Summary
Ribosomal proteins stabilize the folded structure of the rRNA and enable the recruitment of further proteins to the complex. Quantitative hydroxyl radical footprinting was used to measure the extent to which three different primary assembly proteins, S4, S17 and S20, stabilize the 3D structure of the E. coli 16S 5′ domain. The stability of the complexes was perturbed by varying the concentration of MgCl2. Each protein influences the stability of the rRNA tertiary interactions beyond its immediate binding site. S4 and S17 stabilize the entire 5′ domain, while S20 has a more local effect. Multi-stage folding of individual helices within the 5′ domain shows that each protein stabilizes a different ensemble of structural intermediates, that include non-native interactions at low Mg2+. We propose that the combined interactions of S4, S17 and S20 with different helical junctions bias the free energy landscape toward a few RNA conformations that are competent to add the secondary assembly protein S16 in the next step of assembly.
Keywords: ribosome, ribosomal protein, RNA folding, hydroxyl radical footprinting, RNA-protein interactions
Ribosome biogenesis is one of the most important synthetic tasks undertaken by the cell. Among bacteria, short generation times require that several hundreds of ribosomes are produced per minute in order to produce the tens of thousands of ribosomes needed by each new cell 1;2. A critical question is how the RNA and protein components of the ribosome interact to achieve the native subunits and avoid nonfunctional complexes.
Studies on the reconstitution of the 30S ribosomal subunit have shown that binding of ribosomal proteins is coupled to folding of the rRNA 3;4. The addition of the primary assembly proteins, which bind directly to the 16S rRNA, stabilize the helices with which they interact, and enable further assembly of each 30S domain by pre-ordering the binding sites for secondary and tertiary assembly proteins 5;6. The extent to which individual ribosomal proteins stabilize the structure of the rRNA beyond their immediate binding site is thus directly connected to the hierarchy and cooperativity of assembly 4;7.
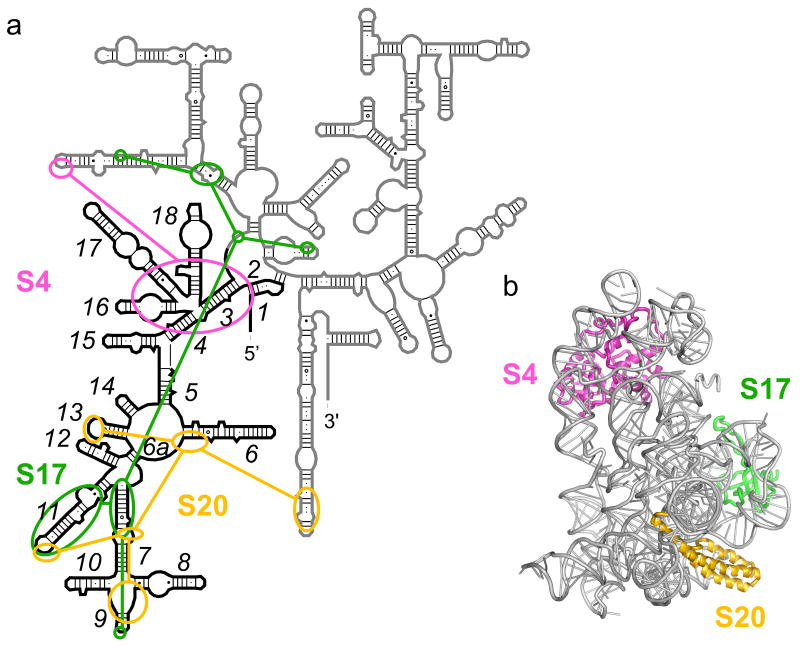
The 5′ domain of the 16S rRNA is the first to be transcribed in vivo and forms a separate tertiary structure that makes up the body and spur of the small subunit (Fig. 1) 8;9. Three primary assembly proteins S4, S17 and S20 each recognize and bind a different helix junction within the 5′ domain (Fig. 1a). Protein S17 binds helices 7 and 11 near the central junction 10. Its flexible loops extend from a β-barrel 11 to contact different regions of the rRNA. Amino acids 13-17 in S17 stabilize a K-turn in helix 11, while aa 29-34 thread between helix 11 and helix 21 to reach the 560-region at the junction with the central domain 12. Mutations in aa 67-68, which stitch together helices 7 and 11, result in defective 30S assembly 13. Protein S20 forms a three-helix bundle that interacts with the lower junction between helices 7-10 and the loop of helix 13 12;14. S20 also contacts helix 44 in the 30S subunit, helping to correctly position the 3′ minor domain14.
Figure 1. Protein interactions in the E. coli 5′ domain.
(a) Secondary structure of the 16S rRNA with 5′ domains nucleotides 21-562 in black. Schematic of protein interactions is redrawn from Ref. 12. (b) Structure of the 5′ domain in the E. coli 30S ribosome (2avy; 60), with S4 in pink, S17, green, S20, yellow. Ribbon made with nuccyl (L. Jovine) and Pymol (Delano Scientific).
Protein S4 binds the junction between 5 helices (3, 4, 16-18) in the “upper” part of the 5′ domain 15;16;17, and makes additional interactions to helix 17 (A496), helix 1 (A9) and the tip of helix 21 12 (Fig. 1a). S4 is one of two 30S proteins required for nucleation of subunit assembly 18. Consistent with this role, a comparison of base chemical modification and backbone protections suggested that S4 strongly influences the structure of the 16S rRNA outside its immediate binding site 19. Temperature-dependent changes in the conformation and stability of the 16S rRNA provided evidence for reorganization of the S4-16S complex 20;21. Reorganization of S4 complexes may be linked to refolding of the N-terminal domain of S4, which is disordered in the free protein 22. By contrast, there is no evidence for temperature-dependent reorganization of the S17 and S20 complexes 23.
We wished to understand in more detail how the three primary binding proteins in the E. coli 16S 5′ domain contribute to assembly of the 30S ribosome. To make the system more amenable to quantitative analysis, we used a transcript that contains only the E. coli 16S 5′ domain. The 5′ domain RNA forms a stable complex with proteins S4, S16, S17 and S20 24. Proteins S12 and S5 also interact with the 5′ domain residues within the 16S rRNA at a later stage of 30S assembly 19;25, but do not stably bind the 5′ domain complexes 24.
The 5′ domain RNA forms all of its predicted tertiary backbone contacts in 20 mM MgCl2 in the absence of proteins 26. However, tertiary interactions in helices 15 and 17 require more than 5 mM magnesium to form completely, while other helices are protected less strongly in the naked RNA than in native 30S ribosomes, suggesting that the RNA tertiary structure is dynamic in the absence of proteins. By contrast, the RNP containing ribosomal proteins S4, S16, S17 and S20 assembles cooperatively in less than 5 mM Mg2+ 27. Thus, the 5′ domain proteins stabilize the folded rRNA under physiological conditions.
Previous footprinting experiments on the E. coli 16S 5′ domain showed that assembly passes through at least two equilibrium intermediates when S4, S17 and S20 are bound. In the ‘native-like’ intermediate, the core of the 5′ domain RNA including helix 15 has the same structure as in the mature 30S subunit, but helix 3 is out of contact with helices 18 and 12. In the ‘non-native’ intermediate, helix 3 packs against helix 18, but the core around helix 6a is misfolded 27. The secondary assembly protein S16 preferentially stabilizes the native-like intermediate in 1 mM MgCl2, smoothing the pathway of assembly. This leads to a conformational switch in 3-5 mM MgCl2 that repositions helix 3 between helices 18 and 12 and stabilizes the helix 18 pseudoknot (or 530-loop). Thus, the ribosomal proteins not only stabilize rRNA tertiary interactions but can also change the path of assembly, avoiding unproductive RNA conformations.
To understand how individual proteins in the 5′ domain stabilize the rRNA tertiary structure and shape the early steps of assembly, we probed the stability of the RNA tertiary interactions in single protein complexes using hydroxyl radical footprinting. The extent of hydroxyl radical cleavage correlates with the solvent accessibility of the RNA backbone 28;29, which in turn reflects the sum of folding equilibria that expose or protect an individual ribose. In general, we expect the RNA tertiary interactions to form in less Mg2+ if the bound protein stabilizes the RNA structure, because stable RNAs typically require less Mg2+ to fold than unstable RNAs 30;31. A correlation between RNA stability and protein binding has been observed for other proteins that bind structured RNAs, such as group I ribozymes 32;33;34. Therefore, under equilibrium conditions, the concentration of Mg2+ required to protect each segment of the RNA reveals how the protein changes the folding free energy of the rRNA. It also provides information about the structures of the most stable (or highly populated) assembly intermediates.
Here, we report that proteins S4 and S17 alone stabilize RNA interactions throughout the the 5′ domain of the 16S rRNA. Thus, individual primary assembly proteins increase the global stability of the 16S 5′ domain. However, tertiary interactions distant from the bound protein form less cooperatively than those that are in direct contact with the protein. Comparison with our previous results on complexes containing all three proteins and S16 suggests that non-native intermediates are preferentially depopulated as more proteins join the complex. Such synergistic interactions between RNA binding proteins may be an important mechanism for increasing the fidelity of RNP assembly.
Results
Magnesium Dependence of S17 binding
We first sought to confirm that the 30S 5′ domain proteins interact more strongly with the folded rRNA, by investigating the Mg2+-dependence of binding under the conditions of our experiments. Geobacillus stearothermophilus S4 was previously shown to bind the 16S rRNA more tightly in 4-8 mM MgCl2 than in KCl alone, consistent with its binding to a folded form of the rRNA 21. We also found that S4 can bind a minimal rRNA substrate containing the five helix junction comprising its binding site 35. The S4 binding site is more preorganized in the minimal RNA than in the 5′ domain RNA (16S nt 21-562), and this correlates with slightly stronger affinity for the minimal RNA 35.
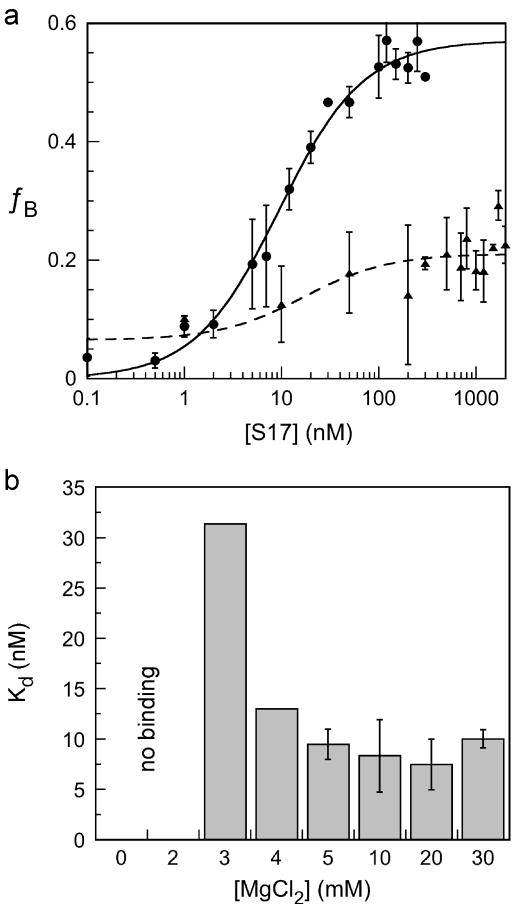
To determine whether Mg2+ also increased the affinity of S17 for the 5′ domain RNA, nitrocellulose filter binding assays were performed in 0 to 30 mM Mg2+. When the Mg2+ concentration was ≤ 2 mM, the maximum extent of binding was 20% (Figure 2a). Above 2 mM MgCl2, filter retention increased up to 55% and the apparent Kd dropped from 31 nM in 3 mM MgCl2 to 8.3 nM in 10 mM MgCl2 (Figure 2b). Non-ribosomal RNAs such as polyA or tRNA were unable to compete with the radiolabeled complex, indicating that the interaction with the 5′ domain is sequence-specific. We were unable to measure the dependence of S20 binding on Mg2+ because of poor retention on nitrocellulose filters and a small gel mobility shift.
Figure 2. Mg2+-dependent binding of protein S17.
Protein S17 was incubated with 1 nM 5′ domain at 37°C in 0-30 mM MgCl2 (see Methods). (a) Fraction RNA retained on nitrocellulose filters versus S17 concentration. Circles, 30 mM MgCl2; triangles, no MgCl2. Symbols and error bars represent the mean and standard deviation of three trials. (b) Equilbrium dissociation constants in 3-30 mM Mg2+ concentration. Less than 20% RNA was bound in 0 and 2 mM MgCl2.
We next tested different 16S fragments in competitive binding assays to determine what regions contribute to S17 binding. Dissociation constants determined by competition of 32P-labeled 5′ domain RNA with either unlabeled 16S rRNA or 5′ domain RNA were similar (Kd ≈ 6 nM; data not shown). Thus, regions of the 16S rRNA outside the 5′ domain do not contribute significantly to the stability of the S17 complex. An rRNA fragment that includes the S17 binding site in helices 7 and 11 (nt 111-313) was bound less strongly by S17, however. In direct titrations with this rRNA fragment, half the RNA formed stable S17 complexes with a Kd = 12 nM, while the other half formed unstable complexes (Kd > 400 nM; data not shown). Because the minimal RNA contains nearly all the residues that directly contact S17, these results suggested that other helices in the 5′ domain contribute to S17 binding, perhaps by correctly orienting helices 7 and 11. Thus, the S17 (and S4) complexes are influenced by a wide network of rRNA interactions that are Mg2+-dependent. Conversely, we expected protein binding to have a similarly distributed effect on folding of the 5′ domain.
Proteins stabilize the rRNA tertiary structure
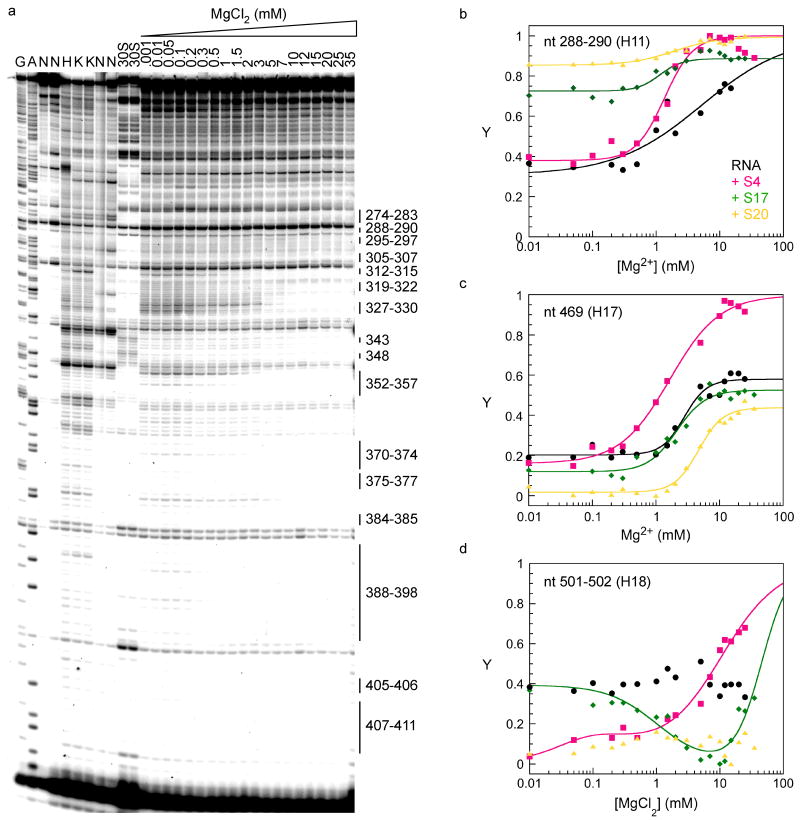
To determine whether single ribosomal proteins stabilize the structure of the 16S 5′ domain outside their immediate binding sites, complexes with S4, S17 or S20 were probed by hydroxyl radical footprinting in 330 mM KCl and 0 to 20 mM MgCl2 (Fig. 3a and Methods). The difference between these experiments and previous footprinting experiments 16;20 is that the Mg2+ concentration was varied to perturb the folding free energy of the RNA, allowing the stabilities of various complexes to be compared.
Figure 3. Fe(II)-EDTA footprinting of single protein complexes.
Complexes with the 16S 5′ domain were formed at 37°C in 0-35 mM MgCl2 before cleavage with hydroxyl radical (see Methods). (a) Representative data with S17. Lanes N, no treatment; H, RNA in 80 mM K-Hepes; K, lane H plus 330 mM KCl; A, G, sequence ladders; 30S, native 30S ribosomes. Extension with primer annealing at nt 433 is shown. Protections explained by predicted RNA-RNA contacts in the 30S subunit are shown. (b-d) Fractional saturation of backbone protection (Y) versus Mg2+ concentration, relative to RNA in Hepes (Y = 0) and native 30S ribosomes (Y = 1). The data were fit to two or four-state models (Methods). Black, RNA only; pink, +S4, green, +S17; yellow, +S20. (b) nt 288-290 (helix 11); (c) nt 481-483 (helix 17); (d) nt 501-502 (helix 18).
The footprinting results on individual protein complexes were compared to previous footprinting experiments on the naked 5′ domain RNA under the same conditions 27. The extent of protection was normalized to control reactions on the RNA in 80 mM Hepes (unfolded) and native 30S subunits (completely folded). Many residues throughout the 5′ domain were protected in less Mg2+ when bound by a ribosomal protein than in the naked RNA (Fig. 3b-d). Thus, each primary binding protein stabilizes the folded structure of the 5′ domain to a large extent.
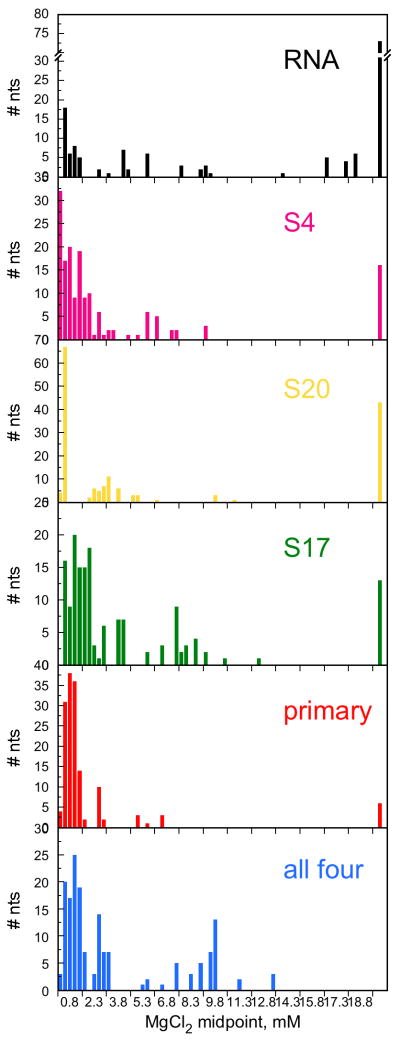
The domain-wide effects of each protein on the stability of the tertiary interactions can be appreciated from histograms for Mg2+-induced protection of the RNA backbone (Fig. 4). In the absence of protein, most folding midpoints are above 13 mM MgCl2 (Fig. 4a). (The 5′ domain RNA is less well folded in 330 mM KCl than in 120 mM NH4Cl, which we used in a previous study 26.) In the presence of S4, however, only 10% of nucleotides require this much Mg2+ to be protected (Fig. 4b). Overall, S4 had the most dramatic effect on the stability of the folded RNA, with S17 and 20 being somewhat less stabilizing (Fig. 4c,d). S4, S17 and S20 together stabilized the RNA more than each protein alone (Fig. 4e), while the addition of S16 to the complex allowed nearly all the RNA interactions to form in low Mg2+ (Fig. 4f).
Figure 4. Protein-dependent perturbation of rRNA tertiary interactions.
The midpoints for Mg2+-dependent saturation of individual contacts along the RNA backbone were sorted into 0.3 mM bins. For residues with two transitions, the highest midpoint was used. Individual parameters are listed in Table S1. Black, RNA only; pink, S4; green, S17; yellow, S20; purple, S4+S17+S20; blue, S4+S16+S17+S20. Data for three and four protein complexes are described elsewhere 27.
Global stabilization by protein S4
To determine which interactions are stabilized by each primary binding protein, the midpoints for protection of the RNA backbone in Mg2+ were projected onto the 2D and 3D structures of the 16S 5′ domain within the 30S ribosome (Figure 5). In the absence of protein, a stable core of protein-independent RNA interactions were observed adjacent to the upper five helix junction, the central junction (helix 11a) and along the interface between helices 6 and 7 (Figure 5a; 27). The functionally important pseudoknot in helix 18 was partly structured in the naked rRNA, but the pattern of backbone protection was inconsistent with the structure of the pseudoknot in the 30S ribosomes 8;9.
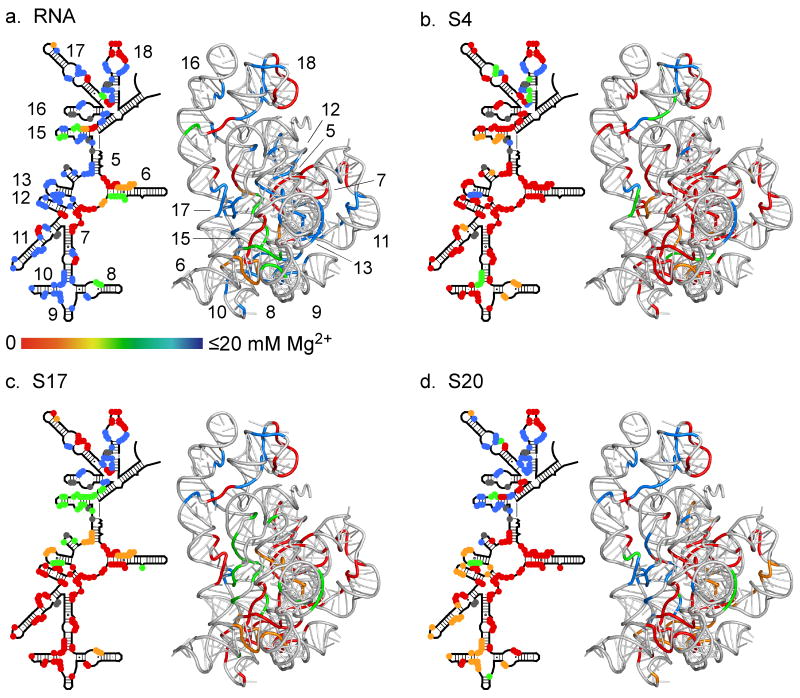
Figure 5. Stabilization of rRNA tertiary structure by ribosomal proteins.
Protected regions are colored according to the midpoint (Cm) of the folding transition: red, 0-2.3 mM; orange, 2.3-4.9 mM; green, 4.9-13.4 mM; blue, >13 mM. Residues having more than one transition are clustered according to the one at highest Mg2+. 2D schematics and 3D ribbons as in Figure 1. See Supplemental Figure S1 and Table S1 for further data. (a) 5′ domain RNA only; (b) RNA plus S4; (c) RNA plus S17; (d) RNA plus S20. Data in (a) are from 27.
As expected, S4 stabilized tertiary interactions around the five helices that comprise its binding site 19 (Figure 5b). For example, tertiary interactions between helices 16 and 18 (nt 427-429) were 80% protected in the presence of S4, but unprotected with S17 or S20 (Figure S1). However, the Mg2+ titrations showed that tertiary interactions in the lower half of the 5′ domain are also stabilized indirectly by binding of S4, accounting for its importance in 30S assembly 18. Binding of S4 allowed the formation of RNA tertiary contacts in helices 10, 11, 12, 15 and 17, which are not stable in the naked RNA (e.g., Figure 3c,d and Figure S1).
In addition, S4 binding enhanced hydroxyl radical cleavage of nt 78-80 in helix 6 relative to the naked RNA (data not shown). Helix 6 forms a spur which protrudes from the base of 30S subunit 8;9. Cleavage of helix 6 is enhanced even more when other proteins are added to the complex, suggesting that the spur becomes increasingly exposed to solvent as the helices in the 5′ domain become more structured. This observation supports the idea, discussed below, that the ensemble of RNA conformations becomes smaller as more proteins join the complex.
Global stabilization by protein S17
Protein S17 stabilized RNA tertiary interactions around its binding site in helices 7 and 11 (Fig. 5c), including the central junctions between helices 6, 6a, 7, 11 and 12, and a kink turn in helix 11 that allows the tip of helix 11 to dock against helix 7 (Figure 3b and Figure S1). S17 indirectly stabilized the lower junction between helices 7, 8, 9 and 10 that overlaps the binding site for S20. Thus, binding of these two proteins may be weakly cooperative. S17 also perturbed RNA interactions in helices 15, 17, and 18, which are distant from its binding site (Fig. 5c). This was unexpected, because, unlike S4, S17 has not been implicated in the nucleation of 30S assembly.
Regional stabilization by protein S20
In comparison with S4 and S17, specific effects of protein S20 were more localized to nucleotides surrounding its binding site between helices 6, 7, 8, 9, 11, and 13 (Figure 5d). Although many nucleotides were partially protected in low Mg2+, 40% of protected nucleotides have a Mg2+ midpoint for complete saturation of backbone contacts that is higher than 13 mM, compared with 10% for S4 (Fig. 4d). S20 failed to stabilize tertiary interactions in the upper half of the domain where S4 binds (helices 15-18; Fig. 5d and S1). Thus, S20 has a more modest influence on the overall structure of the 5′ domain than S4 and S17. The footprinting results are consistent with previous base probing studies and cleavage by tethered Fe(II)-EDTA complexes, which show that S20 interacts with helices at the bottom of the 5′ domain 36.
Single proteins stabilize partially folded intermediates
For each single protein complex, the pattern of hydroxyl radical protection in 20-30 mM Mg2+ was largely consistent with the conformation of the native 5′ domain RNP expected from crystal structures of 30S subunits 8;9, although the pseudoknot in helix 18 and the tip of helix 5 (nt 364) did not fold completely. Many residues, however, were semi-protected from hydroxyl radical cleavage in intermediate concentrations of Mg2+ (Figure 3).
Comparison of the footprinting results on single protein complexes and the naked RNA showed that the primary binding proteins perturb the structure of the 5′ domain RNA in very low Mg2+. Thus, the proteins must interact with the 5′ domain even though they do not form stable complexes under these conditions, presumably because the RNA is not stably folded. Each protein greatly increased protection of helices 6, 7, and 8 in 0.01 mM Mg2+, relative to the RNA alone (Figure S1). However, these interactions did not necessarily progress to the fully native domain. For example, when S4 was present, residues in helices 6 and 7 were 40-60% protected in 0.01 mM MgCl2 relative to 30S controls. However, these backbone protections did not saturate, even when the MgCl2 concentration was raised to 30 mM (Table S1). These results suggested that S4 stabilizes RNA conformations that never achieve the native structure. By contrast, when S17 was bound, the same core helices became fully protected in 20 mM MgCl2 (Table S1 and Figure S1). Thus, S17 may stabilize the native conformation of the core helices more selectively than S4.
Multi-stage folding of the 5′ domain RNA
A comparison of titration curves in different regions of the 5′ domain provided further evidence that each protein stabilizes an ensemble of intermediates which can include both native and non-native RNA structures. Not only did different regions of the rRNA fold in different Mg2+ concentrations, but many nucleotides folded in two or three stages (Figure 3b-d). For example, protection of nt 481-483 in the presence of S4 goes from 30% to 50% in 0.1 mM MgCl2 and then up to 100% between 2 and 20 mM MgCl2 (Figure 3c). Residues in helices 15 and 17, which extend down from the upper junction to pack against the core of the 5′ domain, exhibited multiphasic folding in all three of the primary binding proteins (Figure S1). The lower junction between helices 7, 8, 9 and 10 also folded in two distinct stages, particularly when S4 was bound. S17 induced two-stage folding of helices 5, 6 6a, 13 and 14, which all surround the central junction.
Interestingly, S4 and S17 had opposite effects on the kink in helix 17 (nt 481-485) that packs against a C-loop in helix 15 in the mature 30S subunit 8;9. When S4 was present, nt 481-483 was protected in several phases while nt 484-485 remained unprotected (Figure S2). When S17 was present, nt 484-485 were 30% protected in low Mg2+ and 100% in 5 mM MgCl2, while nt 481-483 were only lightly protected in 20 mM Mg2+. These results illustrate how each protein selectively stabilizes specific conformations of the rRNA.
Transient unfolding of the 5′ domain RNA
In addition to the multi-stage increase in protection described above, we observed transient exposure or unfolding of certain residues during the Mg2+ titration. The most dramatic example is the partial protection of helix 18 in no Mg, exposure in 2-10 mM MgCl2, and reprotection in ≥ 20 mM MgCl2, when S17 is bound to the RNA (Figure 3d, Figure S1, S3). These results suggest that S17 stabilizes an intermediate that is remodeled during folding of the 5′ domain. As discussed below, these results are consistent with a previously proposed conformational switch at helix 3 during assembly of the 16S 5′ domain 27.
We also observed slight unfolding and refolding of the lower helix junction in the presence of S4, S17 and S20. Transient exposure of C217 in 0.5 mM MgCl2 was clearly supported by the footprinting data with S20, and to a lesser extent, with S4 and S17 (Table S1, Figure S1). C217 lies in helix 10 and contacts the tip of helix 17. Other residues that may become transiently exposed are nt 196-200 on the opposite side of helix 10 which contacts helix 6, nts 66-72 (helix 6), nt 151-153 (helix 8), nt 327 (helix 13) and nt 379-380 (helix 15), although the data in low Mg2+ for these residues are noisy relative to the small degree of exposure. All of these residues cluster around helix 6 at the “bottom” of the 5′ domain. Thus, S20 likely stabilizes more than one conformation of the 5′ domain RNA. The idea that the core and the lower junction can adopt a conformation that is different from the one in the 30S ribosome is also supported by the protection of nt 94 in helix 6, which is accessible to solvent in the 30S ribosome.
Discussion
Stabilization of RNA structure by RNA-binding proteins
By using hydroxyl radical footprinting to measure the stability of individual tertiary contacts, we find that ribosomal proteins S4 or S17 stabilize the rRNA structure far beyond their previously defined binding sites 19, while the effects of S20 are more local. Indirect stabilization of entire RNA domains by a single protein has been observed in several large ribozymes. For example, the yeast CBP2 protein is required for splicing of the mitochondrial bI5 group I intron in physiological conditions 37. The naked bI5 intron requires 40-50 mM MgCl2 to fold and be active 38;39. Similarly, other group I splicing factors 34;40 and the protein component of bacterial RNase P 41;42 enhance the catalytic activity of their cognate ribozymes by stabilizing the active conformation of the RNA or by interacting with substrates.
Like the ribozyme examples above, the 5′ domain of the 16S rRNA is rich in RNA interactions, and capable of folding independently in high salt and Mg2+ 26. Thus, the tertiary structure of the 16S 5′ domain is itself sufficiently cooperative that a single protein can stabilize the entire domain. The intrinsic stability of the 16S 5′ domain also helps explain why three of the 5′ domain proteins bind the 16S rRNA without requiring other 30S proteins. As discussed below, however, S4, S17 and S20 each play a distinct role in stabilizing specific tertiary structures within the 5′ domain. Consequently, the three proteins together have the capacity to increase the specificity of the assembly pathway at physiological Mg2+ concentrations by constraining the structure of the rRNA.
Primary binding proteins stabilize different RNA intermediates
The changes in RNA backbone accessibility which are revealed by the footprinting data show that each protein stabilizes the tertiary structure of the 16S rRNA in different ways. Protein S4, which binds the upper five helix junction, strongly stabilizes the tertiary interactions in the upper half of the domain, and by extension, interactions between helices 15 and 17 with helices 6, 6a and 7 in the lower core of the domain (Figure 6a,b and Figure S2). Conversely, S4 stabilizes interactions with helices 5, 12 and 13 only partially.
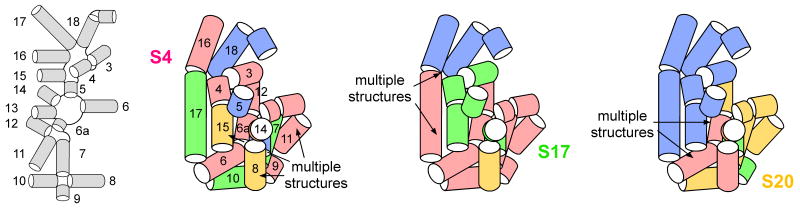
Figure 6. Primary binding proteins stabilize different rRNA intermediates.
In low Mg2+, the 5′ domain forms an ensemble of partly folded states (IC) with a subset of stable tertiary interactions. Individual proteins stabilize partly folded RNAs that may contain non-native interactions. Helices are shown as cylinders (red, most stable; gold, medium stable; green, less stable; blue, disordered). (a) Schematic showing location of helices in the 5′ domain secondary structure; (b) S4; (c) S17; (d) S20.
In contrast, protein S17 strongly stabilizes the tertiary interactions around its binding site in helices 7 and 11 and around the central junction, so that these interactions form at very low (0.01 mM) Mg2+ in S17 complexes (Figure 6c). Some interactions in the upper helix junction (such as with helix 16) are not stabilized at all by S17 (Figure S2e), while those in the lower junction are moderately stabilized. The footprinting data also indicate that S4 and S17 stabilize different conformations of helix 17, where it bends to make a key tertiary contact with helix 15.
S20 has the strongest effect on helices 8 and 14 that surround its binding site, so that tertiary interactions between these helices appear at much lower Mg2+ than in S4 and S17 complexes (Figure 6d). S20 also stabilizes tertiary interactions in helices 6, 7, 11 and 12, but not tertiary interactions in the upper half of the domain.
Ribosomal proteins stabilize more than one RNA conformation
The Mg2+-dependence of the footprinting results shows that each ribosomal protein forms one or more intermediate complexes with the rRNA. Not only do different residues fold at different Mg2+ concentrations, but many residues fold in two or three stages or become transiently exposed to cleavage. Multi-stage folding can be explained by a competition between intermediates with different structures, whose populations change as the Mg2+ concentration is raised 27. The observed “footprint” represents the weighted average solvent accessibility of the populated intermediates 43. A plateau in the level of backbone protection occurs when two intermediates have similar stabilities at a particular Mg2+ concentration, but different susceptibilities to cleavage by hydroxyl radical.
Protein binding lowers (or raises) the free energies of some RNA conformations but not others, driving the RNP population into newly stabilized states. Thus, in this model, the proteins direct the path of assembly by altering the free energy landscape for RNA folding 27;44;45. This type of statistical mechanical model has been used to interpret the effects of ligands on the rates of hydrogen exchange in proteins 46. As we have previously noted 27, multi-stage protection and transient exposure of specific residues all point toward the presence of non-native structures that are remodeled during assembly of the 16S 5′ domain RNP.
The presence of equilibrium assembly intermediates agrees with data on the kinetics of assembly. Time-resolved footprinting of the 16S 5′ domain showed that the less stable tertiary contacts in helices 15 and 17 form slowly in the absence of proteins, indicating that about half the RNA population is kinetically trapped in metastable intermediates 26. The kinetics of 30S reconstitution also showed that assembly occurs in stages with individual complexes taking different routes to the final structure 44;47. While primary assembly proteins such as S4 contact their binding sites in 100 ms or less, other tertiary interactions in the 5′ domain form in 1-100 s47.
Specificity of assembly
An important observation is that the proteins perturb the 5′ domain structure even at very low Mg2+ concentrations, in which the RNA is poorly folded. For example, S17 changes the footprint of the 5′ domain in less than 0.1 mM MgCl2, yet a stable complex needs 4 mM MgCl2 (Figure 2). S4 enables indirect protection of core helices in less Mg2+ than protection of helices 16 and 18 within its own binding site. Thus, the ribosomal proteins are not merely capturing the natively folded rRNA, but interacting with early folding intermediates that are structurally disordered or dynamic.
In agreement with this observation, CBP2 protein has been shown to interact with a near-native collapsed state of bI5 RNA and drive it into the native state 32;48, and single molecule FRET experiments showed that non-specific binding increases the structural dynamics of the bI5 RNA 49. Single molecule FRET experiments on other large ribozymes such as RNase P and a group II intron have uncovered multiple folding pathways for each RNA 50;51;52;53, consistent with fluctuations between different I-states. Time-resolved footprinting of the 16S rRNA suggests a similarly diverse set of conformational states are available to the 5′domain RNA during assembly of the 30S ribosome 47.
Synergy between primary assembly proteins
Although each 5′ domain protein stabilizes rRNA tertiary interactions far from its binding site, they each bind a different helix junction, and favor different folding intermediates. Consequently, we expect fewer RNA conformations to be allowed when all three primary assembly proteins are bound. This is exactly what we find when S4, S17 and S20 are added to the 5′ domain RNA before hydroxyl radical footprinting 27. When three proteins are present, the pattern of backbone cleavage and protection becomes more coherent in low Mg2+, leading to pre-organization of tertiary interactions within the S16 binding site 27.
Thus, the combined interactions of the primary binding proteins can make assembly more specific by driving the RNA population into a smaller number of structures that are competent for subsequent steps of 30S assembly. A similar synergistic effect may exist between S8 and S15 in the central domain of the 16S rRNA, as tethered hydroxyl radical footprinting experiments showed that the structural environment of S15 changes when S8 is bound, even though both proteins bind independently to separate helical junctions 54.
With three proteins bound, we still observe a competition between native-like and non-native configurations of the core helices, that is only resolved in favor of the native-like intermediate when S16 is added to the complex 27. In this context, ‘native-like’ only means that the conformation is similar to that in the mature subunit; we do not yet know whether such intermediates form naturally in the cell. Formation of the native-like intermediate correlates with displacement of helix 3 from its normal position between helix 18 and the tip of helix 12. Raising the Mg2+ a bit further flips helix 3 back into place 27, stabilizing the helix 18 pseudoknot which forms part of the 30S decoding site.
This conformational switch at helix 3 requires coordination between residues in the core (helices 6, 8, 10, 11, 13), the central junction (helices 5, 6a, 12), and the upper junction (helices 3, 17 and 18). Although these structural changes are observed in single protein complexes, they are weakly coupled. Thus, S4, S17 and S20 in combination may stiffen the switch by discriminating against unproductive rRNA conformations.
Conclusions
In summary, these studies reveal that 5′ domain assembly involves a diverse landscape of conformational states that are bound and stabilized by single proteins. Understanding how the dynamics of competition between alternative conformations produce more stable and presumably productive configurations leading toward the final steps of 5′ domain assembly is of particular interest. These last steps include binding of secondary protein S16 to the 5′ domain and native tertiary interactions between helices 3, 12 and 18 27. Because these helices interact with S12, and S12 binding depends on the presence of S16, these conformational steps may form a checkpoint for incorporation of S12 into 30S ribosomes and further interactions between the 5′, central and 3′ minor domains.
In the cell, ribosome assembly is coupled to transcription, allowing the 5′ domain to fold before the 3′ end of the 16S rRNA is synthesized. Thus, RNA and protein interactions within the 5′ domain may correpond reasonably well with assembly intermediates formed in vivo. A caveat is that metastable interactions with the 16S leader, which interacts with the 5′ domain, may change the balance between alternative conformations of the 5′ domain or introduce new conformations 55;56. Nonetheless, our results show that ribosomal proteins stabilize the structure of the rRNA far beyond their immediate binding sites and in specific ways, contributing to the cooperativity of subunit assembly.
Methods
Purification of recombinant ribosomal proteins
E. coli ribosomal proteins S4, S17 and S20 were over-expressed, purified by ion exchange chromatography and assayed for binding activity as previously described 27 57.
Purification of 5′ domain
The 542 nt 5′ domain RNA was transcribed in vitro with T7 RNA polymerase from pRNA1 26 using standard methods 58 and purified by denaturing 4% (w/v) polyacrylamide gel electrophoresis. The RNA concentration was determined by UV absorption at 260 nm and ε260 = 5.4 × 106 M-1cm-1.
Nitrocellulose filter binding
S17 binding reactions were carried out in Fisher low-retention tubes. Uniformly 32P-labeled 16S rRNA 5′ domain was diluted to approximately 1 nM (10,000-25,000 cpm per 50 μL reaction) in reconstitution buffer (80 mm K-Hepes 7.6, 330 mM KCl, 20 mM MgCl2, 0.01% Nikkol detergent, and 6 mM β-mercaptoethanol) except where noted. Serial dilutions of the protein in reconstitution buffer containing 0, 2, 3, 4, 5, 10, 20 or 30 mM MgCl2 were kept on ice until needed. The RNA and protein were coincubated at 37°C for one hour before filtration on 25 mm nitrocellulose filters (Schleicher and Schuell BA85) as previously described 21. The extent of binding reached saturation in 5 min (data not shown). Dried filters were counted, and the data were fit to a Langmuir binding isotherm. For competitive binding assays, 32P-labeled 5′ domain and S17 were incubated 20-30 minutes at 37°C as described above, then challenged with increasing concentrations of competitor RNA for 30 minutes at 37°C. The data were fit as previously described 59.
Hydroxyl radical footprinting
The 5′ domain RNA was folded 30–40 min at 37°C in reconstitution buffer containing 0-20 mM MgCl2 before treatment with Fe(II)-EDTA as previously described 26. The 5′ domain RNA was previously shown to fold in less than 5 minutes 26. Protein-RNA complexes were prepared by incubating 12 pmol RNA in reconstitution buffer containing the desired MgCl2 concentration, 15–20 minutes at 37°C, before addition of 5 molar equivalents of S17 or S20 or 4 molar equivalents of S4 in a total volume of 56 μl. The RNA was coincubated with the proteins for an additional 45 minutes at 37 °C. The optimum protein:RNA ratios and incubation time were determined as described elsewhere (P.R. and S.W., manuscript submitted).
Following assembly of 5′ domain RNP complexes, 14 μl containing 3 pmol 5′ domain RNA was removed from each reaction and treated with Fenton reaction reagents 1 min on ice as previously described 29. Control reactions were carried out in parallel with each titration on native E. coli 30S ribosomal subunits and on 5′domain RNA in 80 mM K-Hepes, 80 mM K-Hepes plus 330 mM KCl, or reconstitution buffer. Samples were analyzed by primer extension and AMV reverse transcriptase as described previously 26.
Data analysis
Protected regions of the RNA backbone were compared with the solvent-accessible surface area for each C4′ atom in the 5′ domain of the 16S rRNA using coordinates from the structure of E. coli 30S ribosome 60 and the program Calc-surf 61. The extent of cleavage at each position in the 5′ domain RNA was quantified with a Molecular Dynamics Phosphorimager and normalized to a reference band whose intensity did not change over the Mg2+-titration. The fractional saturation (Ȳ) was determined by normalizing to the maximum extent of cleavage (Ȳ = 0) and the cleavage of 30S complexes (Ȳ = 1). The extent of cleavage was frequently but not always greatest in 80 mM K-Hepes.
Assuming that the extent of hydroxyl radical cleavage reflects the equilibrium between an “open” and “closed” state at each nucleotide, the fractional saturation (Ȳ) of each backbone protection vs. Mg2+ concentration (C) was fit to an isotherm for a cooperative two-state equilibrium, Ȳ = Ȳ0 + (C/Cm)n/[1+(C/Cm)n], in which Cm is the midpoint, n is the Hill coefficient and Y0 is the extent of protection in 330 KCl and no MgCl2. Multiphasic transitions were fit to a four-state model in which the statistical weight of each term was taken from the equilibrium constant for the open and closed state, Ki = (C/Cm,i)n,i 31;62,
| (1) |
The initial state (Icore) and one intermediate are assumed to be completely exposed, while the second intermediate and the final state (N) are assumed to be fully protected. In some cases, the data could be described equally well by two sequential transitions,
| (2) |
in which A1 and A2 are the magnitude of change in protection with each step.
Histograms
Individual transitions were classified into four groups by sorting midpoints into 0.3 mM bins. Weakly protected positions for which a folding transition with respect to [MgCl2] could not be determined were arbitrarily assigned a Cm = 20 mM and n = 1. For biphasic transitions, the midpoint of the second transition was assumed to represent formation of the native complex and was used in the cluster analysis. 3-D ribbons were made in Pymol (Delano Scientific, http://pymol.org/educational.html) using the crystallographic coordinates of the E. coli ribosome (2avy) 60.
Supplementary Material
Acknowledgments
The authors thank G. Culver, D. Draper, R. Moss and T. Adilakshmi for reagents and plasmids and A. Cukras, J. Brunelle and R. Green for their help and advice. This work was supported by a grant from the NIH (GM60819).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 2.Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb Symp Quant Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 4.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR. Biophysical studies of bacterial ribosome assembly. Curr Opin Struct Biol. 2008;18:299–304. doi: 10.1016/j.sbi.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culver GM. Assembly of the 30S ribosomal subunit. Biopolymers. 2003;68:234–249. doi: 10.1002/bip.10221. [DOI] [PubMed] [Google Scholar]
- 7.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 8.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 9.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 10.Stern S, Changchien LM, Craven GR, Noller HF. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988;200:291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- 11.Jaishree TN, Ramakrishnan V, White SW. Solution structure of prokaryotic ribosomal protein S17 by high- resolution NMR spectroscopy. Biochemistry. 1996;35:2845–2853. doi: 10.1021/bi951062i. [DOI] [PubMed] [Google Scholar]
- 12.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 13.Herzog A, Yaguchi M, Cabezon T, Corchuelo MC, Petre J, Bollen A. A missense mutation in the gene coding for ribosomal protein S17 (rpsQ) leading to ribosomal assembly defectivity in Escherichia coli. Mol Gen Genet. 1979;171:15–22. doi: 10.1007/BF00274010. [DOI] [PubMed] [Google Scholar]
- 14.Culver GM, Noller HF. Directed hydroxyl radical probing of 16S ribosomal RNA in ribosomes containing Fe(II) tethered to ribosomal protein S20. RNA. 1998;4:1471–1480. doi: 10.1017/s1355838298981201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann RA, Muto A, Fellner P, Ehresmann C, Branlant C. Location of ribosomal protein binding sites on 16S ribosomal RNA. Proc Natl Acad Sci USA. 1972;69:1282–1286. doi: 10.1073/pnas.69.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 17.Vartikar JV, Draper DE. S4-16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J Mol Biol. 1989;209:221–234. doi: 10.1016/0022-2836(89)90274-x. [DOI] [PubMed] [Google Scholar]
- 18.Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–7055. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- 19.Powers T, Noller HF. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 20.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J Biol Chem. 1995;270:1238–1242. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 21.Gerstner RB, Pak Y, Draper DE. Recognition of 16S rRNA by ribosomal protein S4 from Bacillus stearothermophilus. Biochemistry. 2001;40:7165–7173. doi: 10.1021/bi010026i. [DOI] [PubMed] [Google Scholar]
- 22.Davies C, Gerstner RB, Draper DE, Ramakrishnan V, White SW. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998;17:4545–4558. doi: 10.1093/emboj/17.16.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutca LM, Jagannathan I, Grondek JF, Culver GM. Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16 S rRNA. J Mol Biol. 2007;368:853–869. doi: 10.1016/j.jmb.2007.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–180. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 25.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 26.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J Mol Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Ramaswamy P, Woodson SA. S16 throws a conformational switch during assembly of 30S 5′ domain. Nat Struct Mol Biol. 2009;16:438–445. doi: 10.1038/nsmb.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- 29.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 30.Rook MS, Treiber DK, Williamson JR. An optimal Mg(2+) concentration for kinetic folding of the tetrahymena ribozyme. Proc Natl Acad Sci USA. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan J, Thirumalai D, Woodson SA. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc Natl Acad Sci USA. 1999;96:6149–6154. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weeks KM, Cech TR. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell. 1995;82:221–230. doi: 10.1016/0092-8674(95)90309-7. [DOI] [PubMed] [Google Scholar]
- 33.Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-trna synthetase protein induces tertiary folding of the group i intron catalytic core. Journal of Molecular Biology. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 34.Caprara MG, Chatterjee P, Solem A, Brady-Passerini KL, Kaspar BJ. An allosteric-feedback mechanism for protein-assisted group I intron splicing. RNA. 2007;13:211–222. doi: 10.1261/rna.307907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellur DL, Woodson SA. A minimized rRNA-binding site for ribosomal protein S4 and its implications for 30S assembly. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutca LM, Culver GM. Assembly of the 5′ and 3′ minor domains of 16S ribosomal RNA as monitored by tethered probing from ribosomal protein S20. J Mol Biol. 2008;376:92–108. doi: 10.1016/j.jmb.2007.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gampel A, Nishikimi M, Tzagoloff A. CBP2 protein promotes in vitro excision of a yeast mitochondrial group I intron. Mol Cell Biol. 1989;9:5424–5433. doi: 10.1128/mcb.9.12.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gampel A, Cech TR. Binding of the CBP2 protein to a yeast mitochondrial group I intron requires the catalytic core of the RNA. Genes Dev. 1991;5:1870–1880. doi: 10.1101/gad.5.10.1870. [DOI] [PubMed] [Google Scholar]
- 39.Weeks KM, Cech TR. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell. 1995;82:221–230. doi: 10.1016/0092-8674(95)90309-7. [DOI] [PubMed] [Google Scholar]
- 40.Mohr G, Zhang A, Gianelos JA, Belfort M, Lambowitz AM. The neurospora CYT-18 protein suppresses defects in the phage T4 td intron by stabilizing the catalytically active structure of the intron core. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- 41.Gardiner KJ, Marsh TL, Pace NR. Ion dependence of the Bacillus subtilis RNase P reaction. J Biol Chem. 1985;260:5415–5419. [PubMed] [Google Scholar]
- 42.Loria A, Niranjanakumari S, Fierke CA, Pan T. Recognition of a pre-tRNA substrate by the Bacillus subtilis RNase P holoenzyme. Biochemistry. 1998;37:15466–15473. doi: 10.1021/bi9816507. [DOI] [PubMed] [Google Scholar]
- 43.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical “footprinting”. Curr Opin Struct Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 44.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramaswamy P, Woodson SA. S16 throws a conformational switch during 30S 5′ domain assembly. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luque I, Leavitt SA, Freire E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu Rev Biophys Biomol Struct. 2002;31:235–256. doi: 10.1146/annurev.biophys.31.082901.134215. [DOI] [PubMed] [Google Scholar]
- 47.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchmueller KL, Weeks KM. Near native structure in an RNA collapsed state. Biochemistry. 2003;42:13869–13878. doi: 10.1021/bi035476k. [DOI] [PubMed] [Google Scholar]
- 49.Bokinsky G, Nivon LG, Liu S, Chai G, Hong M, Weeks KM, Zhuang X. Two distinct binding modes of a protein cofactor with its target RNA. J Mol Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 51.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci USA. 2003 doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc Natl Acad Sci USA. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci USA. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jagannathan I, Culver GM. Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J Mol Biol. 2003;330:373–383. doi: 10.1016/s0022-2836(03)00586-2. [DOI] [PubMed] [Google Scholar]
- 55.Pardon B, Wagner R. The Escherichia coli ribosomal RNA leader nut region interacts specifically with mature 16S RNA. Nucleic Acids Res. 1995;23:932–941. doi: 10.1093/nar/23.6.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besancon W, Wagner R. Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res. 1999;27:4353–4362. doi: 10.1093/nar/27.22.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Culver GM, Noller HF. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–843. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weeks KM, Crothers DM. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- 60.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 61.Gerstein M. A resolution-sensitive procedure for comparing protein surfaces and its application to the comparison of antigen-combining sites. Acta Cryst A. 1992;48:271–276. [Google Scholar]
- 62.Fang X, Pan T, Sosnick TR. A thermodynamic framework and cooperativity in the tertiary folding of a Mg(2+)-dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.