Chang C, Cawthraw S, Newell DG, Miller JF, Falkow S. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol Microbiol. 2004;54:1269–1286. doi: 10.1111/j.1365-2958.2004.04371.x. [DOI] [PubMed] [Google Scholar]
- Manning G, Duim B, Wassenaar T, Wagenaar JA, Ridley A, Newell DG. Evidence for a genetically stable strain of Campylobacter jejuni. Appl Environ Microbiol. 2001;67:1185–1189. doi: 10.1128/AEM.67.3.1185-1189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Shinagawa E, Adachi O, Ameyama M. Membrane-bound d-gluconate dehydrogenase from Pseudomonas aeruginosa. Purification and structure of cytochrome-binding form. J Biochem (Tokyo) 1979;85:1173–1181. [PubMed] [Google Scholar]
- Matsushita K, Shinagawa E, Ameyama M. d-Gluconate dehydrogenase from bacteria, 2-keto-d-gluconate-yielding, membrane-bound. Methods Enzymol. 1982;89(Pt D):187–193. doi: 10.1016/s0076-6879(82)89033-2. [DOI] [PubMed] [Google Scholar]
- McIntire W, Singer TP, Ameyama M, Adachi O, Matsushita K, Shinagawa E. Identification of the covalently bound flavins of d-gluconate dehydrogenases from Pseudomonas aeruginosa and Pseudomonas fluorescens and of 2-keto-d-gluconate dehydrogenase from Gluconobacter melanogenus. Biochem J. 1985;231:651–654. doi: 10.1042/bj2310651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Mandrell RE. Prevalence of. Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection. In: Ketley JM, Konkel ME, editors. Campylobacter: Molecular and Cellular Biology. Norfolk: Horizon Bioscience; 2005. pp. 101–163. [Google Scholar]
- Myers JD, Kelly DJ. A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology. 2005;151:233–242. doi: 10.1099/mic.0.27573-0. [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Allos BM, Ho TW. Campylobacter jejuni infection and the association with Guillain–Barré syndrome. In: Nachamkin I, Blaser MJ, editors. Campylobacter. Washington, DC: American Society for Microbiology Press; 2000. pp. 155–175. [Google Scholar]
- Nachamkin I, Engberg J, Gutacker M, Meinersmann RJ, Li CY, Arzate P, et al. Molecular population genetic analysis of Campylobacter jejuni HS: 19 associated with Guillain–Barré syndrome and gastroenteritis. J Infect Dis. 2001;184:221–226. doi: 10.1086/322008. [DOI] [PubMed] [Google Scholar]
- Nadeau E, Messier S, Quessy S. Prevalence and comparison of genetic profiles of Campylobacter strains isolated from poultry and sporadic cases of campylobacteriosis in humans. J Food Prot. 2002;65:73–78. doi: 10.4315/0362-028x-65.1.73. [DOI] [PubMed] [Google Scholar]
- Newell DG, Wagenaar JA. Poultry infections and their control at the farm level. In: Nachamkin I, Blaser MJ, editors. Campylobacter. Washington, DC: American Society for Microbiology Press; 2000. pp. 497–509. [Google Scholar]
- Palmer T, Sargent F, Berks BC. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 2005;13:175–180. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Campylobacter stress responses during survival in the food chain and colonization. In: Ketley JM, Konkel ME, editors. Campylobacter: Molecular and Cellular Biology. Norfolk: Horizon Bioscience; 2005. pp. 311–330. [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Peekhaus N, Conway T. What's for dinner? Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Burucoa C, Grignon B, Baqar S, Huang XZ, Kopecko DJ, et al. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect Immun. 1998;66:938–943. doi: 10.1128/iai.66.3.938-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucl Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman MS, Kelly DJ. Electron transport through nitrate and nitrite reductases in Campylobacter jejuni. Biochem Soc Trans. 2005;33:190–192. doi: 10.1042/BST0330190. [DOI] [PubMed] [Google Scholar]
- Rappuoli R, Arico B, Scarlato V. Thermoregulation and reversible differentiation in Bordetella: a model for pathogenic bacteria. Mol Microbiol. 1992;6:2209–2211. doi: 10.1111/j.1365-2958.1992.tb01395.x. [DOI] [PubMed] [Google Scholar]
- Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- Rodrigue A, Chanal A, Beck K, Muller M, Wu LF. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schleissner C, Reglero A, Luengo JM. Catabolism of d-glucose by Pseudomonas putida U occurs via extracellular transformation into d-gluconic acid and induction of a specific gluconate transport system. Microbiology. 1997;143(Pt 5):1595–1603. doi: 10.1099/00221287-143-5-1595. [DOI] [PubMed] [Google Scholar]
- Shinagawa E, Chiyonobu T, Matsushita K, Adachi O, Ameyama M. Distribution of gluconate dehydrogenase and ketogluconate reductases in aerobic bacteria. Agric Biol Chem. 1978;42:1055–1057. [Google Scholar]
- Smith MA, Finel M, Korolik V, Mendz GL. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch Microbiol. 2000;174:1–10. doi: 10.1007/s002030000174. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Schwan TG, Rosa PA. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J Bacteriol. 2003;185:2009–2016. doi: 10.1128/JB.185.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Whitworth L. Investigation of the Campylobacter jejuni cold-shock response by global transcript profiling. Genome Lett. 2003;2:18–27. [Google Scholar]
- Straley SC, Perry RD. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995;3:310–317. doi: 10.1016/s0966-842x(00)88960-x. [DOI] [PubMed] [Google Scholar]
- Swanson BL, Hager P, Phibbs P, Jr, Ochsner U, Vasil ML, Hamood AN. Characterization of the 2-ketogluconate utilization operon in Pseudomonas aeruginosa PAO1. Mol Microbiol. 2000;37:561–573. doi: 10.1046/j.1365-2958.2000.02012.x. [DOI] [PubMed] [Google Scholar]
- Sweeney NJ, Klemm P, McCormick BA, Moller-Nielsen E, Utley M, Schembri MA, et al. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996a;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NJ, Laux DC, Cohen PS. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun. 1996b;64:3504–3511. doi: 10.1128/iai.64.9.3504-3511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies FL, Karch H, Hartung HP, Giegerich G. Cloning and expression of the dnaK gene of Campylobacter jejuni and antigenicity of heat shock protein 70. Infect Immun. 1999a;67:1194–1200. doi: 10.1128/iai.67.3.1194-1200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies FL, Karch H, Hartung HP, Giegerich G. The ClpB protein from Campylobacter jejuni: molecular characterization of the encoding gene and antigenicity of the recombinant protein. Gene. 1999b;230:61–67. doi: 10.1016/s0378-1119(99)00054-2. [DOI] [PubMed] [Google Scholar]
- Thies FL, Weishaupt A, Karch H, Hartung HP, Giegerich G. Cloning, sequencing and molecular analysis of the Campylobacter jejuni groESL bicistronic operon. Microbiology. 1999c;145:89–98. doi: 10.1099/13500872-145-1-89. [DOI] [PubMed] [Google Scholar]
- Truesdell SJ, Sims JC, Boerman PA, Seymour JL, Lazarus RA. Pathways for metabolism of ketoaldonic acids in an Erwinia sp. J Bacteriol. 1991;173:6651–6656. doi: 10.1128/jb.173.21.6651-6656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunedomi R, Izu H, Kawai T, Matsushita K, Ferenci T, Yamada M. The activator of GntII genes for gluconate metabolism, GntH, exerts negative control of GntR-regulated GntI genes in Escherichia coli. J Bacteriol. 2003;185:1783–1795. doi: 10.1128/JB.185.6.1783-1795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J, Jones MA, Barrow PA, Kelly DJ. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J, Kelly DJ. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology. 2002;148:685–694. doi: 10.1099/00221287-148-3-685. [DOI] [PubMed] [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang Y, Taylor DE. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, van der Zeijst BA, Ayling R, Newell DG. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J Gen Microbiol. 1993;139:1171–1175. doi: 10.1099/00221287-139-6-1171. [DOI] [PubMed] [Google Scholar]
- White D. The Physiology and Biochemistry of Prokaryotes. New York: Oxford University Press; 2007. Electron transport; pp. 120–148. [Google Scholar]
- Woodall CA, Jones MA, Barrow PA, Hinds J, Marsden GL, Kelly DJ, et al. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect Immun. 2005;73:5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- Yum DY, Lee BY, Hahm DH, Pan JG. The yiaE gene, located at 80.1 minutes on the Escherichia coli chromosome, encodes a 2-ketoaldonate reductase. J Bacteriol. 1998;180:5984–5988. doi: 10.1128/jb.180.22.5984-5988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yum DY, Lee YP, Pan JG. Cloning and expression of a gene cluster encoding three subunits of membrane-bound gluconate dehydrogenase from Erwinia cypripedii ATCC 29267 in Escherichia coli. J Bacteriol. 1997;179:6566–6572. doi: 10.1128/jb.179.21.6566-6572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Proteome analysis comparing protein profiles of wild-type 81-176 and mutant 81-176cj0415. Although the downstream cj0415 gene was insertionally inactivated, both the Cj0415 and Cj0414 proteins are missing in the cj0415 mutant.
Figure S2. GADH activity of C. jejuni 81-176 cell fractions. C. jejuni 81-176 cells grown at 42°C were fractionated as described (Myers & Kelly, 2005), and the presence of GADH activity was determined by a coupled enzyme assay. The samples were total membranes (M), cytoplasm (C), periplasm (P), whole cells (WC), or no cell lysate (N) as a negative control.
Figure S3. Growth at 42°C of C. jejuni 81-176 supplemented with electron donors. Growth of C. jejuni in defined αMEM media containing the electron donors gluconate and formate was monitored by optical density at 600 nm. Electron donors added were: 5 mM formate (open circles), 20 mM d-gluconate (open squares), or no added electron donor (open diamonds). Cultures were inoculated at initial OD600 of approximately 0.02 (left panel), 0.06 (center panel), or 0.1 (right panel). Addition of electron donors did not stimulate C. jejuni growth.
Figure S4. In vitro growth profiles of 81-176 and 81-176cj0415. Wild-type and cj0415 mutant strains were grown individually or mixed in equal concentration prior to inoculation; growth of the three cultures was monitored by OD600 (Panel A) and by viable count of the in vitro competition (Panel B). Results showed no differences in the growth of the strains either individually or during in vitro competition.
TABLE S1. Primers used in this study


