Abstract
Resolvin D1 (RvD1) and Protectin D1 (Neuroprotectin D1, PD1/NPD1) are newly identified anti-inflammatory lipid mediators biosynthesized from docosahexaenoic acid (DHA). In this report, the spectra-structure correlations and fragmentation mechanisms were studied using electrospray low-energy collision-induced dissociation tandem mass spectrometry (MS/MS) for biogenic RvD1 and PD1, as well as mono-hydroxy-DHA and related hydroperoxy-DHA. The loss of H2O and CO2 in the spectra indicates the number of functional group(s). Chain-cut ions are the signature of the positions and numbers of functional groups and double-bonds. The observed chain-cut ion is equivalent to a hypothetical homolytic-segment (cc, cm, mc, or mm) with addition or extraction of up to 2 protons (H). The α-cleavage ions are equivalent to: [cc + H], with H from the hydroxyl through a β-ene or γ-ene rearrangement; [cm - 2H], with 2H from hydroxyls of PD1 through a γ-ene rearrangement, or one H from the hydroxyl and the other H from the α-carbon of mono-HDHA through an α-H-β-ene rearrangement; [mc – H], with H from hydroxyl through a β-ene or γ-ene rearrangement, or from the α-carbon through an α-H-β-ene rearrangement; or [mm] through charge-direct fragmentations. The β-ene or γ-ene facilitates the H shift to γ position and α-cleavage. Deuterium labeling confirmed the assignment of MS/MS ions and the fragmentation mechanisms. Based on the MS/MS spectra and fragmentation mechanisms, we identified RvD1, PD1, and mono-hydroxy-DHA products in human neutrophils and blood, trout head-kidney, and stroke-injury murine brain-tissues.
Introduction
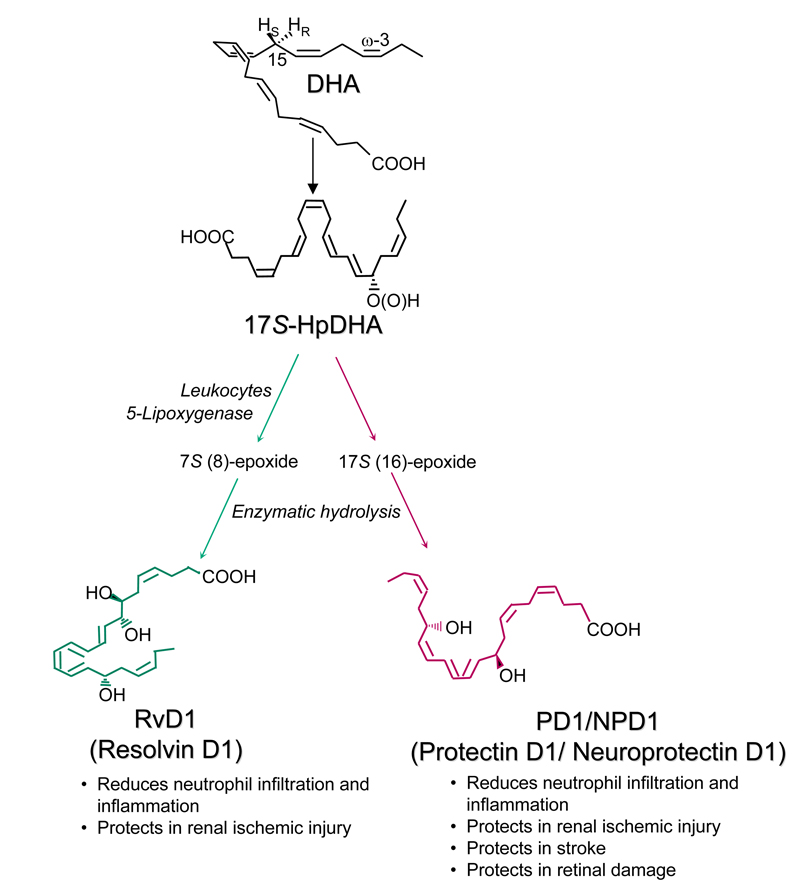
Numerous reports demonstrate the beneficial effects of fish oil on human diseases such as arthritis, Alzheimer’s disease, lung fibrosis, and inflammatory bowel diseases [1,2,3]. As an essential omega-3 polyunsaturated fatty acid, docosahexaenoic acid (DHA) is a major component of fish oil [4]. Novel bioactive oxygenated DHA products biosynthesized in resolving inflammatory exudates and tissues were recently identified and their structures and bioactions were elucidated [5–8]. They were named as the D series of Resolvins (resolution phase interaction products) and Protectins (protecting brain and other organs against inflammatory diseases) because their biosynthetic pathways display potent anti-inflammatory and immunoregulatory properties (Scheme 1) [5–9]. As illustrated in Scheme 1, DHA is initially converted to 17S-hydroperoxy-DHA (HpDHA), then further enzymatically transferred to Resolvin D1(RvD1) and Protectin D1 (Neuroprotectin D1, PD1/NPD1) via epoxide intermediates. With aspirin treatment, the aspirin-acetylated cyclooxygenase type II (COX-2) converts DHA into 17R-HpDHA, which is further converted to 17R-RvD1 and 17R-PD1 [5].
Scheme 1.
Biosynthetic pathways for Resolvin D1 (RvD1) and Protectin D1 (PD1).
To obtain the temporal and spatial profiles and regulation of biosynthesis of RvD1 and PD1 as well as other DHA products during physiopathological processes, their identification needs to be accurate, sensitive, and fast. One suitable approach is liquid chromatography coupled to an ultra-violet spectrometer and a low collision-energy tandem mass spectrometer (LC-UV-MS/MS) with atmospheric pressure ionization, specifically electrospray ionization (ESI) [5–8]; this is used on full product ion scan bases for structure elucidation and identification of Resolvins and Protectins at low nanogram levels [5–8]. Authentic standards of Resolvins and Protectin D1 prepared via total organic synthesis are used for confirmation during the identification. The low levels (under a few nanograms/sample) of these mediators present in vivo make other instrumentation, such as nuclear magnetic resonance or optical spectrometers, inappropriate for the analysis. In comparison, different types of mass spectrometry were used in the earlier studies of DHA-derived products. These included gas chromatography-MS with electron-impact ionization was used by Murphy et al. to analyze the structures of mono-hydroxy-DHA products after silanization and methylation [9]. Kim et al. conducted preparation and structure analysis of HDHAs and HpDHAs using LC-positive ion thermospray MS/MS [10]. They also developed an analytical method and conducted stereochemistry studies of all the HDHAs produced by human platelets and rat brain homogenates using chiral LC-thermospray-MS and GC/MS (electron-impact ionization) [11].
Low-energy ionization primarily generates molecular (or pseudo-molecular) ions for collision-induced dissociation (CID) MS/MS analysis, through which the MS/MS spectra obtained are used widely to identify and elucidate the structures of lipid mediators derived from polyunsaturated fatty acids [12–20]. The low-energy CID of eicosanoids includes charge-remote and charge-directed fragmentations [12,21], many of which occur through “α-hydroxy-β-ene like rearrangement” as referred to by Murphy, i.e., α-cleavage of the carbon-carbon bond (α position to hydroxy group), facilitated by a double-bond (ene) in the β position [22]. For polyunsaturated fatty acids and their derivatives, ions formed in low energy CID MS/MS via the loss of only H2O and CO2 are usually much more abundant than ions formed through cleavage of the carbon-chain. In this report, the former are called “peripheral-cut ions” and the latter are called “chain-cut ions”. Peripheral-cut ions provide information about the number of hydroxys and carboxylates in a compound; chain-cut ions are the signatures for the positions of hydroxys and double-bonds. The ions formed via a combination of chain-cut and peripheral-cut processes are called “chain-plus-peripheral-cut” ions [23]. The structure of a compound is deduced from the integration of the structures assigned to its MS/MS ions. Thus, the fragmentation mechanisms for the formation of MS/MS ions are very important for the identification using the MS/MS ions of a novel and/or known molecular structure. These mechanisms form the basis for lipidomic databases, which consist of lipid mass spectra and other analytical data, and search algorithms [23].
Here, we report the analysis of Resolvin D1 and Protectin D1, as well as other DHA-derived products without derivatization, using low-collision-energy tandem mass spectra acquired on anions generated from electrospray ionization of molecules eluted from LC. For structure elucidation and identification, the ion structures and MS/MS fragmentation mechanisms are proposed and confirmed via deuterium-labeled isotopomers of these compounds. RvD1 and PD1, as well as mono-hydroxy-DHA products, were found to be generated by human neutrophils, whole blood [6], trout head-kidney [25], and stroke-injury murine brain tissues [7], using this LC-UV-MS/MS approach.
Experimental
Instrumentation
The MS/MS spectrum was acquired on each chromatographic peak eluted from a column of a LC-UV-MS/MS system. This system includes a HPLC (P4000) coupled to a photo-diode-array UV detector and an ion trap (LCQ) MS/MS (Thermoelectron, San Jose, CA). Approximately 10 nanograms of each authentic standard were injected individually into the column (LUNA C18-2 150 mm × 2 mm × 5 m, Phenomenex, Torrance, CA), which was eluted at 0.2 ml/min and 23 C. The mobile phase flowed as C (water : methanol : acetic acid = 65% : 35% : 0.01%) from 0 to 50 min; ramped to 100% methanol from 50.1 to 110 mins; maintained as 100% methanol for 10 mins, then back to C. The UV detector scanned from 200 to 400 nm, providing additional spectral confirmation to ensure the purity of the authentic standards. For RvD1, PD1, mono-hydroxy-DHAs (HDHAs), and HpDHA, the maximum UV absorption wavelengths are 301 nm, 270 nm, and 235 nm, respectively. Conditions for the mass spectrometer are: electrospray voltage, 4.3 kV; heating capillary, −39 V; tube lens offset, 60 V; sheath N2 gas, 1.2 L/min; and auxiliary N2 gas, 0.045 L/min; collision energy was 35% to 45% (a relative collision energy of 0 to 100% corresponds to a high frequency alternating voltage for resonance excitation from 0 to 5 V maximum); helium gas, 0.1 Pa as a collision gas; and scan range, m/z 95 to 390 [23].
Materials and Sample Preparation
Biogenic syntheses of RvD1, 17S-HDHA, and 17-HpDHA were conducted via incubation of DHA (Cayman Chemical, Ann Arbor, MI) [5,6,24] with isolated enzyme(s), i.e., soybean 15-lipoxygenase and/or potato 5-lipoxygenase (Sigma Co., St. Louis, MO) Following the same procedures but replacing DHA with d5-DHA (21,21,22,22,22-d5-DHA) (Cayman Chemical), we prepared d5-RvD1, d5-PD1, d5-17S-HDHA, and d5-17-HpDHA, and acquired their spectra and chromatograms on LC-UV-MS/MS. The structures of biogenically synthesized RvD1 and PD1 were further confirmed via matching the spectra of MS/MS and UV (coupled to a LC), GC/MS, and chromatographic retention times of RvD1 and PD1 which were prepared by total organic synthesis [24, 26]. HDHAs were purchased from Cayman Chemical. All other reagents were from available commercial sources at the highest grade.
Some of the deuterium-labeled compounds were obtained via deuterium-exchange of labile hydrogens in the hydroxy(s) and carboxylate, which include O,O,O,O-d4-RvD1, O,O,O,21,21,22,22,22-d8-PD1 (from deuterium exchange of labile hydrogens in the two hydroxys and one carboxylate of d5-PD1), O,O,O-d3-PD1, and O,O-d2-HDHAs. The deuterium-exchange was achieved when the samples were analyzed on LC-UV-MS/MS using the same gradient as described above except substituting MeOD, D2O, and d-acetic acid for methanol, water, and acetic acid, respectively.
Samples of human whole blood (venous) and PMN were obtained and prepared as described elsewhere [6]. Stroke-injury murine brain tissues were provided by Professor N. Bazan’s group at Louisiana State University Health Sciences Center. The surgery procedures and sample preparation have also been described [7]. Samples of trout brain and head-kidney were prepared as in [25]. All samples were added with two volumes of cold methanol [6]. Briefly, this procedure is tailored as follows. The samples were centrifuged (3,000 rpm, 4°C, 15 min) to remove cellular and protein materials. After the supernatants were decanted, the supernatants were diluted with 10 volumes of Milli-Q water. The pH was adjusted to 3.5 with 1 M HCl for C18 solid-phase extraction (SPE). After washing with 15 ml of H2O and then 8 ml of hexane, the SPE cartridges (C18, 3 ml, Milford, Waters, MA) were eluted with 8 ml of methyl formate, and the effluent was reconstituted into methanol for lipidomic analysis using LC-UV-MS/MS.
Nomenclature
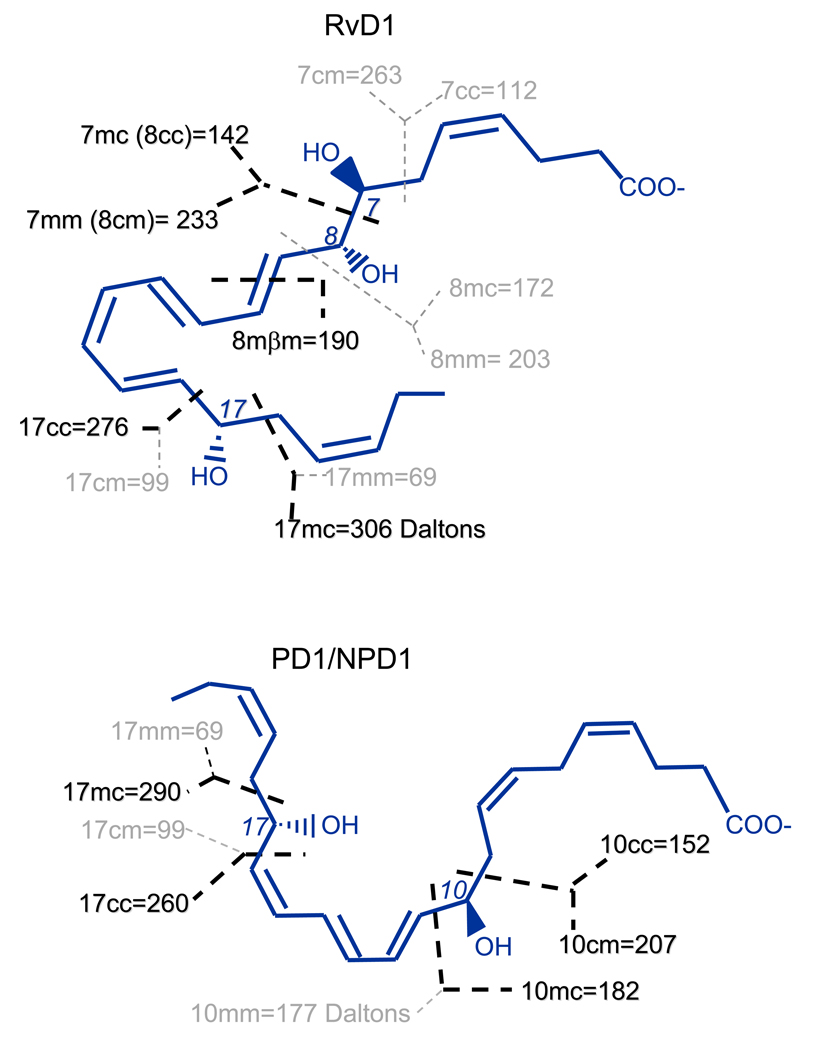
To illustrate the interpretation of MS/MS ions generated from RvD1, PD1/NPD1, and other related DHA-derived products, we use the nomenclature recently proposed [23] to name the specific “segments”, which are fragments formed through hypothetic homolytic-cleavages without hydrogen or group migration. This is depicted in Scheme 2 using RvD1 and PD1 structures. A number is used to designate the position of a hydroxy or another functional group on the carbon-chain where the cleavage occurs. The letter immediately following the number indicates the side of the functional group on which the cleavage occurs: “c” is for cleavage on the carboxyl side of the functional group and “m” is for cleavage on the methyl side of the functional group. Each cleavage can directly generate two segments. The second letter indicates the side of the cleavage on which the segment forms: “c” is for a segment formed on the carboxyl side of the cleavage and “m” is for a segment formed on the methyl side of the cleavage. Segments formed through β-cleavage or γ-cleavage along the functional group are named by adding β or γ between the two “c”/“m” letters [23].
Scheme 2.
Naming hypothetic-homolytic-segments of lipid mediators with RvD1 and PD1/NPD1 as an example.
Results
The electrospray ionization in negative mode generated primarily deprotonated molecular ions [M-H or D]− from Resolvin D1 (Figure 1), Protectin D1, other related DHA-derived compounds, and their deuterium isotopomers eluted from the LC column. In MS/MS experiments, [M-H]− ions were selected and fragmented via CID. Cleavage on the carbon chain was frequently observed on the α locations toward hydroxy and hydroperoxy. This is similar to the CID fragmentation of derivatives from arachidonic acid and eicosapentaenoic acid [5,6,13,27].
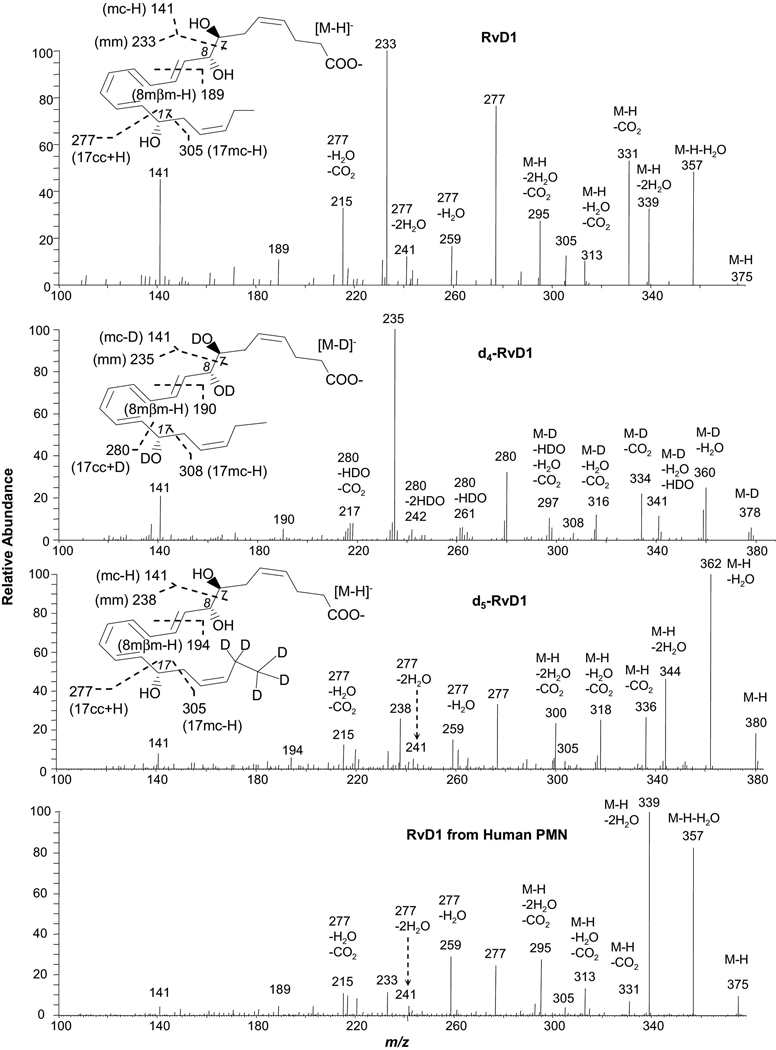
Figure 1.
Low energy MS/MS spectra acquired via CID of the electrospray-generated carboxylate anion [M-H or D]− from the LC peak of Resolvin D1 or deuterated RvD1. RvD1, O,O,O,O-d4-RvD1, and 21,21,22,22,22-d5-RvD1 were obtained from isolated-enzyme reactions with or without deuterium-exchange, as well as from human PMN, as detailed in the “Experimental” section. Each inset depicts the structure and CID fragmentation.
RvD1, O,O,O,O-d4-RvD1, and 21,21,22,22,22-d5-RvD1 (Figure 1 and Scheme 3)
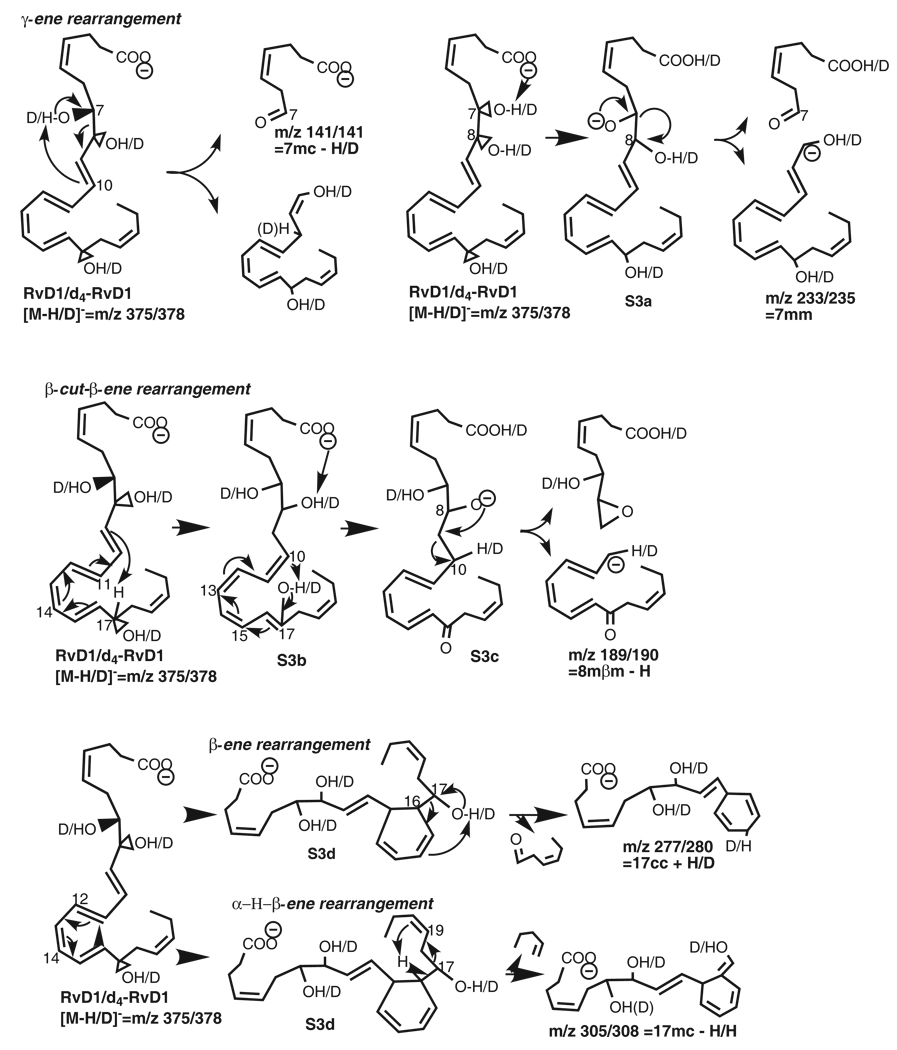
Scheme 3.
Fragmentation mechanisms for RvD1/d4-RvD1 ([M-H/D]−=m/z 375/378).
RvD1 is 7S,8R,17S-trihydroxy-docosa-4Z,9E,11E,13Z,15E,19Z-hexaenoic-acid, which has three hydroxy groups and six double-bonds, four of which are conjugated and bracketed by the 8-hydroxy and 17-hydroxy (see inset of Figure 1). The low collision-energy MS/MS spectrum of the LC peak of the RvD1 shows peripheral-cut ions at m/z 357 [M-H-H2O] (relative intensity is 47%), 339 [M-H-2H2O] (32%), 331 [M-H-CO2] (52%), 313 [M-H-H2O-CO2] (10%), and 295 [M-H-2H2O-CO2] (26%) (Figure 1). The assignment of these ions is consistent with O,O,O,O-d4-RvD1 (d4-RvD1), ions: 360 [M-D-H2O] (27%), 341 [M-D-H2O-HDO] (14%), 334 [M-D-CO2] (22%), 316 [M-D-H2O-CO2] (14%), and 297 [M-D-HDO-H2O-CO2] (12%); 21,21,22,22,22-d5-RvD1 (d5-RvD1) ions: 362 [M-H-H2O] (100%), 344 [M-H-2H2O] (46%), 336 [M-H-CO2] (27%), 318 [M-H-H2O-CO2] (25%), and 300 [M-H-2H2O-CO2] (24%); and d4-RvD1 also possesses ions 359 [M-D-HDO] (14%), 342 [M-D-2H2O] (4%), 315 [M-D-HDO-CO2] (8%), and 298 [M-D-2H2O-CO2] (8%). These peripheral-cut ions support the existence of the carboxyl group (for CO2 loss) and hydroxy groups (for H2O loss) in RvD1. However, these ions do not provide the specific information or clues for the positions of the hydroxy(s) and double-bonds. The loss of H2O from d4-RvD1, whose hydroxy hydrogens were completely deuterated, indicates that the deuteroxy deuterium is exchanged with the carbon-chain-bonded hydrogen. The chain-cut MS/MS ions of RvD1 are as follows; some of them further transfer to chain-plus-peripheral-cut ions via loss of water or CO2. Chain-cut and chain-plus-peripheral-cut ions provide the signatures for the positions of functional groups and double-bonds.
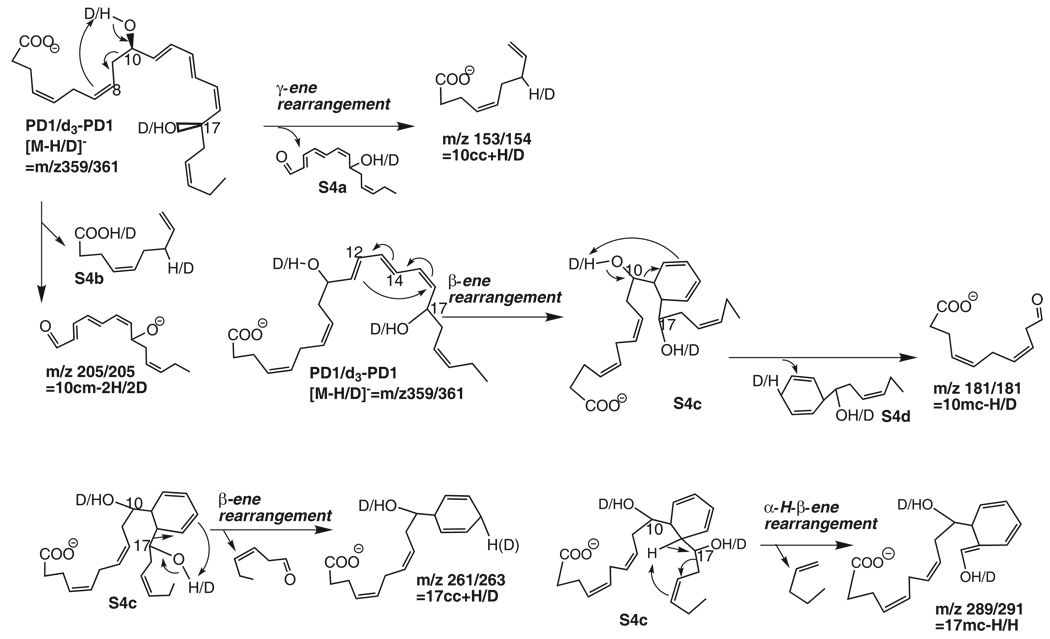
The ion m/z 141 (44%) of RvD1 is equal to segment 7mc minus the 7-hydroxy proton (H from 7-OH or 7-OH H), namely [7mc – H from 7-OH] corresponding to the following mechanism (Scheme 3). When the 7-OH H is extracted, through a transit six-membered ring, to C10 by the double-bond C9,10, which is at γ position to 7-OH, 7-OH converts to a carbonyl group, the allylic single-bond C7,8 at α position to 7-OH cleaves, and the C8,9 double-bond forms, yielding ion m/z 141 and an enol. We refer to such process having γ-ene facilitated α-OH H migration and α-cleavage as a “γ-ene rearrangement”. The mechanism was confirmed by ion m/z 141 as [7mc – deuteron from 7-deuteroxy] (i.e., [7mc – D from 7-OD]) from d4-RvD1 and as [7mc – H from 7-OH] from d5-RvD1.
In competition with the formation of ion m/z 141, 7-OH H shifts to the carboxyl anion internally, forming 7-alkoxide anion S3a. The negative charge of S3a directs the cleavage of C7,8 allylic single-bond bond at α position, resulting in ion m/z 233 (100%) {corresponding to ions m/z 235 (100%) for d4-RvD1 and 238 (26%) for d5-RvD1} (Figure 1), which is equivalent to segment [7mm]. Additionally, the deuterium labeling on C21 and C22 of d5-RvD1 changed the base-peak from ion m/z 233 of RvD1 or m/z 235 (equivalent to [7mm]) of d4-RvD1 to ion m/z 362 [M-H-H2O].
Ion m/z 189 is equal to [8mm – H] for RvD1, which is equivalent to ion m/z 190 [8mβm – H] for d4-RvD1. Their formation corresponds to the following mechanism. Initially, H from C17 of the parent ion is extracted to C9 by C9,10 double-bond at β position to 8-OH and the conjugated tetraene Δ9,11,13,15 converts to Δ10,12,14,16 in S3b. When 17-OH H (or D 17-OD D) moves to C10, this tetraenol S3b changes to tetraenone S3c; meanwhile, the 8-OH H (or 8-OD D for d4-RvD1) migrates to the carboxylate anion, resulting in the 8-alkoxide anion S3c. Then the negative charge of the S3c directs the breakage of the C9,10 allylic single-bond at the β position to the 8-alkoxide group, yielding ion m/z 189 (14%) from RvD1, m/z 190 (6%) from d4-RvD1, and m/z 194 (7%) from d5-RvD1. We refer to such process with β-cleavage facilitated by α-OH and β-ene as a “β-cut-β-ene rearrangement”.
Ion m/z 277 (75%) of RvD1 and ion m/z 280 (32%) of d4-RvD1 are equivalent to [17cc + H from 17-OH] and [17cc+D from 17-OD], respectively. They correspond to the α-cleavage of the C16,17 bond of their parent ions, of which the β double-bond C15,16 facilitates the cleavage. This is a typical α-hydroxy-β-ene-like rearrangement (abbreviated as β-ene rearrangement) [22]. When the conjugated triene Δ11,13,15 of the parent ion forms a six-membered ring in S3d, the β double-bond C15,16 shifts to γ position, forming a C14,15 double-bond, and the vinyl single-bond C16,17 becomes the allylic single-bond C16,17 in S3d. Then the 17-OH H (or 7-OD D in d4-RvD1) shifts to C14 of the γ double-bond C14,15, a 17-carbonyl group forms, and the C16,17 allylic single-bond cleaves, producing ion m/z 277 of RvD1 and d5-RvD1[or 280 of d4-RvD1]. Thus, this β-ene rearrangement includes conversion of β-ene to γ-ene and αvinyl single-bond to α allylic single-bond, and subsequently the γ-ene rearrangement, similar to the case of the ion m/z 141 from RvD1 discussed above.
When the C16-proton (H) in S3d shifts to C20, instead of the shift of H from 17-OH to C20 (which would have generated ion m/z 307 for d5-RvD1), the C17,18 allylic single-bond fragments, yielding ion m/z 305 (11%, equivalent to [17mc – H from C16]) from RvD1. This mechanism is confirmed by ion m/z 308 (5%) from d4-RvD1 (Scheme 3) and ion m/z 305 (6%) from d5-RvD1. We refer to this process as “α-H-β-ene rearrangement”.
The 17-OH in S3d is competitively deprotonated internally by its carboxyl anion to form a C17-alkoxide anion, of which the negative charge directs the breakage of C16,17 allylic single-bond, generating ion m/z 277; similar to that discussed for S3d above, a charge-remote process for this alkoxide anion, also involving the migration of C16-H to C20, cleaves allylic single-bond C17,18 and generates ion m/z 305 [12] (Scheme 3). This is also an α-H-β-ene rearrangement. The α-H is less active than H from α-OH because the C-H bond is much less polarized than the O-H bond; thus the abundance of ion m/z 277 is higher than that of ion m/z 305 for RvD1 and d5-RvD1. The same pattern is observed: ion m/z 280 is more abundant than ion m/z 308 for d4-RvD1, and ion m/z 277 is more abundant than ion m/z 305 for d5 RvD1.
The loss of water and/or CO2 generated ions 259 [277-H2O] for RvD1(15%) and d5-RvD1(16%) , 241 [277-2H2O] for RvD1 (11%) and d5-RvD1 (6%), 215 [277-H2O-CO2] for RvD1 (32%) and d5-RvD1 (15%) , and 217 [280 –HDO-CO2] (7%) for d4-RvD1, which further confirmed the structure assignment of ion m/z 277. Using this LC-UV-MS/MS analysis, RvD1 was found to be biosynthesized by human neutrophils. The MS/MS spectrum of a chromatographic peak acquired from the samples of human neutrophils matches the spectrum of standard RvD1 (bottom panel, Figure 1), as do the UV spectrum and chromatographic retention time (data not shown).
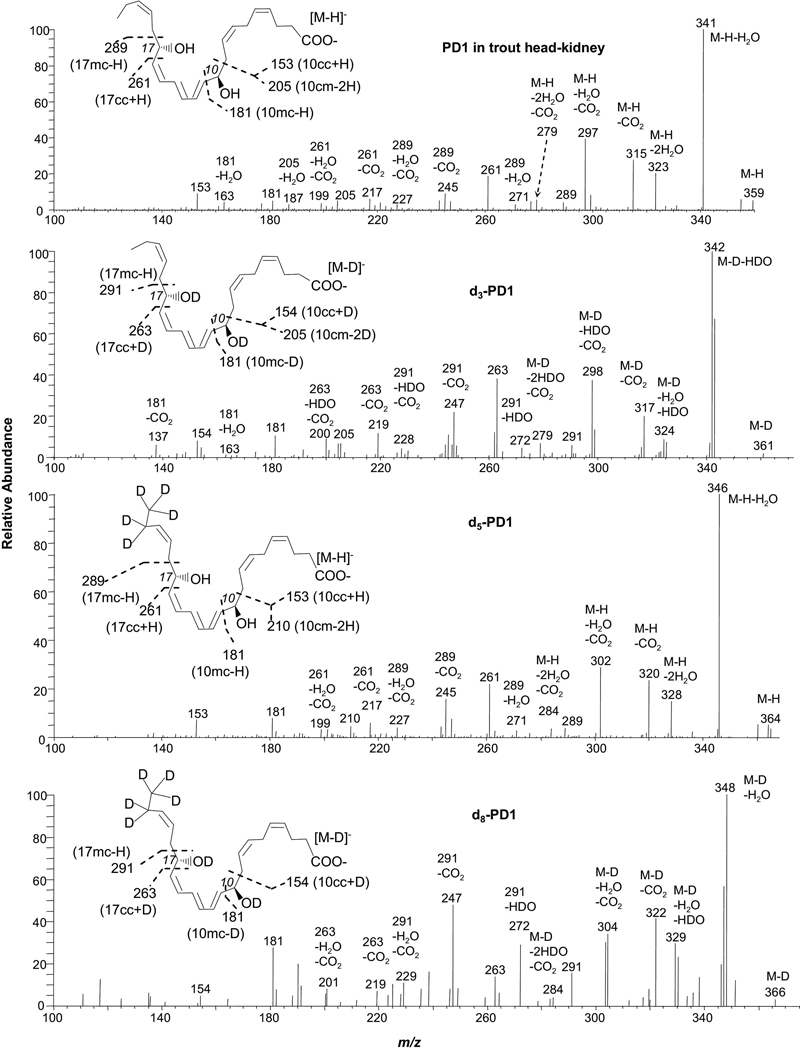
PD1, O,O,O-d3-PD1, 21,21,22,22,22-d5-PD1, and O,O,O,21,21,22,22,22-d8-PD1 (Figure 2 and Scheme 4)
Figure 2.
Low-energy MS/MS spectra acquired via CID of electrospray-generated carboxylate anion [M-H or D]− from the LC peak of PD1 or deuterated PD1. PD1 was obtained from trout head kidney; O,O,O-d3-PD1, 21,21,22,22,22-d5-PD1, and O,O,O,21,21,22,22,22-d8-PD1 were obtained from isolated-enzyme reactions with or without deuterium-exchange, as detailed in the “Experimental” section. Each inset depicts the structure and CID fragmentation.
Scheme 4.
Fragmentation mechanisms for PD1/d3-PD1([M-H/D]−=m/z 359/361).
Among the six double-bonds of PD1 (Protectin D1/Neuroprotectins D1: 10R,17S-dihydroxy-docosa-4Z,7Z,11E,13E,15Z,19Z-hexaenoic acid), three are conjugated between 10-OH and 17-OH (see Figure 2 inset). Negative electrospray ionization generated ion m/z 359, a deprotonated molecular ion [M-H]−, from PD1. The MS/MS spectrum at m/z 359 of PD1 from trout head-kidney matches to that acquired from synthetic PD1 (see Figure 1 of reference 25). There are peripheral-cut ions at m/z of 341 [M-H-H2O] (100%), 323 [M-H-2H2O] (20%), 315 [M-H-CO2] (29%), 297 [M-H-H2O-CO2] (39%), and 279 [M-H-2H2O-CO2] (7%), consistent with the PD1 structure of one carboxylic group and two hydroxy groups. These ions are equivalent to those at m/z 342 [M-D-HDO] (100%), 324 [M-D-H2O-HDO] (7%), 317 [M-D-CO2] (18%), 298 [M-D-HDO-CO2] (37%), and 279 [M-D-2HDO-CO2] (8%) from MS/MS of O,O,O-d3-PD1, respectively. They are further confirmed by ions in MS/MS of d5-PD1 (21,21,22,22,22-d5-PD1) at m/z 346 [M-H-H2O] (100%), 328 [M-H-2H2O] (13%), 320 [M-H-CO2] (23%), 302 [M-H-H2O-CO2] (27%), and 284 [M-H-2H2O-CO2] (6%), respectively; and along with ions from MS/MS of d8-PD1 (O,O,O,21,21,22,22,22-d8-PD1) at m/z 348 [M-D-H2O] (100%), 329 [M-D-H2O-HDO] (30%), 322 [M-D-CO2] (41%), 304 [M-D-H2O-CO2] (35%), and 284 [M-D-2HDO-CO2] (3%), respectively. It is interesting that some of the water loss was as H2O rather than HDO. Loss of HDO is expected for d3-PD1 because D has replaced every hydroxy H. D in the deuteroxy group exchanged with the hydrogen on the carbon chain when the [M-D]− ion of d3-PD1 was selected and activated for the MS/MS fragmentation in the ion trap, similar to d4-RvD1 (see above). The MS/MS chain-cut ions and the formation mechanisms are shown in Figure 2 and Scheme 4. Those ions formed via loss of H2O from d3-PD1 also manifest the exchange of deuteroxy deuterium with carbon-chain-bonded hydrogen.
The MS/MS ion m/z 153 of PD1 is consistent with the fragmentation mechanism of a γ-ene rearrangement: when the 10-OH H shifts to the C7 at γ double-bond C7,8 through a 6-membered ring, a carbonyl group forms (in S4a) and the C9,10 allylic single-bond, at α-position to 10-OH, cleaves, yielding the ion at m/z 153 in MS/MS of PD1 (11%) and d5-PD1 (7%). The equivalent ion is at m/z 154 for d3-PD1 (7%) and d8-PD1 (5%). Ions m/z 153 and 154 are equal to [10cc + H from 10-OH] (for PD1 and d5-PD1) and [10cc + D from 10-OD] (for O,O,O-d3-PD1 and d8-PD1), respectively. If additionally the 17-OH H or 17-OD D shifts to the carboxyl anion, it yields the neutral molecule S4b and ion m/z 205, equal to [10cm - 2H from hydroxys] for PD1 (7%) or [10cm - 2D from deuteroxys] for d3-PD1 (7%). Its equivalent ion is at m/z 210 (6%) for d5-PD1. The assignment of ions m/z 205 and 210 is consistent with ion 187 [205-H2O] (4%) of PD1.
Ion m/z 181 is equal to [10mc - H from OH] for PD1 (5%) and d5-PD1 (10%), or to [10mc - D from OD] for d8-PD1 (29%) and d3-PD1 (10%). The corresponding fragmentation mechanism is a charge-remote β-ene rearrangement with α-OH as 10-OH and β-ene as C11,12 double-bond of the parent ion: the conjugated triene Δ11,13,15 in PD1 forms a six-membered ring in intermediate S4c via the Diels-Alder_process, changing the vinyl single-bond C10,11 at position to an allylic single-bond (Scheme 4); then the H from 10-OH moves to C13 on the newly formed γ-ene (at C12,13), the allylic single-bond C10,11 breaks, and 10-carbony forms, generating S4d and ion m/z 181. Additional evidence for the composition of ion m/z 181 is ion m/z 163 [181-H2O] (5%) in the MS/MS spectrum of PD1 (Figure 2, Scheme 4).
Ion m/z 261 (20%) in the MS/MS spectra of PD1 is equivalent to [17cc + H from 17-OH] (Scheme 4), generated through a β-ene rearrangement analogous to the formation mechanism for ion m/z 277 from RvD1 (Figure 1 and Scheme 3). When 17-OH H shifts to C14 in S4c, the allylic single-bond C16,17 breaks, resulting in a carbonyl group in the neutral loss hexen-3-al and ion m/z 261. This is verified by ion m/z 263 from d3-PD1 (37%) and d8-PD1 (15%), equal to [17cc + D from17-OD]; and ion m/z 261 (23%) from d5-PD1, equal to [17cc + H from 17-OH]. This fragmentation mechanism is further confirmed by ions m/z 217 [261-CO2] and 199 [261-H2O-CO2] from both PD1 (9%, 5%) and d5-PD1 (7%, 5%), as well as by ions m/z 219 [263-CO2] (12%) and 200 [263-HDO-CO2] (9%) from d3-PD1. Ions m/z 219 [263-CO2] and 201 [263-H2O-CO2] from d8-PD1 (10%, 11%) are also consistent with this mechanism.
The formation of ion m/z 289 of PD1 corresponds to the α-H-β-ene rearrangement, which is analogous to that for ion m/z 305 from RvD1 (Figure 1 and Scheme 3), with the shift of C16 H instead of 17-OH H, where α-OH is 17-OH, and γ-ene is C19,20 double-bond. When C16 H shifts to C20 in S4c, the allylic single-bond C17,18 cleaves, yielding a pentaene (neutral loss) and ion m/z 289 (5%) with a 17-enol and a six-membered ring (Figure 2, Scheme 4). This fragmentation process was confirmed by ion m/z 289 from d5-PD1 (4%) and ion m/z 291 from d3-PD1 (9%) and d8-PD1 (16%) (Figure 2). Therefore ions m/z 289 and 291 are equal to [17mc – H from C16] of PD1 and d3-PD1, respectively (Figure 2 and Scheme 4). This process was consistent with the chain-plus-peripheral-cut ions formed from m/z 289 and 291 via loss of water or/and CO2: 271 [289-H2O], 245 [289-CO2], and 227 [289-H2O-CO2] from PD1 (4%, 10%, 4%) and d5-PD1 (5%, 18%, 4%); 272 [291-HDO] (5%), 247 [291-CO2] (23%), and 228 [291-HDO-CO2] (4%) from d3-PD1; 272 [291-HDO] (29%), 247 [291-CO2] (47%), and 229 [291-H2O-CO2] (15%) from d8-PD1.
The stereoisomers of PD1 were obtained through total organic synthesis [26], and are indistinguishable based only on their MS/MS spectra [24 and data not shown]. However, most can be separated via reversed-phase LC [24]. Their bioactivities were found to depend on their stereo-structures [24].
HDHAs and O,O-d2-HDHAs (Table 1, Supplemental Figure 1 and Scheme 5)
Table 1.
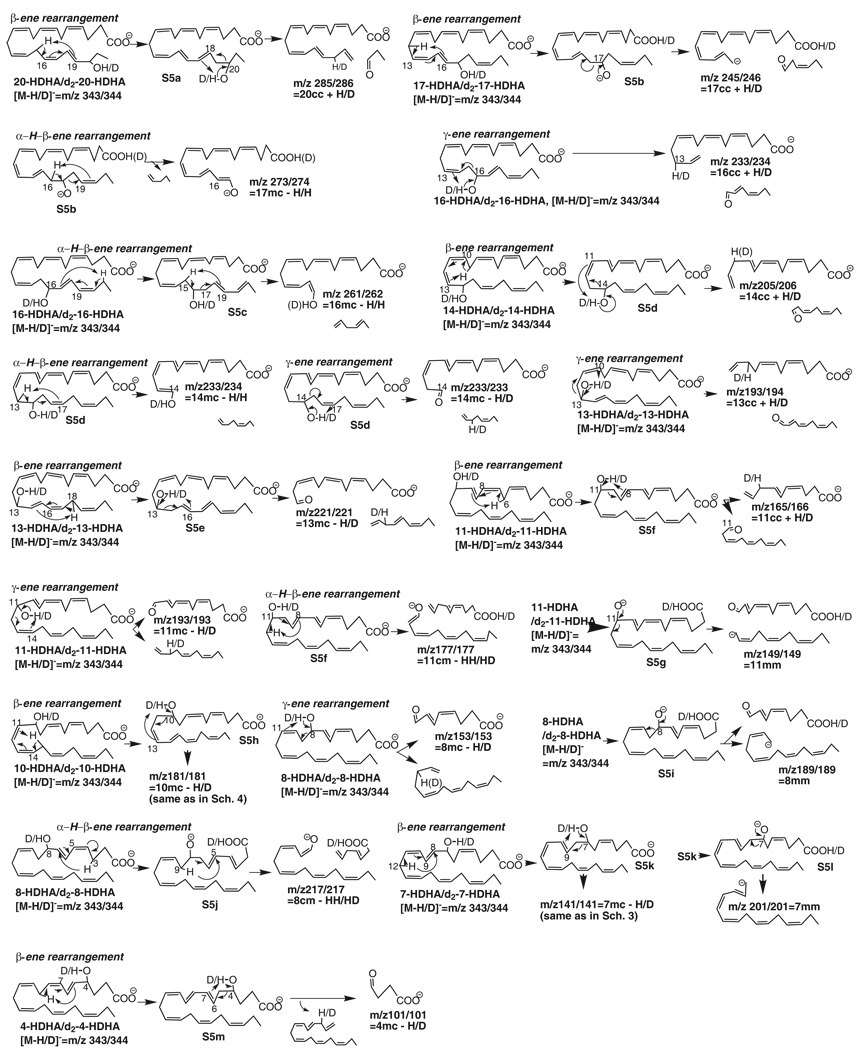
Chain-cut ions and fragmentation mechanisms for low energy MS/MS spectra obtained via CID of electrospray-generated carboxylate anion [M-H or D]− from the LC peak of a mono-HDHA or O,O-d2-mono-HDHA.
| -HDHA |
Ion m/z |
Algorithm | H is from | Fragmentation rearrangemnt |
Confirmation by Deuterium-labeling | ||||
|---|---|---|---|---|---|---|---|---|---|
| labeled- HDHA |
ion m/z |
Algorithm | D/H is from | ||||||
| cc* | 20- | 285 | 20cc+H | 20-OH | β-ene | d2-20- | 286 | 20cc+D | 20-OD |
| 17- | 245 | 17cc+H | 17-OH | β-ene | d2-17- | 246 | 17cc+D | 17-OD | |
| 16- | 233 | 16cc+H | 16-OH | γ-ene | d2-16- | 234 | 16cc+D | 16-OD | |
| 14- | 205 | 14cc+H | 14-OH | β-ene | d2-14- | 206 | 14cc+D | 14-OD | |
| 13- | 193 | 13cc+H | 13-OH | γ-ene | d2-13- | 194 | 13cc+D | 13-OD | |
| 11- | 165 | 11cc+H | 11-OH | β-ene | d2-11- | 166 | 11cc+D | 11-OD | |
| 10- | 153 | 10cc+H | 10-OH | γ-ene | d2-10- | 154 | 10cc+D | 10-OD | |
| cm | 11- | 177 | 11cm-2H | 11-OH,12C | α-H-β-ene | d2-11- | 177 | 11cm-D-H | 11-OD, H from 12C |
| 8- | 217 | 8cm-2H | 8-OH,9C | α-H-β-ene | d2-8- | 217 | 8cm-D-H | 8-OD, H from 9C | |
| mc | 17- | 273 | 17mc-H | 16C | α-H-β-ene | d2-17- | 274 | 17mc-H | H from 16C |
| 16- | 261 | 16mc-H | 15C | α-H-β-ene | d2-16- | 262 | 16mc-H | H from 15C | |
| 14- | 233 | 14mc-H | 13C | α-H-β-ene | d2-14- | 234 | 14mc-H | H from 13C | |
| 13- | 221 | 13mc-H | 13-OH | β-ene | d2-13- | 221 | 13mc-D | 13-OD | |
| 11- | 193 | 11mc-H | 11-OH | γ-ene | d2-11- | 193 | 11mc-D | 11-OD | |
| 10- | 181 | 10mc-H | 10-OH | β-ene | d2-10- | 181 | 10mc-D | 10-OD | |
| 8- | 153 | 8mc-H | 8-OH | γ-ene | d2-8- | 153 | 8mc-D | 8-OD | |
| 7- | 141 | 7mc-H | 7-OH | β-ene | d2-7- | 141 | 7mc-D | 7-OD | |
| 4- | 101 | 4mc-H | 4-OH | β-ene | d2-4- | 101 | 4mc-D | 4-OD | |
| mm | 11- | 149 | 11mm | n.a.** | charge-direct | d2-11- | 149 | 11mm | n.a. |
| 8- | 189 | 8mm | n.a. | charge-direct | d2-8- | 189 | 8mm | n.a. | |
| 7- | 201 | 7mm | n.a. | charge-direct | d2-7- | 201 | 7mm | n.a. | |
hypothetical homolytic-segment
not applicable.
Scheme 5.
Fragmentation mechanisms for mono-HDHA/d2-HDHA ([M-H/D]−=m/z 343/344).
The hydroxy of each mono-hydroxy docosahexaenoic acid (HDHA) reported here is at the α position relative to its conjugated-diene. The electrospray-generated molecular ion for each HDHA eluted from the LC column is at m/z 343 [M-H]. The peripheral-cut ions from low energy MS/MS at m/z 343, generated via neutral losses, are at m/z 325 [M-H-H2O], 299 [M-H-CO2], and 281 [M-H-H2O-CO2]. These ions are consistent with ions generated from MS/MS at m/z 344 [M-D], for any labile-hydrogen-deuterated HDHA (O,O-d2-HDHA or d2-HDHA), namely, m/z 325 [M-D-HDO] or 326 [M-D-H2O], 300 [M-D-CO2], and 281 [M-D-HDO-CO2] and 282 [M-D-H2O-CO2], respectively, where ions m/z 326 [M-D-H2O] and 282 [M-D-H2O-CO2] demonstrated that the deuteroxy D was exchanged with carbon-chain hydrogens prior to the water loss in MS/MS processes. The LC-UV-MS/MS analysis also showed that the LC peak of each HDHA has an asymmetric band at λmax 233–236 nm in the UV spectrum (data not shown), indicating the presence of a conjugated-diene in each HDHA.
20-HDHA and O,O-d2-20-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
The first HDHA eluted from the LC column is 20-HDHA (20-hydroxy-4Z,7Z,10Z,13Z,16Z,18E-docosahexaenoic acid). MS/MS ion m/z 285 (35%) or 286 (50%) results from segment 20cc, equivalent to [20cc + H from 20-OH from 20-HDHA] or [20cc + D from 20-OD of O,O-d2-20-HDHA], via a β-ene rearrangement with cleavage of the C19,20 bond. Prior to the cleavage of the C19,20 bond, the C15 H shifts to C19, and conjugated double-bond Δ16,18 changes to Δ15,17. Then the 20-OH H migrates to C17 (S5a) and the newly-formed allylic single-bond C19,20 cleaves.
17-HDHA and O,O-d2-17-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
Ion m/z 245 (41%) is equivalent to 17cc plus H from 17-OH of 17-HDHA (17-hydroxy-4Z,7Z,10Z,15E,19Z-docosahexaenoic acid). This is further confirmed by ion m/z 246 [17cc + D from 17-OD] (60%) from O,O-d2-17-HDHA (Table 1, Supplemental Figure 1). The migration of C12-H to C16 leads to the transfer of conjugated double-bonds Δ13,15 to Δ12,14 in S5b. A 17-alkoxide anion is generated when 17-OH H (or 17-OD D) shifts to the carboxyl in S5b. The negative charge of the 17-alkoxide anion directs the cleavage of the C16,17 bond, resulting in ion m/z 245 of 17-HDHA (246 of d2-17-HDHA). This ion also forms through a charge-remote β-ene rearrangement similar to that for ion m/z 277 from RvD1 (Figure 1 and Scheme 3) and ion m/z 153 from PD1 (Figure 2, Scheme 4): the 17-alkoxide in S5b is in 17-OH form, of which the H shifts to C14, then the double-bond C14,15 shifts to C15,16 and the allylic single-bond C16,17 cleaves, resulting in ion m/z 245.
An α-H-β-ene rearrangement, similar to that for ion m/z 305 from RvD1 (Figure 1 and Scheme 3) and ion m/z 289 from PD1 (Figure 2, Scheme 4), also occurs for 17-HDHA: when C16-H shifts to C20 through a six-membered ring in S5b, of which the 17-alkoxide anion is also in 17-OH form, the C17,18 bond cleaves, yielding ion m/z 273 (54%) equivalent to [17mc – H from C16]. This ion is equivalent to ion m/z 274 [17mc – H from C16] (50%) from O,O-d2-17-HDHA. Chain-plus-peripheral ions, resulting from the above ions via loss of H2O and/or CO2, are 201 [245-CO2] (12%), 229 [273-CO2] (19%), and 255 [273-H2O] (13%) of 17-HDHA; and 202 [246-CO2] (13%), 230 [274-CO2] (21%), and 256 [274- H2O] (10%) of O,O-d2-17-HDHA. The appearance of these chain-plus-peripheral ions further confirms the presence of a carboxyl and/or hydroxy in the originating ions.
16-HDHA and O,O-d2-16-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
The ion m/z 233 (85%) or 234 (100%) in the MS/MS spectrum of 16-HDHA (16-hydroxy-4Z,7Z,10Z,13Z,17E,19Z-docosahexaenoic acid) or O,O-d2-16-HDHA is equivalent to [16cc + H from 16-OH of 16-HDHA] or [16cc + D from 16-OD of O,O-d2-16-HDHA], respectively. Furthermore, ion m/z 261 (60%) or 262 (35%) is equivalent to [16mc – H from C15]. When H from 16-OH shifts to C13 in 16-HDHA, the C15,16 allylic single-bond cleaves, yielding ion m/z 233 in a γ-ene rearrangement. The same mechanism occurs for O,O-d2-16-HDHA, producing ion m/z 234. On the other hand, an α-H-β-ene rearrangement also takes place: the C21 H migrates to C17 and conjugated double-bonds Δ17,19 convert to Δ18,20 in S5c; then the C15 H shifts to C19 and the C16,17 allylic single-bond in S5c cleaves, generating ion m/z 261 for 16-HDHA or 262 for O,O-d2-16-HDHA (Scheme 5). Chain-plus-peripheral ions m/z 189 [233-CO2] (9%) of 16-HDHA and 190 [234-CO2] (5%) of O,O-d2-16-HDHA are consistent with the formation mechanism for ions [16cc + H from OH of 16-HDHA] or [16cc + D from OD of O,O-d2-16-HDHA].
14-HDHA and O,O-14-d2-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
Ions m/z 205 (34%) and 233 (87%) are equivalent to [14cc + H] and [14mc – H], respectively, from 14-HDHA (14-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid). The former corresponds to ion m/z 206 (68%, equivalent to [14cc + D from 14-OD]) in O,O-14-d2-HDHA, and the latter is consistent with ions m/z 233 (93%, equivalent to [14mc – D from 14-OD]) and 234 (97%, equivalent to [14mc – H from C13]) in O,O-14-d2-HDHA (Table 1, Supplemental Figure 1). Thus the H for [14cc + H] was from 14-OH (Scheme 5); and the H for [14mc – H] was from both C13 and 14-OH, indicating the involvement of two competing fragmentation mechanisms.
Ion m/z 205 forms through a β-ene rearrangement with the cleavage of the C13,14 bond and shift of 14-OH H to C11 in S5d, which results from migration of C9 H to C13 and conversion of conjugated double-bonds Δ10,12 to Δ9,11. When C13 H in S5d shifts to C17 through an α-H-β-ene rearrangement, the resulting ion is at m/z 233 for 14-HDHA or 234 for O,O-d2-14-HDHA. Meanwhile, ion m/z 233 also forms for both isotopomers through a γ-ene rearrangement when 14-OH H in 14-HDHA or 14-OD D in O,O-d2-14-HDHA shifts to C17, generating a carbonyl and breaking the C14,15 allylic single-bond. The existence of carboxyl and/or hydroxy groups in these four ions is further confirmed by ions m/z 161 [205-CO2] (13%), 189 [233-CO2] (7%), and 215 [233-H2O] (4%) of 14-HDHA and 162 [206-CO2] (11%), 216 [234-H2O] (5%), and 190 [234-CO2] (15%) of O,O-d2-14-HDHA.
13-HDHA and O,O-d2-13-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
The chain-cut ions in the MS/MS spectrum of 13-HDHA (13-hydroxy-4Z,7Z,10Z,14E,16Z,19Z-docosahexaenoic acid) are at m/z 193 (34%) and 221 (31%), equivalent to [13cc + H] and [13mc – H], respectively (Table 1, Supplemental Figure 1). They are consistent with ions m/z 194 (50%, [13cc + D from 13-OD]) and 221(78%, [13mc – D from 13-OD]) in the MS/MS spectrum of d2-13-HDHA. Therefore, H in [13cc + H] or [13mc – H] is from 13-OH. These ions correspond to the following MS/MS fragmentation mechanisms: through a γ-ene rearrangement, the 13-OH H (or 13-OD D) shifts to C10, and the C12,13 allylic single-bond cleaves, resulting in ion m/z 193 for 13-HDHA (194 for 13-d2-HDHA); in parallel, through a β-ene rearrangement the C13,14 vinyl single-bond converts to an allylic single-bond in S5e when the C18 H migrates to C14, then 13-OH H (or 13-OD D) shifts to C16 in S5e, and the C13,14 bond breaks, generating ion m/z 221 from both 13-HDHA and 13-d2-HDHA, equivalent to [13mc – H] and [13mc – D], respectively. Additionally, the chain-plus-peripheral ions m/z 203 [221-H2O] (5%) and 177 [221-CO2] (8%) of 13-HDHA as well as ion 177 [221-CO2] (8%) of O,O-d2-13-HDHA further confirm the composition of ion m/z 221.
11-HDHA and O,O-d2-11-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
Ions m/z 165, 177, 193, and 149 in the MS/MS spectrum of 11-HDHA (11-hydroxy-4Z,7Z,9E,13Z,16Z,19Z-docosahexaenoic acid) are equivalent to [11cc + H], [11cm – 2H], [11mc – H], and [11mm], respectively. The assignments are confirmed by ions m/z 166 [11cc + D from 11-OD], 177 [11cm – D from 11-OD – H from carbon-chain], 193 [11mc – D from 11-OD], and 149 [11mm] in the MS/MS spectrum of O,O-d2-11-HDHA. These indicate the following fragmentation mechanisms in the MS/MS processes: through a β-ene rearrangement, the C10,11 bond in 11-HDHA changes to an allylic single-bond and intermediate S5f forms when C6-H shifts to C10, then 11-OH H (or 11-OD D) shifts to C8 through a 6-membered ring in S5f and the C10,11 allylic single-bond cleaves, yielding ion m/z 165 (14%) for 11-HDHA (or 166 (12%) for O,O-d2-11-HDHA) and an aldehyde; through a γ-ene rearrangement, 11-OH H in 11-HDHA or 11-OD D in d2-11-HDHA shifts to C14, and the C11,12 allylic single-bond cleaves, generating ion m/z 193 (22% for 11-HDHA, 21% for O,O-d2-11-HDHA) with a carbonyl group; through an α-H-β-ene rearrangement, the C12-H shifts to C8 in S5f, the 11-OH H or 11-OD D migrates to the carboxylic group, and the C11,12 bond cleaves, yielding ion m/z 177 (8% for 11-HDHA or 7% for d2-11-HDHA). Additionally, when 11-OH H from 11-HDHA and 11-OD D of O,O-d2-11-HDHA shifts to the carboxyl group, an 11-alkoxide anion (S5g) forms and the negative charge directs the cleavage of the C11,12 allylic single-bond, producing ion m/z 149 (26% for 11-HDHA, 28% for d2-11HDHA). The composition and formation mechanism for ion m/z 165 are further confirmed by ion 121 [165-CO2] (4%) of 11-HDHA, and those for ion m/z 166 of d2-11-HDHA are verified by 122 [166-CO2] (4%) of d2-11-HDHA.
10-HDHA and O,O-d2-10-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
In contrast to the C10,11 bond in 11-HDHA, the C9,10 bond in 10-HDHA (10-hydroxy-4Z,7Z,11E,13Z,16Z,19Z-docosahexaenoic acid) is already an allylic single-bond as in PD1 (Figure 2 and Scheme 4). It cleaves through a -ene rearrangement when 10-OH H shifts to C7 and a carbonyl group forms, yielding ion m/z 153 (6%), which is equivalent to [10cc + H from 10-OH], following the same pathway as that for ion m/z 153 from PD1 (Scheme 4), except that the neutral loss here is a 2,4,7,10-tridecatetraenal versus the neutral loss S4a from PD1. This is consistent with ion m/z 154 [10cc + D from 10-OD] (5%) in the MS/MS spectrum of O,O-d2-10-HDHA. Ion m/z 153 in the MS/MS spectra of the 10 series of Neuroprostanes is also likely to have been generated through this mechanism although their segment 10cm is different from that of PD1 (Figure 2 and Scheme 4) or 10-HDHA [28].
In competition, a β-ene rearrangement occurs along 10-OH similar to that for ion m/z 181 from PD1 (Figure 2 and Scheme 4): the migration of C15 H to C11 through a six-membered ring converts the C10,11 vinyl bond to an allylic single-bond in S5h; then 10-OH H in 10-HDHA or 10-OD D in O,O-d2-10-HDHA shifts to C13, 10-OH or 10-OD changes to 10-carbony, and the C10,11 single-bond cleaves, generating ion m/z 181, equivalent to [10mc – H from 10-OH in 10-HDHA] (13%) or [10mc – D from 10-OD of O,O-d2-10-HDHA] (37%).
8-HDHA and O,O-d2-8-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
The chain-cut ions in the MS/MS spectrum of 8-HDHA (8-hydroxy-4Z,6E,10Z,13Z,16Z,19Z-docosahexaenoic acid), and O,O-d2-8-HDHA are at m/z 153 (10%, 8%), 189 (51%, 22%), and 217 (13%, 7%), which correspond to [8mc – H/D], [8mm], and [8cm –HH/HD], respectively. Via a -ene rearrangement, the migration of 8-OH H in 8-HDHA or 8-OD D of O,O-d2-8-HDHA to C11 generates 8-carbony, dissociates the C8,9 allylic single-bond, and yields ion m/z 153, equivalent to [8mc – H from 8-OH of 8-HDHA or D from 8-OD of O,O-d2-8-HDHA]. On the other hand, the migration of 8-OH H of 8-HDHA or 8-OD D of O,O-d2-8-HDHA to the carboxyl anion results in alkoxide anion S5i, of which the negative charge directs the cleavage of the C8,9 allylic single-bond, yielding an 8-carbonyl group and ion m/z 189 [8mm]. Ion m/z 217 is equivalent to [8cm - 2H] for 8-HDHA or [8cm – HD] for d2-HDHA, generated through an α-H-β-ene rearrangement: when C3 H migrates to C7 through a six-membered ring, C7,8 becomes an allylic single-bond in S5j; in the meantime, the 8-OH H or 8-OD D shifts to the carboxyl anion; then the C9 H in S5j shifts to C5 and the C7,8 bond breaks, yielding ion m/z 217 in the MS/MS spectrum of 8-HDHA or O,O-d2-8-HDHA. After loss of CO2, ion m/z 153 was transferred to ion m/z 109 for 8-HDHA (29%) or O,O-d2-8-HDHA (18%).
7-HDHA and O,O-d2-7-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
The chain-cut ions in the MS/MS spectrum of 7-HDHA (7-hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid) or O,O-d2-7-HDHA are at m/z 141 (13%, 18%) and 201 (8%, 7%), which are equivalent to [7mc – H or D] and [7mm], respectively, and correspond to the following fragmentation mechanisms. The generation of ion m/z 141 is via a β-ene rearrangement: the C12 H shifts to C8, changing the C7,8 vinyl single-bond to an allylic single-bond in S5k; then the H from 7-OH or D from 7-OD in S5k migrates to C10 and the C7,8 bond cleaves, yielding ion m/z 141. Competitively, the 7-OH H in S5k migrates to the carboxyl, yielding S5l; the negative charge of the 7-alkoxide anion S5l directs the cleavage of C7,8 bond, resulting in ion m/z 201.
4-HDHA and O,O-d2-4-HDHA (Table 1, Supplemental Figure 1 and Scheme 5)
The chain-cut ion is m/z 101 (14%) for 4-HDHA (4-hydroxy-5E,7Z,10Z,13Z,16Z,19Z-docosahexaenoic acid) (27% for O,O-d2-4-HDHA), which is equivalent to [4mc – H from 4-OH of 4-HDHA or D from 4-OD of O,O-d2-4-HDHA] (Table 1, Supplemental Figure 1). It is generated through a β-ene rearrangement: when C9 H shifts to C5, the C4,5 bond changes to an allylic single-bond (S5m); C 4,5 bond in S5m cleaves upon the migration of 4-OH H of 4-HDHA or 4-OD D of O,O-d2-4-HDHA to C7, generating ion m/z 101.
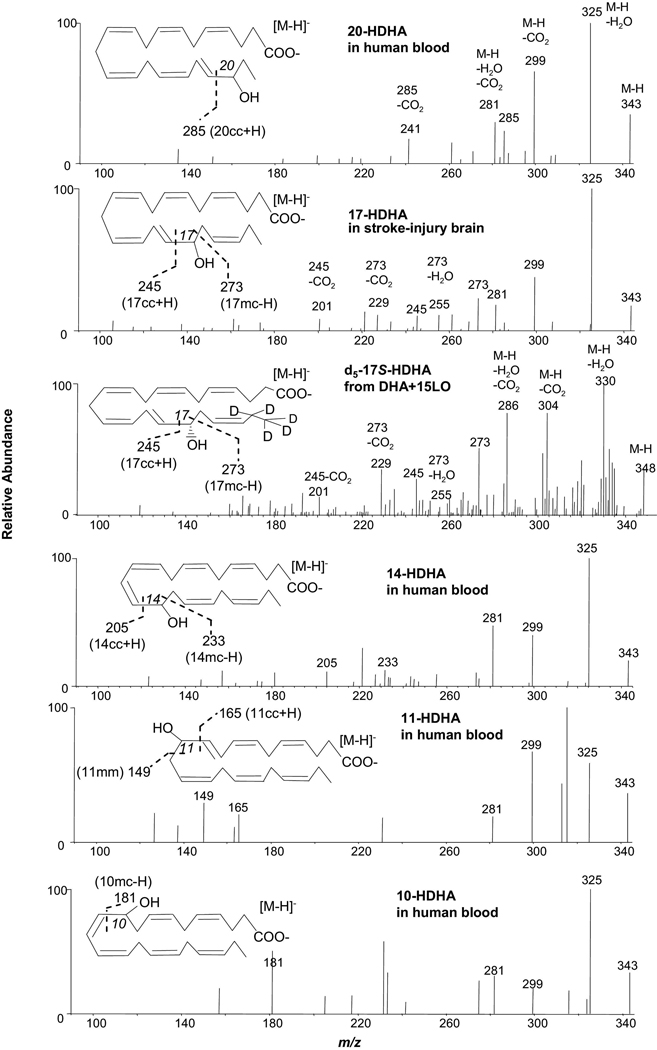
Identification of HDHAs from biogenic samples (Figure 3)
Figure 3.
Identification of 20-HDHA, 14-HDHA, 11-HDHA and 10-HDHA in human blood, 17-HDHA from stroke-injury murine brain tissues, and 21,21,22,22,22-d5-17S-HDHA in the incubations of 21,21,22,22,22-d5-DHA and soybean 15-LO as demonstrated with low collision energy MS/MS spectra acquired in the LC-UV-electrospray MS/MS analysis.
Based on the above MS/MS spectra of HDHAs, 20-HDHA, 14-HDHA, 11-HDHA, and 10-HDHA were found in human whole blood via LC-UV-MS/MS detailed here as well as 17-HDHA found in stroke-injury murine brain tissues. All the peripheral-cut ions in these spectra match the ions in the spectra acquired from standard compounds. There are one or two chain-cut ions from α-cleavage in these spectra that match the ions in standard spectra (Table 1, Supplemental Figure 1). The LC retention times and UV spectra of each identified mono-HDHA also match those of the corresponding standard compound (data not shown). Additionally, 21,21,22,22,22-d5-17S-HDHA (d5-17S-HDHA) was identified in the incubation of d5-DHA with 15-LO. It has the same position of double-bonds and the position of hydroxy as 17-HDHA, except its C21 and 22 were deuterated. In comparison with the MS/MS spectrum of 17-HDHA in Table 1, Supplemental Figure 1, the corresponding ions shifted 5 Daltons, e.g., MS/MS ions m/z 348 [M-H], 330 [M-H-H2O], 304 [M-H-CO2], and 286 [M-H-H2O-CO2], consistent with d5-17S-HDHA. Ions m/z 245 [17cc + H] (27%), m/z 273 [17mc –H] (50%), 255 [273 – H2O] (7%), 229 [273 – CO2] (32%), and 201 [245 – CO2] (14%) revealed that this MS/MS spectrum is of d5-17S-HDHA. The LC retention time and UV spectrum (λmax 235 nm) of d5-17S-HDHA matched those of 17S-HDHA (data not shown).
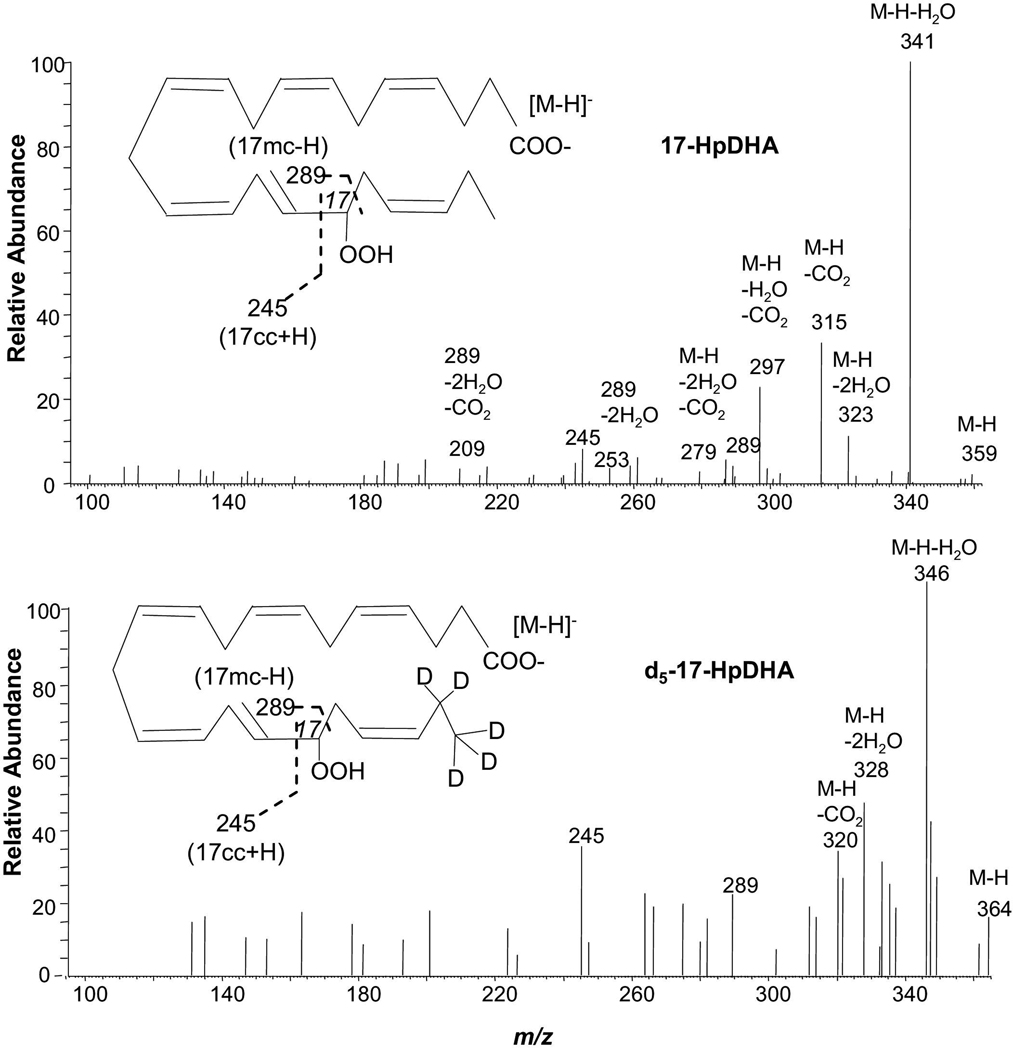
17-HpDHA and 21,21,22,22,22-d5-17-HpDHA (Figure 4, Scheme 6)
Figure 4.
Low energy MS/MS spectra acquired via CID of electrospray-generated carboxylate anion [M-H]− from the LC peak of 17-HpDHA and 21,21,22,22,22-d5-17-HpDHA, which were obtained from isolated-enzyme reactions, as detailed in the “Experimental” section. Each inset depicts the structure and CID fragmentation.
Scheme 6.
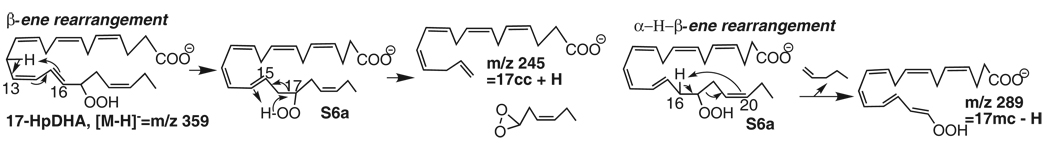
Fragmentation mechanisms for 17-HpDHA ([M-H]−=m/z 359).
There is a conjugated double-bond in 17-HpDHA. Collision-induced dissociation of the molecular ion m/z 359 [M-H] from 17-HpDHA generated the peripheral-cut ions m/z 341 [M-H-H2O], 323 [M-H-2H2O], 315 [M-H-CO2], 297 [M-H-H2O-CO2] and 279 [M-H-2H2O-CO2]. The corresponding ions in the MS/MS spectrum of d5-17-HpDHA are 346 [M-H-H2O], 328 [M-H-2H2O], and 320 [M-H-CO2]. Ions m/z 245 (9%) and 289 (6%) are equivalent to [17cc + H] and [17mc - H] for 17-HpDHA. Ion m/z 245 forms through a -ene rearrangement: 17-hydroperoxyl H shifts to C14 in S6a, a dioxirany ring forms on C17, and the C16,17 bond breaks. Ion m/z 289 forms through an α-H-β-ene rearrangement when C16 H in S6a shifts to C20 and the C17,18 allylic single-bond breaks. Ion 289 further transfers to ions m/z 253 [289-2H2O] (5%) and 209 [289-2H2O-CO2] (4%). The presence of ions m/z 245 (36%) and 289 (23%) in the MS/MS spectrum of d5-17-HpDHA further confirms this fragmentation mechanisms.
Discussion
Impact of a functional group and enes on MS/MS fragmentation of different lipid mediators
Under the same MS/MS conditions as for RvD1, Lipoxin A4 (5S,6R,15S-trihydroxy-7E,9E,11Z,13E-eicosatetraenoic acid) produces chain-cut ions m/z 115 [5mc – H], m/z 235 [5mm], and m/z 251 [15cc + H] [27,29], equivalent to ions m/z 141[7mc – H], m/z 233 [7mm], and m/z 277 [17cc + H], respectively, from RvD1 (Figure 1,Scheme 3). This corresponds to the same structure of Lipoxin A4 between C5 and C15 as that of RvD1 between C7 and C17, the conjugated tetraene bracketed by two vicinal hydroxys on the carboxyl (c)-side and one hydroxy on the methyl terminus (m)-side. However the ion m/z 279 [15mc – H] was not observed for Lipoxin A4. There is no double-bond at γ position to 15-OH on the m side of Lipoxin A4, i.e., in segment 15mm, to facilitate generation of this ion through an α-H-β-ene rearrangement that yields ion [17mc – H] m/z 305 from RvD1. The same phenomenon was observed for 15-hydroxy-eicosatetraenoic acid (15-HETE) vs 17-HDHA and PD1. 15-HETE has no [15mc – H] ion (m/z 247), but 17-HDHA and PD1 have [17mc – H] ions as m/z 273 and 289, respectively. Therefore, ion [xmc – H] indicates double-bond(s) in xmm segment, with x representing the position of the functional group.
7-HDHA also has the same chain-cut ions as RvD1 does: [7mc – H] and [7mm] around 7-OH (Figure 1, Table 1, Supplemental Figure 1 and Scheme 3 and Scheme 5). The γ-ene rearrangement for the cleavage of C7,8 allylic single-bond in RvD1 for producing ion m/z 141 (Scheme 3) is analogous to that for S5k, a transient state of 7-HDHA in MS/MS. However, the shift of C12 H to C8 prior to such cleavage for 7-HDHA is unnecessary for RvD1 because the C7,8 bond in RvD1 is already an allylic single-bond. The [7mm] ions m/z 233 and 201 are generated through the same mechanism from RvD1 and 7-HDHA, respectively. Analogously, 5-HETE has the chain-cut ions m/z 115 ([5mc – H]) and 203 ([5mm]) around 5-OH; this is similar to what LXA4 has [27,29,30].
Although there is an 8-OH in both RvD1 and 8-HDHA, there are no MS/MS ions from RvD1 analogous to ions [8cm – 2H], [8mc – H], and [8mm] from 8-HDHA (Figure 1, Table 1, Supplemental Figure 1, and Scheme 3 and Scheme 5). Because of the 7-OH and the conjugated tetraene 9,11,13,15 of RvD1, the 8cm (equal to 7mm) segment of RvD1 is involved in formation of ion m/z 233 as 7mm, not as 8cm, namely, it follows the fragmentation mechanism to produce ion [7mm] instead of [8cm – 2H]. The ion m/z 141, coming from segment 8cc (equivalent to 7mc) of RvD1, is equivalent to [7mc – H] instead of [8cc + H]. RvD1 has the [8mm - H] ion (m/z 189) because the ion can form a conjugated tetraenonyl structure to disperse the carbon anion charge for greater stabilization. In contrast, 8-HDHA does not have the [8mβm - H] ion (would be m/z 174) due to lack of such stabilization.
The structural similarity among 17-HDHA and 17-HpDHA is reflected by some common features of their MS/MS spectra (Table 1, Supplemental Figure 1, Figure 3 and Figure 4). The conversion of the C16,17 vinyl single-bond to an allylic single bond via shift of C12 H to C16 is likely to be the first step of the MS/MS fragmentation (Scheme 5 and Scheme 6). They all have [cc + H] ion m/z 245, although the H is from 17-OH of 17-HDHA and 17-OOH of 17-HpHDHA. They all have [mc – H] ions with the H migrating from C16 to C20. Such similarity was also observed among 15-HETE, 15-hydroperoxy-eicosatetraenoic acid, and 15-oxo-eicosatetraenoic acid [30,31].
[cc + H] ions in MS/MS of lipid mediators
[cc + H] ions are generated with the shift of H from the hydroxy or hydroperoxy to the γ position in segment cc through a β-ene rearrangement if the segment cc has a β double-bond, an α vinyl single-bond, and an allylic hydrogen (for 20-HDHA and 17-HDHA) or conjugated-triene (for 17cc in RvD1 and PD1, 12cc of LTB4 [19], 15cc of LXA4 [19]) that can facilitate the migration of π electrons from the β double-bond to the γ-bond and convert the α vinyl single-bond to an α allylic single-bond. Examples are ions m/z 277 from RvD1 (Figure 1 and Scheme 3), 261 from PD1 (Figure 2, Scheme 4), 245 from 17-HpDHA (Figure 4 and Scheme 6), 195 from LTB4 and 251 from LXA4 [27,29], as well as ions 285 from 20-HDHA, 245 from 17-HDHA, 205 from 14-HDHA, and 165 from 11-HDHA (Table 1, Supplemental Figure 1 and Scheme 5). These fragmentation processes can also occur through γ-ene-rearrangement if the cc segment has a γ double-bond, which is at least the second double-bond counted from the carboxyl group (for ion-trap MS/MS, unnecessary for triple-quadrople MS/MS, such as for 8cc of 8,9-dihydroxy-eicosatrienoic acids [27]), and an α allylic single-bond (Scheme 3 to 6). Examples are ions m/z 153 from PD1 (Figure 2, Scheme 4), as well as 153 from 10-HDHA, 233 from 16-HDHA, and 193 from 13-HDHA (Table 1, Supplemental Figure 1 and Scheme 5). Additional examples are ions m/z 167 did167 from 11,12-diHEtrE (dihydroxy eicosatrienoic acid), 207 from 14,15-diHETrE, 195 from 5,12-diHETE (dihydroxyeicosatetraenoic acid), 235 from 5,15-dihydroxy-eicosatetraenoic acid, and 221 from lipoxin B4 [27,29].
There is no double-bond in 4cc of 4-HDHA and no allylic hydrogen that can facilitate the migration of the π electrons of the β double-bond to the γ-bond in 8cc of 8-HDHA. The double-bond in 7cc of 7-HDHA and RvD1 is the first double-bond counted from the carboxyl group. Therefore these hydroxys have no [cc + H] ions (for ion-trap mass spectrometer LCQ, ThermoElectron).
[mc - H] ions in MS/MS of lipid mediators
[mc – H] ions are generated with shift of the hydroxy H to the γ position in segment mm, through a β-ene rearrangement if the mm has a β double-bond, an α vinyl single-bond, and an allylic hydrogen (for 13-HDHA, 10-HDHA, 7-HDHA, and 4-HDHA in Scheme 5) or conjugated-triene (for 10mm in PD1) that can facilitate the migration of π electrons of the β double-bond to the γ-bond and convert the α vinyl single-bond to an α allylic single-bond. Examples include ions m/z 221 from 13-HDHA, 181 from 10-HDHA, 141 from 7-HDHA, and 101 from 4-HDHA (Table 1, Supplemental Figure 1 and Scheme 5), as well as ion m/z 181 from PD1 (Figure 2, Scheme 4). This process also takes place through γ-ene-rearrangement if the segment mm has a γ-double-bond and an α allylic single-bond. Examples are ions 141 from RvD1 (Figure 1, Scheme 3), 193 from 11-HDHA, and 153 from 8-HDHA (Table 1, Supplemental Figure 1 and Scheme 5), as well as ions 145 of 5,6-diHETrE, 185 of 8,9-diHETrE, and 115 of both 5,6-diHETE and LXA4 [27,29,30]. [mc – H] ions are also produced through an α-H-β-ene rearrangement with shift of the α-H from segment mc to the γ position in segment mm whose α-bond is a vinyl single-bond and β-bond is a double-bond, such as ions m/z 261 from 16-HDHA (Table 1, Supplemental Figure 1 and Scheme 5), m/z 305 from RvD1 (Figure 1, Scheme 3), m/z 289 from PD1 (Figure 2, Scheme 4), m/z 233 from 14-HDHA, 273 from 17-HDHA (Table 1, Supplemental Figure 1 and Scheme 5), and m/z 289 from 17-HpDHA (Figure 4, Scheme 6).
[cm - 2H] and [mm] ions in MS/MS of lipid mediators
Only three [cm – 2H] ions were observed: m/z 205 from PD1 (Figure 2, Scheme 4), m/z 177 from 11-HDHA and m/z 217 from 8-HDHA (Table 1, Supplemental Figure 1 and Scheme 5), which are generated through a γ-ene rearrangement and an α-H-β-ene rearrangement, respectively. For PD1, the 17-OH H was extracted from cm to cc by the carboxyl group and 10-OH H was extracted to C7 of the cc. For 11-HDHA and 8-HDHA, the α-H is in segment cm and is extracted to segment cc that has a γ-ene (the π electrons come from the β-ene of the molecular ion); the OH H is extracted from cm to cc by the carboxyl group. The [mm] ions detected are m/z 233 ([7mm]) from RvD1 (Figure 1, Scheme 3), as well as ions m/z 149 ([11mm]) from 11-HDHA, 189 ([8mm]) from 8-HDHA, and 201 ([7mm]) from 7-HDHA (Table 1, Supplemental Figure 1 and Scheme 5). They are generated through charge-direct fragmentation mechanisms, where the negative charge of the transient alkoxide anion, resulted from extraction of the hydroxy H by the carboxylic group, directing the cleavage of the α allylic single-bond in mm.
Conclusions
ESI-MS/MS analysis of underivatized ESI-deprotonated molecules of Resolvin D1, Protectin D1 and other DHA-derived products reveals the structures, especially the locations of hydroxy, hydroperoxy, or carbonyl groups, as well as the double-bonds. The definitive fingerprints are the CID MS/MS ions formed via cleavage of the carbon-chain, namely chain-cut ions. These ions are generated via charge-remote and/or charge-directed fragmentation mechanisms as depicted in Scheme 2 to Scheme 6. The CID fragmentation usually consists of several serial and parallel reactions. Each chain-cut ion is equivalent to the corresponding hypothetically homolytic segment (cc, cm, mc, or mm) with addition or extraction of up to 2 protons. Deuterium labeling facilitated the structural analysis of MS/MS ions and the corresponding fragmentation mechanisms. The molecular ion structures determine the fragmentation mechanisms, which in turn provide the rationale for the assignment of the MS/MS ion structures, and consequently the molecular ion structures of the DHA-derived compounds. This paper demonstrates a simple approach for comprehensive structure analysis of DHA-derived products without the need for derivatizing the compounds. The fragmentation rules reported here were used for the development of a database and search algorithm for the theoretic MS/MS spectra of novel compounds derived from fatty acids [23].
Supplementary Material
Acknowledgements
We are very grateful to Professor Andrew F. Rowley’s group in the Department of Biological Sciences, University of Wales, Swansea, UK for providing trout head-kidney and Professor N. Bazan’s group in the Louisiana State University Health Sciences Center for providing the stroke-injury murine brain tissues. Many thanks to Ms. Mary H. Small, Dr. Barry Torem and Ms. Stacey Lindberg for assistance with manuscript preparation. This work was supported in part by P50-DE016191 (S.H., Y.L., C.N.S.) and 5R37GM038765 (C.N.S.) from the National Institutes of Health.
Abbreviations
- RvD1
Resolvin D1 7S,8R,17S-trihydroxy-docosa-4Z,9E,11E,13Z,15E,19Z-hexaenoic-acid
- d4-RvD1
O,O,O,O-d4-RvD1
- d5-RvD1
21,21,22,22,22-d5-RvD1
- PD1
Protectin D1/Neuroprotectins D1; 10R,17S-dihydroxy-docosa-4Z,7Z,11E,13E,15Z,19Z-hexaenoic acid
- d3-PD1
O,O,O-d3- PD1
- d5-PD1
21,21,22,22,22-d5- PD1
- d8-PD1
O,O,O,21,21,22,22,22-d8-PD1
- d5-17S-HDHA
21,21,22,22,22-d5-17S-HDHA
- 17-HpDHA
17-hydroperoxy-4Z,7Z,10Z,15E,19Z-docosahexaenoic acid
- d5-17-HpDHA
21,21,22,22,22-d5-17-HpDHA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gruppo Italiano per lo Studio della Streptochinasi nell’Infarto Miocardico (GISSI)-Prevenzione Investigators. Dietary supplementation with n-3 polyunsaturated fatty acids and vitamin E after myocardial infarction: results of the GISSI-Prevenzione trial. Lancet. 1999;354:447–455. [PubMed] [Google Scholar]
- 2.Freedman SD, Blanco PG, Zaman MM, Shea JC, Ollero M, Hopper IK, Weed DA, Gelrud A, Regan MM, Lapostata M, et al. Association of Cystic Fibrosis with Abnormalities in Fatty Acid Metabolism. N. Engl. J. Med. 2004;350:560–569. doi: 10.1056/NEJMoa021218. [DOI] [PubMed] [Google Scholar]
- 3.Teitelbaum JE, Walker WA. The role of omega 3 fatty acids in intestinal inflammation. J. Nutr. Biochem. 2001;12:21–32. doi: 10.1016/s0955-2863(00)00141-8. [DOI] [PubMed] [Google Scholar]
- 4.Bazan NG. Nestle Nutrition Workshop Series. 1992;28:121–133. doi: 10.1159/000351395. [DOI] [PubMed] [Google Scholar]
- 5.Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, Moussignac RL. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J. Exp. Med. 2002;196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong S, Gronert K, Devchand PR, Moussignac RL, Serhan CN. Novel docosatrienes and 17S-resolvins generated from docosahexaenoic acid in murine brain, human blood and glial cells: autacoids in anti-inflammation. J. Biol. Chem. 2003;278:14677–14687. doi: 10.1074/jbc.M300218200. [DOI] [PubMed] [Google Scholar]
- 7.Marcheselli LV, Hong S, Lukiw WJ, Tian XH, Gronert K, Musto A, Hardy M, Gimenez JM, Chiang N, Serhan CN, Bazan NG. Novel Docosanoids Inhibit Brain Ischemia-Reperfusion-mediated Leukocyte Infiltration and Pro-inflammatory Gene Expression. J. Biol. Chem. 2003;278:43807–43817. doi: 10.1074/jbc.M305841200. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee PK, Marcheselli VL, Serhan CN, Bazan NG. Neuroprotectin D1: a docosahexaenoic acid –derived docosatrienes protects human retinal pigment epithelial cells from oxidative stress. Proc Natl Acad Sci USA. 2004;101:8491–8496. doi: 10.1073/pnas.0402531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VanRollins M, Murphy RC. Autooxidation of docosahexaenoic acid: analysis of ten isomers of hydroxydocosahexaenoate. Journal of Lipid Research. 1984;25:507–517. [PubMed] [Google Scholar]
- 10.Kim HY, Salem N., Jr Preparation and the structural determination of hydroperoxy derivatives of docosahexaenoic acid and other polyunsaturates by thermospray LC/MS. Prostaglandins. 1989;37:105–119. doi: 10.1016/0090-6980(89)90035-x. [DOI] [PubMed] [Google Scholar]
- 11.Kim HY, Karanian JW, Shingu T, Salem N., Jr Stereochemical analysis of hydroxylated docosahexaenoates produced by human platelets and rat brain homogenate. Prostaglandins. 1990;40:473–490. doi: 10.1016/0090-6980(90)90110-h. [DOI] [PubMed] [Google Scholar]
- 12.Cheng C, Gross ML. Applications and mechanisms of charge-remote fragmentation. Mass Spectrometry Reviews. 2000;19:398–420. doi: 10.1002/1098-2787(2000)19:6<398::AID-MAS3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 13.Murphy RC, Fiedler J, Hevko J. Analysis of nonvolatile lipids by mass spectrometry. Chem. Rev. 2001;101:479–526. doi: 10.1021/cr9900883. [DOI] [PubMed] [Google Scholar]
- 14.Lee SH, Williams MV, DuBois RN, Blair LA. Targeted lipidomics using electron capture atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:2168–2176. doi: 10.1002/rcm.1170. [DOI] [PubMed] [Google Scholar]
- 15.Hsu FF, Turk J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000;11:797–803. doi: 10.1016/S1044-0305(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths WJ. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom. Rev. 2003;22:81–152. doi: 10.1002/mas.10046. [DOI] [PubMed] [Google Scholar]
- 17.Moe MK, Strom MB, Jensen E, Claeys M. Negative electrospray ionization low-energy tandem mass spectrometry of hydroxylated fatty acids: a mechanistic study. Rapid Commun. Mass Spectrom. 2004;18:1731–1740. doi: 10.1002/rcm.1545. [DOI] [PubMed] [Google Scholar]
- 18.James PF, Perugini MA, O'Hair RA. Sources of artifacts in the electrospray ionization mass spectra of saturated diacylglycerophosphocholines: from condensed phase hydrolysis reactions through to gas phase intercluster reactions. J. Am. Soc. Mass Spectrom. 2006;17:384–394. doi: 10.1016/j.jasms.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Ham BM, Jacob JT, Keese MM, Cole RB. Identification, quantification and comparison of major non-polar lipids in normal and dry eye tear lipidomes by electrospray tandem mass spectrometry. J. Mass Spectrom. 2004;39:1321–1336. doi: 10.1002/jms.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekroos K, Ejsing CS, Bahr U, Karas M, Simons K, Shevchenko A. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. Journal of Lipid Research. 2003;44:2181–2192. doi: 10.1194/jlr.D300020-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Tomer KB, Crow FW, Gross ML. Location of double bond position in unsaturated fatty acids by negative ion MS/MS. J. Am. Chem. Soc. 1983;105:5487–5488. [Google Scholar]
- 22.Murphy RC, Barkely RM, Berry KZ, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, Zarini S. Electrospray ionization and tandem mass spectrometry of eicosanoids. Analytical Biochemistry. 2005:1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y, Hong S, Tjonahen E, Serhan CN. Mediator-lipidomics: databases and search algorithms for PUFA-derived mediators. J. Lipid Research. 2005;46:790–802. doi: 10.1194/jlr.D400020-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Serhan CN, Gotlinger K, Hong S, Lu Y, Siegelman J, Baer T, Yang R, Colgan SP, Petasis NA. Anti-Inflammatory Actions of Neuroprotectin D1/Protectin D1 and Its Natural Stereoisomers: Assignments of Dihydroxy-Containing Docosatrienes. J Immunol. 2006;176:1848–1859. doi: 10.4049/jimmunol.176.3.1848. [DOI] [PubMed] [Google Scholar]
- 25.Hong S, Tjonahen E, Morgan EL, Yu L, Serhan CN, Rowley AF. Rainbow trout (Oncorhynchus mykiss) brain cells biosynthesize novel docosahexaenoic acid-derived resolvins and protectins -- mediator lipidomic analysis. Prostaglandins Other Lipid Mediat. 2005;78:107–116. doi: 10.1016/j.prostaglandins.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Yang R. Doctoral thesis. Los Angeles, CA, USA: Department of Chemistry, University of Southern California; 2005. Synthetic Study of Lipid mediators. [Google Scholar]
- 27.Wheelan P, Zirrolli JA, Murphy RC. Electrospray Ionization and Low Energy Tandem Mass Spectrometry of Polyhydroxy Unsaturated Fatty Acids. J. Am. Soc. Mass Spectrom. 1996;7:140–149. doi: 10.1016/1044-0305(95)00628-1. [DOI] [PubMed] [Google Scholar]
- 28.Yin H, Musiek ES, Gao L, Porter NA, Morrow JD. Regiochemistry of Neuroprostanes Generated from the Peroxidation of Docosahexaenoic Acid in Vitro and in Vivo. Biol. Chem. 2005;280:26600–26611. doi: 10.1074/jbc.M503088200. [DOI] [PubMed] [Google Scholar]
- 29.Chiang N, Takano T, Clish CB, Petasis NA, Tai HH, Serhan CN. Aspirin-triggered 15-epi-LXA4 generation by costimulation of human peripheral blood cell types and in a mouse acute peritonitis model: Development of a specific 15-epi-LXA4 ELISA. Journal of Pharmacology and Experimental Therapeutics. 1998;287:779–790. [PubMed] [Google Scholar]
- 30. http://www.lipidmaps.org/data/classification/fa.html.
- 31.MacMillan DK, Murphy RC. Analysis of Lipid Hydroperoxides and Long-Chain Conjugated Keto Acids by Negative Ion Electrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1995;6:1190–1201. doi: 10.1016/1044-0305(95)00505-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.