Abstract
Although infection with Campylobacter jejuni is one of the leading causes of gastroenteritis worldwide, relatively little is known about the factors that are required to elicit a protective immune response. The need for a vaccine against this pathogen is well recognized and a number of vaccine candidates have been tested with varying degrees of success; however, there is still a lack of a suitable vaccine. To gain a better understanding of the outer-membrane protein components of this organism, a ‘gold standard’ method to purify the outer membrane is needed. Therefore, we attempted to develop a robust and reliable method which resulted in a pure outer-membrane fraction. A total of nine methodologies were examined and analysed by SDS-PAGE and immunoblotting using subcellular markers for the cytoplasm, cytoplasmic membrane and outer membrane. We found that glycine extraction, differential detergent extraction using Triton X-100, serial extraction using 1 M Tris pH 7, spheroplasting by lysozyme and sonication, and carbonate extraction did not produce pure outer-membrane preparations. However, we identified three methods that provided outer-membrane fractions free from subcellular contamination. Isopycnic centrifugation using a 30–60 % sucrose gradient produced seven fractions free from cytoplasmic or cytoplasmic membrane contamination; however, these fractions did not correspond as well as expected with the typical outer-membrane-associated peak (e.g. Escherichia coli or Salmonella). The spheroplast method using lysozyme alone also resulted in pure outer-membrane fraction, as did carbonate washing of this sample. The extraction of outer membranes using N-lauroylsarcosine (Sarkosyl) produced the purest and most reproducible sample. These outer-membrane preparations will be useful for future studies aimed at identifying C. jejuni surface proteins as vaccine components.
INTRODUCTION
Infection with Campylobacter jejuni is one of the most common causes of gastroenteritis worldwide (Allos, 2001; Girard et al., 2006, Buzby et al., 1997; Samuel et al., 2004). Disease arises most commonly after consumption of undercooked chicken products, a natural reservoir of C. jejuni, contaminated during processing. Human campylobacteriosis is an acute self-limiting gastrointestinal illness, characterized by (often bloody) diarrhoea, fever and abdominal cramps and usually consists of a 5–7 day course of illness (Butzler, 2004).
The development of a vaccine against Campylobacter constitutes a high priority for the military, for travellers and for infants in the developing world (Girard et al., 2006). To date, an effective vaccine is not available. Some of the problems encountered by researchers trying to develop Campylobacter vaccines range from the relative lack of experimental genetic systems, lack of understanding of the mechanism of pathogenicity and infection and poor animal models, to tremendous antigenic diversity of the organism, poorly defined protective epitopes and a lack of understanding of what constitutes a protective immune response for this organism. A number of vaccine candidates have been tested with varying degrees of success (Baqar et al., 1995, Lee et al., 1999, Sizemore et al., 2006).
Outer-membrane proteins (OMPs) are the most obvious targets for a protective immune response, and therefore for subunit vaccine research, primarily because they are surface exposed. Most of what is known about C. jejuni OMPs is derived from the NCTC 11168 genome (Parkhill et al., 2000). The genome sequence predicts ~20 genes encoding OMPs. However, prediction of OMP genes by the genome sequence is just a starting point toward understanding OMPs that are actually present. Only a handful of C. jejuni proteins have actually been experimentally localized to the outer membrane; they include MOMP (De et al., 2000; Labesse et al., 2001; Zhang et al., 2000), Omp50 (Bolla et al., 2000), FlgE (Kinsella et al., 1997; Luneberg et al., 1998), CadF (Konkel et al., 1997; Monteville & Konkel, 2002; Moser et al., 1997), PEB1 (Pei & Blaser, 1993; Pei et al., 1998), JlpA (Jin et al., 2001), CmeC (Fakhr & Logue, 2007) and OmpH 1 (Meinersmann et al., 1997). It has been estimated that approximately 2–3 % of the Gram-negative genome encodes integral OMPs, and a significant proportion of these are expressed ubiquitously (~0.5 % of the genome, which correlates to about 20 proteins that are abundant in Escherichia coli (Molloy et al., 2000; Wimley, 2003). Because the genome of C. jejuni is ~1.6 Mb in size, at least three times smaller than that of E. coli, 0.5 % would correspond to only eight OMPs in its proteome; this is highly unlikely. Further, computer predictions are certainly imperfect and it is possible that important OMPs may be missed due to the inability of computer algorithms to accurately predict OMPs.
In the past, crude extractions were used by Campylobacter researchers; however, a standard method has not been established. The objective of this work was to identify a reliable, reproducible and sensitive method for isolation of OMPs from C. jejuni. Using a number of isolation techniques commonly employed for bacterial pathogens and monitored by fractionation of marker proteins, we determined that incubation of membranes with N-lauryolsarcosine (Sarkosyl) provided an outer-membrane preparation that contained a large number of OMPs with relatively few contaminating proteins.
METHODS
Bacterial strains, media and growth conditions
Campylobacter jejuni strains 81–176 (serotype HS: 23,26), INP44 (serotype HS: 4), INP59 (serotype HS: 41), HB95-29 (serotype HS: 19), 81116 (serotype HS:6) and 11168 (serotype HS: 2), and Campylobacter coli strain D3088, were used in this study. Stock cultures were maintained at −80 °C in 20 % (v/v) glycerol-Mueller–Hinton broth (MHB) (Difco). Cultures were grown on Tryptic soy agar (Difco) with 5 % defibrinated sheep’s blood (Remel) at 37 °C in an atmosphere containing 5 % O2 and 10 % CO2 produced by means of a gas generation kit for campylobacters (Pack-MicroAero, Mitsubishi Gas Chemical Co.). Plate cultures were passed to MHB and cultivated at 37 °C under microaerophilic conditions (10 % CO2). Escherichia coli strain W3110 (a gift from M. Stephen Trent) was used as a control strain for sucrose gradient centrifugation and was grown in Luria–Bertani broth (Difco) at 37 °C with shaking.
French pressure cell disruption
C. jejuni or C. coli cells were lysed by passing the culture twice through a French press (Thermo Electron Corporation) at 1000 p.s.i. (6.9 MPa; 40K cell), unless otherwise stated. The lysed cell preparation was centrifuged at 10 000 g for 10 min at 4 °C to remove cell debris and unlysed cells.
Isolation of outer membranes using N-lauroylsarcosine
Cells from a 250 ml culture of C. jejuni or C. coli were resuspended in 7 ml 10 mM HEPES, pH 7.4, and lysed by French pressure cell disruption (as described above). The membranes were collected by ultracentrifugation at 100 000 g for 1 h at 4 °C (Beckman, Ti70.1 rotor). The pellet was resuspended in 2 ml 10 mM HEPES, pH 7.4, using an 18-gauge needle, washed in a total volume of 10 ml 10 mM HEPES, pH 7.4, and spun again in the ultracentrifuge (using the conditions described above). The pellet was resuspended in 5 ml 1 % (w/v) N-lauroylsarcosine (Sarkosyl) (Sigma) in 10 mM HEPES, pH 7.4, and incubated at 37 °C for 30 min with shaking. The Sarkosyl-treated membranes were spun at 100 000 g for 1 h at 4 °C (Beckman, Ti70.1 rotor) and the pellet washed with 10 ml 10 mM HEPES, pH 7.4. Following the final ultracentrifugation, the pellet was resuspended in 500 μl 10 mM HEPES, pH 7.4.
Sucrose density-gradient centrifugation
Double-washed membranes were prepared from a 250 ml culture of C. jejuni or C. coli. Cells were lysed by French pressure cell disruption (as described above) and centrifuged at 10 000 g for 10 min (JA17 rotor, Beckman). The supernatant was ultracentrifuged at 100 000 g (Ti70.1 rotor, Beckman) for 60 min at 4 °C to pellet the total membranes. The membrane pellet was washed in 10 ml 10 mM HEPES, 0.05 M EDTA pH 7.5 (HE buffer), and ultracentrifuged again. The final membranes were homogenized in 2 ml HE buffer.
Continuous sucrose gradients were prepared by layering sucrose solutions (prepared in HE buffer) into 14 × 89 mm ultracentrifuge tubes (Beckman) in the following order: 0.4 ml 60 % (w/v), 0.9 ml 55 %, 2.2 ml 50 %, 2.2 ml 45 %, 2.2 ml 40 %, 1.3 ml 35 % and 0.4 ml 30 %. Total membranes were layered on top of each gradient, with no more than 2.5 ml per gradient. Sucrose gradients were centrifuged in an SW41 swinging-bucket rotor (Beckman) at 100 000 g for 60 min at 4 °C. The sucrose gradient tubes were then removed from the rotor buckets and 500 μl fractions were collected from the bottom of each gradient by puncturing with a 26½-gauge needle and allowing them to drip by gravity.
Other extraction methods
Glycine extracts were performed by the method of Logan & Trust (1983). Differential detergent extraction (Triton X-100) of cell wall proteins was performed by the method of Murakami et al. (2002). Cell wall proteins were isolated from C. jejuni or C. coli by a serial extraction method as described by Schaumburg et al. (2004). Spheroplast formation using lysozyme digestion was carried out on C. jejuni or C. coli cultures using the method of Hill & Sillence (1997). Spheroplasting using sonication and lysozyme digestion was carried out as described by Osborn & Munson (1974). Spheroplasts were washed with carbonate buffer as described by Hill & Sillence (1997). Outer membranes were also isolated using the carbonate extraction method of Molloy et al. (2000).
SDS-PAGE and staining
Polyacrylamide gel electrophoresis, using 12 % acrylamide gels, was carried out using the method of Laemmli (1970). Proteins were stained with Silver Stain Plus (Bio-Rad).
Dialysis, protein concentration and protein estimation
Dialysis was performed using 10 000 MWCO Snakeskin Pleated dialysis tubing (Pierce) against distilled H2O at 4 °C with two or three buffer changes over a period of 48 h. If required, proteins were concentrated using Centricon YM-3 (3000 Da cut-off) columns (Millipore). Protein content was determined using the bicinchoninic acid (BCA) assay kit (Pierce), following the manufacturer’s instructions.
Protein purification of recombinant Cj0355
Primers JAF01 (5′-CATATGATGAGAATTTTAGTTATAGAAGATG-3′) and JAF02 (5′-CTCGAGAGATTTTTTAGGGAAGCAGAAACGGTATCC-3′) (restriction sites underlined) were used to amplify a 680 bp fragment from strain 81–176. The PCR product was cloned into pCRII-Topo (Invitrogen) to yield pJAF01. The insert was released by NdeI/XhoI digestion and subsequently cloned into the NdeI/XhoI site of the C-terminally His-tagged expression vector pET20b (Novagen) to yield pJAF02. pJAF02 was transformed into the E. coli expression strain BL21DE3(pLysS) for overexpression by the method of Studier & Moffat (1986). A 2 litre culture was grown in LB supplemented with 50 μg ampicillin ml−1 to an OD600 of 0.5 and induced with 1 mM IPTG for 3 h. The cell pellet was washed with STE buffer (0.1 M NaCl, 10 mM Tris/HCl, pH 8, 1 mM EDTA, pH 8) and resuspended in lysis buffer (50 mM Tris pH 7.8, 2 mM β-mercaptoethanol, 5 % glycerol, 2 mM EDTA, pH 8) and homogenized. Four millilitres of lysis buffer was added to the homogenate, incubated for 30 min then sonicated on full power for ten 10 s pulses. The lysate was centrifuged at 12 000 r.p.m. (Beckman, JA17 rotor) and the supernatant collected. The supernatant was fractionated with ammonium sulfate (0.56 g ml−1) for 1 h and centrifuged as previously mentioned. The pellet was resuspended in CJA buffer (125 mM Tris pH 7.8, 5 mM β-mercaptoethanol, 2.5 % glycerol, 200 mM NaCl) and dialysed against CJA buffer at 4 °C overnight. The dialysed sample was applied to a Ni-Sepharose column (1.6 × 30 cm) under the control of the AKTAprime protein purification system (Amersham Pharmacia) and eluted with CJA buffer containing 300 mM imidazole. Fractions were collected and those containing the purified C-terminally His-tagged Cj0355 were pooled and dialysed against storage buffer. A total of 1 mg of the purified protein was sent to Cocalico Biologicals (Reamstown, PA, USA) for the production of rabbit polyclonal antisera.
Antisera and Western immunoblotting
Antisera raised in rabbits to Cj0355 (C. jejuni cytoplasmic orphan response regulator protein), CetA (C. jejuni inner-membrane energy taxis protein) and MOMP (C. jejuni major OMP) were used as markers to validate the localization of protein fractions. Proteins were transferred to nitrocellulose (Nitrobind, Osmonics) membrane using a Trans-blot semi-dry transfer apparatus (Bio-Rad). The membrane was blocked with 5 % skim milk in Tris-buffered saline, pH 7.4, for 60 min at room temperature or overnight at 4 °C. Transfer of proteins was confirmed by staining the gel with Coomassie brilliant blue (Gelcode, Pierce). Standard immunoblotting methodologies were utilized. Primary antibodies were used at a 1: 1000 dilution in blocking buffer. Secondary antibody (goat anti-rabbit IgG-horseradish perox-idase, Pierce) was used at 1: 10 000) The bands were developed using DAB (stable horseradish peroxidase conjugate, Pierce) development solution at room temperature, until bands were visible. Development was stopped by rinsing the membrane in distilled water.
RESULTS AND DISCUSSION
Subcellular localization using enzymic activity
To evaluate outer-membrane separation techniques, it was necessary to first establish assays to measure levels of cross-subcellular contamination of the fractions obtained. A number of enzymic assays are commonly reported for this function in the literature; these include NADH dehydrogenase (cytoplasmic membrane) (Kubak & Yotis, 1981; Linder et al., 1983), succinate dehydrogenase (cytoplasm) (Johnston & Gotschlich, 1974), alkaline phosphatase (cytoplasmic membrane/periplasmic space) (Alahari et al., 2007), aminopeptidase C/PepC (cytoplasm) (Schaumburg et al., 2004), hippurate hydrolase (cytoplasm) (Steele et al., 2006) and lactate dehydrogenase (cytoplasm) (Bledsoe et al., 1994; Horikiri et al., 2003; Johnston & Gotschlich, 1974; Yamada & Carlsson, 1975). Upon closer inspection of the C. jejuni NCTC 11168 genome sequence (Parkhill et al., 2000), some of these proteins are either not present or are not functionally active. C. jejuni does not utilize NADH as a respiratory substrate, as the NADH dehydrogenase subunit of complex I is not encoded in the genome sequence. Similarly, aminopeptidase C is also not encoded in the genome sequence. Although alkaline phosphatase (Cj0145), hippurate hydrolase (Cj0985c), succinate dehydrogenase (Cj0437-9) and lactate dehydrogenase (Cj1167) are encoded in the NCTC 11168 genome sequence, these enzymic assays are either not sensitive enough to differentiate subcellular fractions (data not shown), are absent in study strain 81–176, or are inhibited by the reagents used to prepare the outer-membrane fractions (for example, HEPES, Tris/HCl and EDTA). Therefore, these proteins were not found to be useful enzymic markers of subcellular contamination (data not shown). Even though succinate dehydrogenase has been used to analyse sub-cellular fractions of C. jejuni, C. coli and C. fetus strains (Page & Taylor, 1988), we did not pursue the use of this enzymic marker because it has been reported that succinate dehydrogenase is not found in some Campylobacter species, including C. coli (Miller 2008), based on genome sequences.
Identification of protein markers for immunoblotting assays
In order to circumvent the problems encountered with enzymic markers, we decided to pursue antibody-based markers. Our aim was to identify markers to be used in identifying the cytoplasm, cytoplasmic membrane and outer-membrane fractions.
The antibodies chosen to detect markers were either prepared in our laboratory or kind gifts from other researchers. The cytoplasmic marker used in this study was Cj0355. Cj0355 is a DNA-binding orphan response regulator and, based on its function and on the algorithms of the bacterial subcellular localization prediction program pSORTb (Gardy et al., 2005), is strongly predicted to be localized within the cytoplasm. Recombinant Cj0355 protein was prepared by expressing the mature form of the protein from the His-tag expression vector pET20b (Novagen). Purification of the recombinant His-tagged protein was performed using standard native purification methodologies (data not shown). Rabbit polyclonal antibodies raised against Cj0355 were used as the cytoplasmic marker. The anti-Cj0355 antiserum, at a dilution of 1: 1000, could detect 10 ng purified recombinant protein (data not shown).
The cytoplasmic membrane marker used in this study was antiserum raised against the energy taxis protein CetA (Cj1190). The predicted protein sequence of CetA contains an N-terminal transmembrane helix domain (TMHMM server, v2.0) and localization to the cytoplasmic membrane is predicted by the pSORTb program (Gardy et al., 2005). Topology analysis has also been performed using phoA and lacZ fusions, which indicated that CetA has two transmembrane helices in a helical hairpin. Localization studies in C. jejuni have also been performed showing that CetA is located within the cytoplasmic membrane fraction (Elliott & DiRita, 2008). The antiserum against CetA was a kind gift from Vic DiRita (University of Michigan).
Due to the fact that PorA (MOMP, Cj1259) is the major OMP of C. jejuni (Albert et al., 2007; Clark et al., 2007), it was an obvious choice as an outer-membrane marker (antibodies against MOMP were a kind gift from Qijing Zhang, Iowa State University). This protein has been well documented in the literature as an OMP.
All three antibodies were tested, at various dilutions ranging from 1: 500 to 1: 10 000, for optimal reactivity against a standard whole-cell preparation of C. jejuni strain 81–176 (10 μg total protein). An antibody dilution of 1: 1000 was determined to be the optimal dilution to use in terms of sensitivity to this sample (data not shown) and was further used in all subsequent immunoblot experiments.
Membrane separation techniques
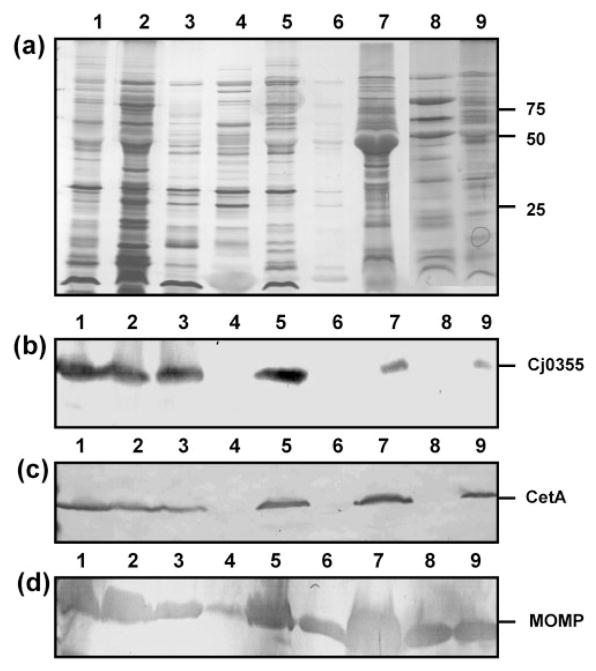
In an effort to determine a suitable method for the purification of OMPs from C. jejuni strain 81–176, a total of nine protocols were analysed for the ability to produce pure outer-membrane preparations free from cytoplasmic and cytoplasmic membrane proteins, which are as complete as possible (i.e. contain the entire repertoire of OMPs) and which are reproducible in terms of the consistent identification of proteins purified. Other methods, however, are available and include techniques such as comparative proteomics (Cordwell et al., 2008). The use of a combination of proteomics and immunoblotting is a very interesting approach in the identification of OMPs; however, it is still limited by the method used to prepare the outer-membrane fraction so that potential false positives are reduced. Preparations obtained by glycine extraction, differential detergent extraction using Triton X-100, serial extraction using 1 M Tris/HCl, pH 7.8, and carbonate extraction were all heavily contaminated with both Cj0355 and CetA, indicating both cytoplasmic and cytoplasmic membrane contamination (Fig. 1, panels b and c, respectively, lanes 1, 2, 3 and 7). Glycine extraction has been previously used to provide fractions containing outer membranes of C. jejuni (Herbrink et al., 1988); however we were attempting to isolate pure outer-membrane fractions and therefore this method was not satisfactory for our needs. The same was found for Triton X-100 extraction in our hands and others (Page & Taylor, 1988). Similarly, outer membranes produced using lysozyme and sonication also contained both cytoplasmic and cytoplasmic membrane contamination (Fig. 1b and c, lane 5). These methods, although producing a large number of proteins and clearly containing OMPs as shown by immunoblot detection of the MOMP protein (Fig. 1d), were not found to be useful primarily due to their significant impurity.
Fig. 1.
Outer-membrane fractions of C. jejuni strain 81–176 from nine purification methods. (a) Silver-stained SDS-PAGE analysis of OMP preparations (5 μg). Lanes: 1, glycine extraction; 2, differential detergent extraction using Triton X-100; 3; serial extraction using 0.1 M Tris/HCl, pH 7.8; 4, spheroplasts isolated using lysozyme treatment; 5, spheroplasts isolated using sonication and lysozyme treatment; 6, carbonate washed spheroplasts; 7, carbonate extraction; 8, Sarkosyl extraction; 9, sucrose gradient fraction 14. Molecular masses are indicated on the right (in kDa). (b–d) Immunoblots of the outer-membrane preparations using the cytoplasmic marker anti-Cj0355 (b), the inner-membrane marker anti-CetA (c) and the outer-membrane marker anti-MOMP (d). All antibodies were used at 1: 1000 dilution. Loading for panels (b)–(d) is the same as for panel (a).
Spheroplast samples isolated using the lysozyme digestion method were washed with carbonate buffer to aid in the removal of soluble proteins and to convert closed membrane vesicles to open membrane sheets, releasing any trapped soluble or peripheral membrane proteins. Carbonate washing produced a sample which was free from cytoplasmic and cytoplasmic membrane protein contamination (Fig. 1b and c, lane 6). However, the majority of the proteins observed from the original lysozyme spheroplasts were lost during this treatment and a 50-fold concentration of the sample was consequently required in order to detect the protein content of this sample, even with silver stain (compare lanes 4 and 6 in Fig. 1a).
The three methods which produced the most encouraging initial results were the carbonate-washed spheroplasts (Fig. 1, lane 6), the Sarkosyl outer-membrane preparation (Fig. 1, lane 8) and the sucrose gradient (Fig. 1, lane 9). These methods were further analysed in more detail.
Sucrose gradient centrifugation does not produce optimal isolation of inner and outer membranes of C. jejuni
A sucrose gradient ranging from 30 to 60 % sucrose (in 5 % increments) was used in this study. Four distinct bands were visible upon inspection of the gradient following 18 h of ultracentrifugation (bands 1– 4, from bottom to top of gradient) (data not shown). The densities of these four bands were not measured, but they were localized within the following sucrose concentrations: band 1, at the 45 %/50 % junction; band 2, halfway between the 45 %/50 % and 40 %/45 % junctions; band 3, at the 40 %/45 % junction; band 4, at the 35 %/40 % junction. In comparison, only two distinct bands were observed in the control gradient (E. coli W3110) and these were localized at the 45 %/50 % sucrose junction and the 50 %/55 % junction (data not shown).
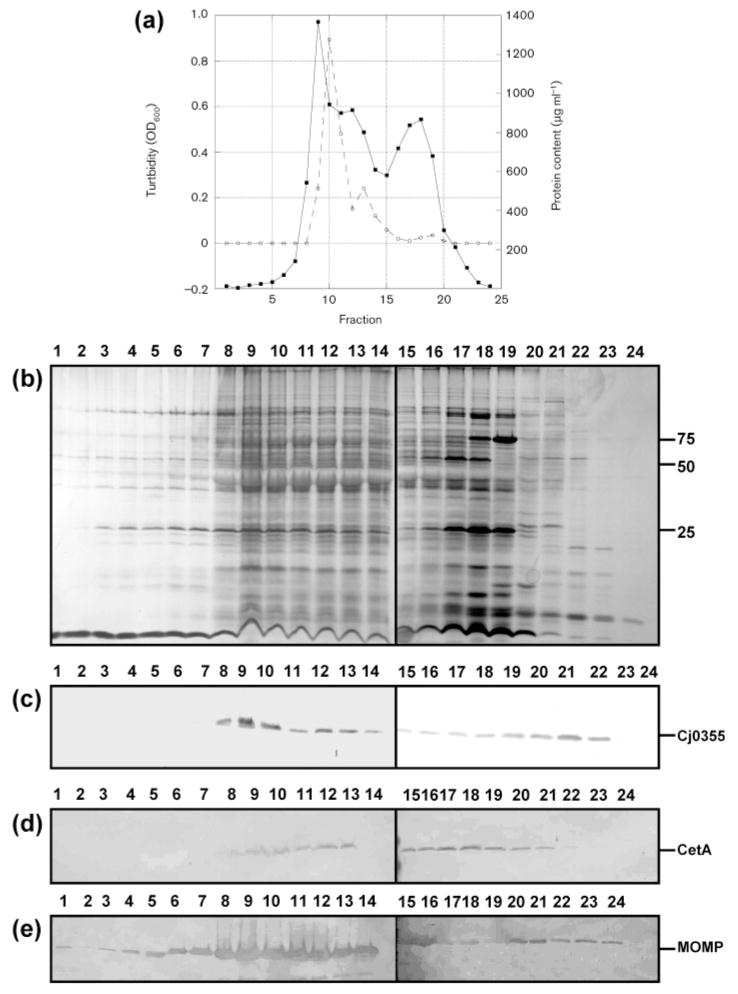
As shown in Fig. 2(a), two major peaks were observed. The first peak was higher in turbidity and protein content than the second and contained a small shoulder peak ranging from fractions 12 to 14. However, the profile of the peak based on protein content was shifted to the right by one to two fractions. Based on work done in E. coli, two distinct peaks are observed upon analysis of protein content of fractions from sucrose gradients and these peaks are usually separated by a distance of over five fractions (Page & Taylor, 1988). However, Osborn & Munson (1974) noted that four peaks were obtained upon sucrose gradient centrifugation of Salmonella; these were designated L1, L2, H and M. Four peaks have also been noted for Pseudomonas aeruginosa isopycnic centrifugation (Hancock & Nikaido, 1978). The L1 and L2 peaks of Salmonella were identified as cytoplasmic membrane, and this fragmentation into two bands was hypothesized by the authors to reflect heterogeneity in the degree of contamination by the outer-membrane material. The H peak was identified as the outer-membrane fraction and, as in our case, was the largest of the peaks and was the first peak to come off the gradient. In addition, the M peak was described as unseparated envelope fragments and was observed at the shoulder of the H peak. Since we also observed a similar peak, it is highly possible that the shoulder peak observed at fraction 10–11 (Fig. 2a), contains unseparated envelope components also.
Fig. 2.
Analysis of fractions obtained from the sucrose gradient of C. jejuni strain 81–176. (a) Graphical representation of the turbidity measured as OD600 (solid line) and protein content measured by BCA assay (dashed line) for fractions 1–24. (b) Silver-stained SDS-PAGE gel of sucrose gradient fractions. Lanes 1–24, fractions 1–24 (10 μl). Molecular masses are indicated on the right (in kDa). (c–e) Immunoblots of sucrose gradient fractions 1–24 using the cytoplasmic marker anti-Cj0355 (c), the inner-membrane marker anti-CetA (d) and the outer-membrane marker anti-MOMP (e). All antibodies were used at 1: 1000 dilution. Loading for panels (c)–(e) is the same as for panel (b).
An attempt to optimize this method was made; this included changing the range of sucrose concentrations used (35–55 % with seven steps, 40–50 % with 11 steps, 30–60 % with seven steps) and altering the volume of each layer of the gradient. Interestingly, these alterations actually resulted in significantly worse separation (data not shown).
A number of factors are known to affect separation of cytoplasmic and outer membranes. These include addition of Mg2+ or other divalent cations and the nature of the outer-membrane LPS, although isopycnic centrifugation is less sensitive to LPS structure than electrophoretic methods. Interestingly, good separation has been reported for LPS mutants lacking the complete core, N-acetylglucosamine and galactose, although heptose-less mutants resulted in incomplete peaks of cytoplasmic and outer membranes (Osborn & Munson, 1974). Therefore, it would not be likely that lipooligosaccharide structure variation is the only reason for the less than ideal separation, in terms of distinct peaks, of the cytoplasmic and outer membranes of C. jejuni using isopycnic centrifugation. Another feature of the campylobacters is that they lack Braun’s lipoproteins (Logan & Trust, 1982) a structural feature of the E. coli and Salmonella typhimurium cell wall involved in anchoring the outer membrane to the peptidogylcan layer. This, in conjunction with the possibility that, as in E. coli (Bayer, 1968), the cytoplasmic and outer membranes of C. jejuni are fused at numerous places and result in greater stabilization of the cell envelope, may affect the ability to easily separate them. Furthermore, higher resolution of the peaks was attempted using a variety of factors including changing the range of sucrose concentrations used and altering the volume of each layer of the gradient, but none of these improved the separation of the cytoplasmic and outer membranes. Further resolution was also attempted by Page & Taylor (1988) with no success.
Sucrose gradient fractions were analysed by SDS-PAGE and subsequently by our subcellular antibody markers. As shown in Fig. 2(b, c), no Cj0355 or CetA was detected in fractions 1–7, indicating that there was no cytoplasmic or cytoplasmic membrane contamination within these fractions. The remaining fractions did, however, contain Cj0355 and CetA. MOMP was detected in all 24 fractions (Fig. 2d). One possibility is that because MOMP is the major OMP and constitutes a large proportion of the OMP profile, cross-contamination with this particular protein is highly probable. However, it is very likely that this method may be inefficient at separation of the cytoplasmic and outer membranes in this species and therefore is not suitable for preparation of pure outer membranes.
Spheroplasting using lysozyme results in isolation of a fraction with no cytoplasmic or inner membrane contamination
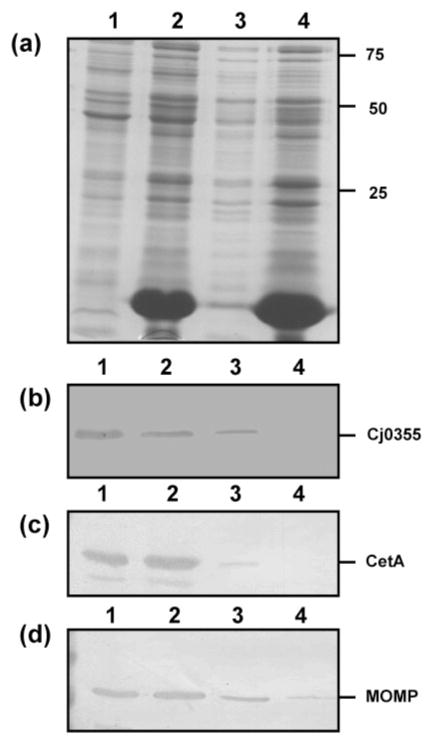
It has been shown that during spheroplast formation, outer membranes of E. coli and Pseudomonas are broken by the lysozyme and EDTA and form complex-coiled structures, while the cytoplasmic membrane remains intact (i.e. the spheroplast) and is pelleted during centrifugation (Birdsell & Cota-Robles, 1967; Costerton et al., 1967). Such procedures were originally developed for use with E. coli; however, other Gram-negative organisms such as Salmonella were found to be more resistant to these methods. The method described here was based on the work of Osborn & Munson (1974) and Hill & Sillence (1997) and produces rapid and efficient spheroplasting of bacteria while the outer membrane tends to peel away during spheroplast formation (Osborn & Munson, 1974). Treating C. jejuni cells with lysozyme produced a protein fraction which contained no cytoplasmic or cytoplasmic membrane contamination, as determined by immunoblotting with anti-Cj0355 (cytoplasmic marker) and anti-CetA (cytoplasmic membrane marker) (Fig. 3b and c, lane 4). MOMP was detected in this sample (Fig. 3d, lane 4) but the amount of MOMP detected in the cytoplasmic membrane fraction was reduced (Fig. 3d, lane 3), indicating that some outer membrane was still attached to the cytoplasmic membrane and would require further purification to remove it as completely as possible. Similar results were obtained with five other C. jejuni and one C. coli strain tested (data not shown). Even though a large number of putative OMPs were obtained using this protocol, the fact that the OMP marker (MOMP) could still be detected in the cytoplasmic membrane fraction may indicate that other, possibly novel OMP targets would fail to be detected. To confirm whether this is the case, further analysis of this fraction is required, possibly utilizing 2D gel electrophoresis and identification of spots via mass spectrometry.
Fig. 3.
Analysis of the preparative steps for spheroplasts of C. jejuni strain 81–176 using the lysozyme method. (a) Silver-stained SDS-PAGE gel of the preparative steps. Lanes: 1, whole-cell lysate; 2, post-lysozyme treatment supernatant; 3, inner membrane isolated from spheroplasts; 4, supernatant following centrifugation to remove unlysed cells (contains mostly outer membrane). Ten microlitres of each of the preparative steps was loaded per lane. Molecular masses are indicated on the right (in kDa). (b–d) Immunoblots of the lysozyme spheroplast preparatory samples using the cytoplasmic marker anti-Cj0355 (b), the inner-membrane marker anti-CetA (c) and the outer-membrane marker anti-MOMP (d). All antibodies were used at 1: 1000 dilution. Loading for panels (b)–(d) is the same as for panel (a).
Interestingly, it has been reported that washing spheroplasts with carbonate buffer (100 mM sodium carbonate, pH 11.5) aided in the removal of soluble proteins and converted closed membrane vesicles to open membrane sheets, releasing trapped proteins and peripheral membrane proteins (Hill & Sillence, 1997). The authors noted that carbonate washing was twice as effective as washing with Tris buffer and aided in the reduction of contaminating proteins. We were not able to confirm whether carbonate washing improved the removal of contaminating proteins. We used the sample obtained from spheroplasting using lysozyme (as described above) but since it was free from cytoplasmic and cytoplasmic membrane contamination this comparison was not possible. However, by analysing the actual proteins identified in each of these samples (spheroplast fraction and carbonate-washed spheroplast fraction) we may be able to determine whether carbonate washing further purifies the sample without OMP loss.
Because of the lower yield of OMPs due to apparently incomplete separation of membranes, the spheroplast method using lysozyme and sonication was not ideal for the purification of the outer membranes of campylobacters.
Isolation of OMPs of C. jejuni using N-lauroylsarcosine produces pure outer membranes
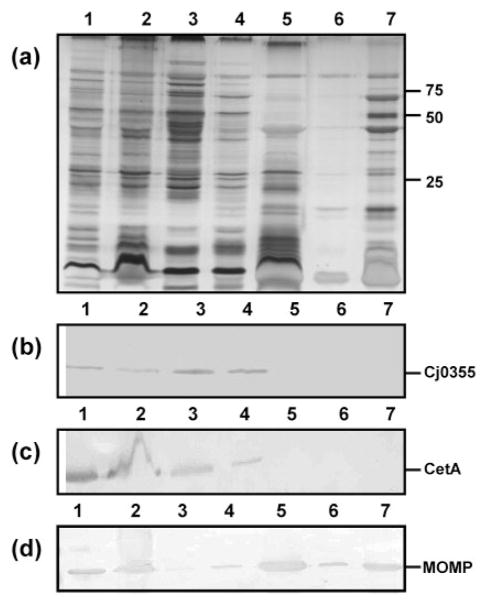
Sarkosyl is commonly used in the purification of OMPs of Gram-negative bacteria. We used a protocol in which lysed C. jejuni cells were washed in 10 mM HEPES buffer, pH 7.4, and incubated once with 1 % Sarkosyl. The Sarkosyl-insoluble fraction contained at least 25 proteins, as determined by silver staining (Fig. 4a, lane 7). Previous attempts at purifying OMPs using this method in our laboratory involved incubating the cells three times with 1 % Sarkosyl with 10 mM HEPES buffer pH 7.4 washes in between. Although this method produced samples free of cytoplasm and cytoplasmic membrane, the number of proteins identified was significantly reduced (data not shown). We suggest that excessive washing of the cells resulted in the loss of many proteins, some of which may contain novel vaccine candidates. Interestingly, other authors have reported a small number of proteins (fewer than 10) recovered following Sarkosyl treatment (Logan & Trust, 1983; Nachamkin & Hart, 1985). The small number of Sarkosyl-insoluble proteins obtained by other investigators may be explained by excessive washing of the pellets after Sarkosyl treatment resulting in loss of protein, or use of cells harvested from plate culture rather than broth culture, as in our case. This may also affect the yield of Sarkosyl-insoluble proteins due to potential lysis of cells as they are scraped from the plate, or cell morphology differences due to cells being in different growth phases.
Fig. 4.
Analysis of the preparative steps for the OMPs of C. jejuni strain 81–176 using Sarkosyl. (a) Silver-stained SDS-PAGE gel of the preparative steps. Lanes: 1, whole-cell lysate (10 μl); 2, lysate following French pressure lysis of whole cells (10 μl); 3, ultracentrifuge supernatant from first wash with 10 mM HEPES, pH 7.4 (10 μl); 4, ultracentrifuge supernatant from second wash with 10 mM HEPES, pH 7.4 (10 μl); 5, supernatant following Sarkosyl incubation (10 μl); 6, supernatant following final wash with 10 mM HEPES, pH 7.4 (10 μl); 7, Sarkosyl-insoluble proteins (OMPs) (5 μg). Molecular masses are indicated on the right (in kDa). (b–d) Immunoblots using the cytoplasmic marker anti-Cj0355 (b), the inner-membrane marker anti-CetA (c) and the outer-membrane marker anti-MOMP (d). All antibodies were used at 1: 1000 dilution.
Immunoblot analysis of the preparative samples from the Sarkosyl OMP preparations using our three subcellular antibody markers revealed that the final outer-membrane sample was free from cytoplasmic and cytoplasmic membrane contamination (Fig. 4b and c, lane 7). In fact, all of the early preparative samples from the whole-cell lysate to the supernatant following Sarkosyl treatment contained Cj0355, CetA and MOMP, as expected (Fig. 4b–d, lanes 1–4). The presence of abundant MOMP and the lack of cytoplasmic and cytoplasmic membrane contamination in the final fractions (Fig. 4, lanes 5–7) indicates highly purified OMPs. Sarkosyl preparations of outer membranes of five additional C. jejuni strains and one C. coli strain were also shown to be free of cytoplasmic and cytoplasmic membrane contamination (see Supplementary Fig. S1, available with the online version of this paper). Thus, Sarkosyl extraction is the ideal method for Campylobacter outer membrane preparation.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health grants AI055715, AI058284 and AI061026 (to S. A. T.). We would like to thank M. Stephen Trent (University of Texas) for helpful discussions and for the kind gift of E. coli W3110. The anti-CetA antibodies were a kind gift from Vic DiRita (University of Michigan), and the anti-MOMP antibodies were kindly supplied by Qijing Zhang (Iowa State University).
Abbreviation
- OMP
outer-membrane protein
Footnotes
A supplementary figure showing the analysis of Sarkosyl preparations of outer membranes from additional C. jejuni and C. coli strains is available with the online version of this paper.
References
- Alahari A, Saint N, Campagna S, Molle V, Molle G, Kremer L. The N-terminal domain of OmpATb is required for membrane translocation and pore-forming activity in mycobacteria. J Bacteriol. 2007;189:6351–6358. doi: 10.1128/JB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MJ, Haridas S, Steer D, Dhaunsi GS, Smith AI, Adler B. Identification of a Campylobacter jejuni protein that cross-reacts with cholera toxin. Infect Immun. 2007;75:3070–3073. doi: 10.1128/IAI.00139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allos BM. Campylobacter infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- Baqar S, Applebee LA, Bourgeois AL. Immunogenicity and protective efficacy of a prototype Campylobacter killed whole-cell vaccine in mice. Infect Immun. 1995;63:3731–3735. doi: 10.1128/iai.63.9.3731-3735.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer ME. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- Birdsell DC, Cota-Robles EH. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J Bacteriol. 1967;93:427–437. doi: 10.1128/jb.93.1.427-437.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledsoe HA, Carroll JA, Whelchel TR, Farmer MA, Dorward DW, Gherardini FC. Isolation and partial characterization of Borrelia burgdorferi inner and outer membranes using isopycnic centrifugation. J Bacteriol. 1994;176:7447–7455. doi: 10.1128/jb.176.24.7447-7455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolla JM, De E, Dorez A, Pages JM. Purification, characterization and sequence analysis of Omp50, a new porin isolated from Campylobacter jejuni. Biochem J. 2000;352:637–643. [PMC free article] [PubMed] [Google Scholar]
- Butzler JP. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect. 2004;10:868–876. doi: 10.1111/j.1469-0691.2004.00983.x. [DOI] [PubMed] [Google Scholar]
- Buzby JC, Allos BM, Roberts T. The economic burden of Campylobacter-associated Guillain–Barre syndrome. J Infect Dis. 1997;176:S192–S197. doi: 10.1086/513785. [DOI] [PubMed] [Google Scholar]
- Clark CG, Beeston A, Bryden L, Wang G, Barton C, Cuff W, Gilmour DL, Ng LK. Phylogenetic relationship of Campylobacter jejuni based on porA sequence. Can J Microbiol. 2007;53:27–38. doi: 10.1139/w06-099. [DOI] [PubMed] [Google Scholar]
- Cordwell SJ, Len AC, Touma RG, Scott NE, Falconer L, Jones D, Connolly A, Grossett D, Djordjevic SP. Identification of membrane-associated proteins from C. jejuni using complementary proteomics technologies. Proteomics. 2008;8:122–139. doi: 10.1002/pmic.200700561. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Forsberg C, Matula TI, Buckmire FLA, Macleod RA. Nutrition and metabolism of marine bacteria. XVI. Formation of protoplasts, spheroplasts, and related forms from a gram-negative marine bacterium. J Bacteriol. 1967;94:1764–1777. doi: 10.1128/jb.94.5.1764-1777.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De E, Jullien M, Labesse G, Pages JM, Molle G, Bolla JM. MOMP (major outer membrane protein) of Campylobacter jejuni; a versatile pore-forming protein. FEBS Lett. 2000;469:93–97. doi: 10.1016/s0014-5793(00)01244-8. [DOI] [PubMed] [Google Scholar]
- Elliott KT, DiRita VJ. Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol Microbiol. 2008;69:1091–1103. doi: 10.1111/j.1365-2958.2008.06357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhr MK, Logue CM. Sequence variation in the outer membrane protein-encoding gene cmeC, conferring multidrug resistance among Campylobacter jejuni and Campylobacter coli strains isolated from different hosts. J Clin Microbiol. 2007;45:3381–3383. doi: 10.1128/JCM.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FSL. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Girard MP, Steele D, Chaignat CL, Kieny MK. A review of vaccine research and development: human enteric infections. Vaccine. 2006;24:2732–2750. doi: 10.1016/j.vaccine.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Hancock REW, Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas auruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978;136:381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrink P, van den Munchkof HA, Bumkens M, Lindeman J, van Dijk WC. Human serum antibody repsonse in Campylobacter jejuni enteritis as measured by enzyme-linked immunosorbent assay. Eur J Clin Microbiol Infect Dis. 1988;7:388–393. doi: 10.1007/BF01962343. [DOI] [PubMed] [Google Scholar]
- Hill RA, Sillence MN. Improved membrane isolation in the purification of beta2-adrenoceptors from transgenic Escherichia coli. Protein Expr Purif. 1997;10:162–167. doi: 10.1006/prep.1997.0732. [DOI] [PubMed] [Google Scholar]
- Horikiri S, Aizawa Y, Kai K, Amachi S. Electron acquisition system constructed from an NAD-independent D-lactate dehydrogenase and cytochrome c2 in Rhodopseudomonas palustris no. 7. Biosci Biotechnol Biochem. 2003;68:516–522. doi: 10.1271/bbb.68.516. [DOI] [PubMed] [Google Scholar]
- Jin S, Joe A, Lynett J, Hani EK, Sherman P, Chan VL. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol Microbiol. 2001;39:1225–1236. doi: 10.1111/j.1365-2958.2001.02294.x. [DOI] [PubMed] [Google Scholar]
- Johnston KH, Gotschlich EC. Isolation and characterization of the outer membrane of Neisseria gonorrhoeae. J Bacteriol. 1974;119:250–257. doi: 10.1128/jb.119.1.250-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella N, Guerry P, Cooney J, Trust T. The flgE gene of Campylobacter coli is under the control of the alternative sigma factor sigma factor 54. J Bacteriol. 1997;179:4647–4653. doi: 10.1128/jb.179.15.4647-4653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Garvis SG, Tipton SL, Anderson J, Cieplak J. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) fom Campylobacter jejuni. Mol Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- Kubak BM, Yotis WW. Analysis of Staphylococcus aureus cytoplasmic membrane proteins by isoelectric focusing. Biochim Biophys Acta. 1981;649:642–650. doi: 10.1016/0005-2736(81)90169-3. [DOI] [PubMed] [Google Scholar]
- Labesse G, Garnotel E, Bonnel S, Dumas C, Pages JM, Bolla JM. MOMP, a divergent porin from Campylobacter jejuni; cloning and primary structural characterization. Biochem Biophys Res Commun. 2001;280:380–387. doi: 10.1006/bbrc.2000.4129. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee LH, Burg E, III, Baqar S, Bourgeois AL, Burr DH, Ewing CP, Trust T, Guerry P. Evaluation of a truncated recombinant flagellin subunit vaccine against Campylobacter jejuni. Infect Immun. 1999;67:5799–5805. doi: 10.1128/iai.67.11.5799-5805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder L, Andersson C, Sund C, Shockmen GD. Protoplast formation and localization of enzymes in Streptococcus mitis. Infect Immun. 1983;40:1146–1154. doi: 10.1128/iai.40.3.1146-1154.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SM, Trust T. Outer membrane characteristics of Campylobacter jejuni. Infect Immun. 1982;38:898–906. doi: 10.1128/iai.38.3.898-906.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SM, Trust T. Molecular identification of surface proteins against Campylobacter jejuni. Infect Immun. 1983;42:675–682. doi: 10.1128/iai.42.2.675-682.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luneberg E, Glenn-Calvo E, Hartmann M, Bar W, Frosch M. The central, surface-exposed region of the flagellar hook protein FlgE of Campylobacter jejuni shows hypervariability among strains. J Bacteriol. 1998;180:3711–3714. doi: 10.1128/jb.180.14.3711-3714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinersmann RJ, Hiett KL, Tarplat A. Cloning of an outer membrane protein gene from Campylobacter jejuni. Curr Microbiol. 1997;34:360–366. doi: 10.1007/s002849900196. [DOI] [PubMed] [Google Scholar]
- Miller WG. Comparative genomics of non-C. jejuni Campylobacter spp. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3. Washington, DC: ASM Press; 2008. pp. 73–95. [Google Scholar]
- Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA. Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x. [DOI] [PubMed] [Google Scholar]
- Monteville MR, Konkel ME. Fibronectin-faciliated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect Immun. 2002;70:6665–6671. doi: 10.1128/IAI.70.12.6665-6671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser I, Schroeder W, Slanikow J. Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membanes. FEMS Microbiol Lett. 1997;157:233–238. doi: 10.1111/j.1574-6968.1997.tb12778.x. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Imai M, Nakamura H, Yoshimura F. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur J Oral Sci. 2002;110:157–162. doi: 10.1034/j.1600-0722.2002.11171.x. [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Hart AM. Western blot analysis of the human antibody response to Campylobacter jejuni cellular antigens during gastrointestinal infection. J Clin Microbiol. 1985;21:33–38. doi: 10.1128/jcm.21.1.33-38.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Munson R. Separation of the inner membrane (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 1974;31:642–653. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- Page WJ, Taylor DE. Comparison of methods used to separate the inner and outer membranes of cell envelopes of Campylobacter spp. J Gen Microbiol. 1988;134:2925–2932. doi: 10.1099/00221287-134-11-2925. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, et al. The genome sequence of the foodborne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Pei Z, Blaser MJ. PEB1, the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in gram-negative nutrient transport systems. J Biol Chem. 1993;268:18717–18725. [PubMed] [Google Scholar]
- Pei Z, Burucoa C, Grignon B, Baqar S, Huang XZ, Kopecko DJ, Bourgeois AL, Fauchere JL, Blaser MJ. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect Immun. 1998;66:938–943. doi: 10.1128/iai.66.3.938-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MC, Vugia D, Shallow S, Marcus R, Segler S, McGivern T, Kassenborg H, Reilly K, Kennedy M, et al. Epidemiology of sporadic campylobacter infection in the United States and declining trend in incidence, FoodNet, 1996–1999. Clin Infect Dis. 2004;38:S165–S174. doi: 10.1086/381583. [DOI] [PubMed] [Google Scholar]
- Schaumburg J, Diekmann O, Hagendorff P, Bergmann S, Rohde M, Hammerschidt S, Jansch L, Wehland J. The cell wall subproteome of Listeria monocytogenes. Proteomics. 2004;4:2991–3006. doi: 10.1002/pmic.200400928. [DOI] [PubMed] [Google Scholar]
- Sizemore DR, Warner B, Lawrence J, Jones A, Killeen KP. Live, attenuated Salmonella typhimurium vectoring Campylobacter antigens. Vaccine. 2006;24:3793–3803. doi: 10.1016/j.vaccine.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Steele M, Marcone M, Gyles C, Chan VL, Odumeru J. Enzymatic activity of Campylobacter jejuni hippurate hydrolase. Protein Eng Des Sel. 2006;19:17–25. doi: 10.1093/protein/gzi071. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffat BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Yamada T, Carlsson J. Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J Bacteriol. 1975;124:55–61. doi: 10.1128/jb.124.1.55-61.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Meitzler JC, Huang S, Morishita T. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter jejuni major outer membrane protein. Infect Immun. 2000;68:5679–5689. doi: 10.1128/iai.68.10.5679-5689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.