Abstract
Multidrug efflux pumps are major contributors to intrinsic antibiotic resistance in Gram-negative pathogens. The basic structure of these pumps comprises an inner membrane transporter, a periplasmic membrane fusion protein and an outer membrane channel. However, the architecture and composition of multidrug efflux complexes vary significantly because of the topological and functional diversity of the inner membrane transporters. This article presents the current views on architecture and assembly of multicomponent drug efflux transporters from Gram-negative bacteria.
Keywords: antibiotic resistance, membrane fusion protein, multidrug efflux transporter, TolC
Drug resistance presents an ever-increasing threat to public health and encompasses all major microbial pathogens and antimicrobial drugs [1]. Some pathogens have acquired resistance to multiple antibiotics and cause infections that are effectively untreatable. Among pathogenic Gram-negative Acinetobacter and Pseudomonas, species have emerged that are resistant to all good antibiotics (i.e., with low side effects) and many Enterobacteriaceae are resistant to all antibiotics except carbapenems [2]. Thus, there is a strong need for new antibiotics, particularly those directed against multidrug-resistant Gram-negative bacteria.
For a long time, the outer membrane (OM) was believed to be the major reason why Gram-negative pathogens are recalcitrant to antibiotic treatment [3]. The seminal finding that Escherichia coli cells contain multidrug efflux transporters dramatically changed our view of how these bacteria resist antibiotics [4,5]. Current models postulate that intrinsic multidrug resistance of Gram-negative bacteria is the result of synergy between reduced uptake of drugs across the OM and active drug efflux from the inner membrane (IM) [6–8]. This synergy is possible because multidrug efflux transporters of Gram-negative bacteria function together with proteins belonging to the membrane fusion protein (MFP) family [9]. MFPs are located in the periplasm and act on both membranes to enable drug efflux across the whole cell envelope directly into the medium. On the IM, MFPs associate with drug efflux transporters and stimulate their activity [10–12]. On the OM, they recruit OM channels and possibly enable expulsion of drugs into the medium [13,14]. The three components form large multiprotein assemblies that traverse both the IMs and OMs of Gram-negative bacteria. Working together as a well-coordinated team, they achieve the direct extrusion of substrates from the cytoplasm and/or the periplasm and into the medium.
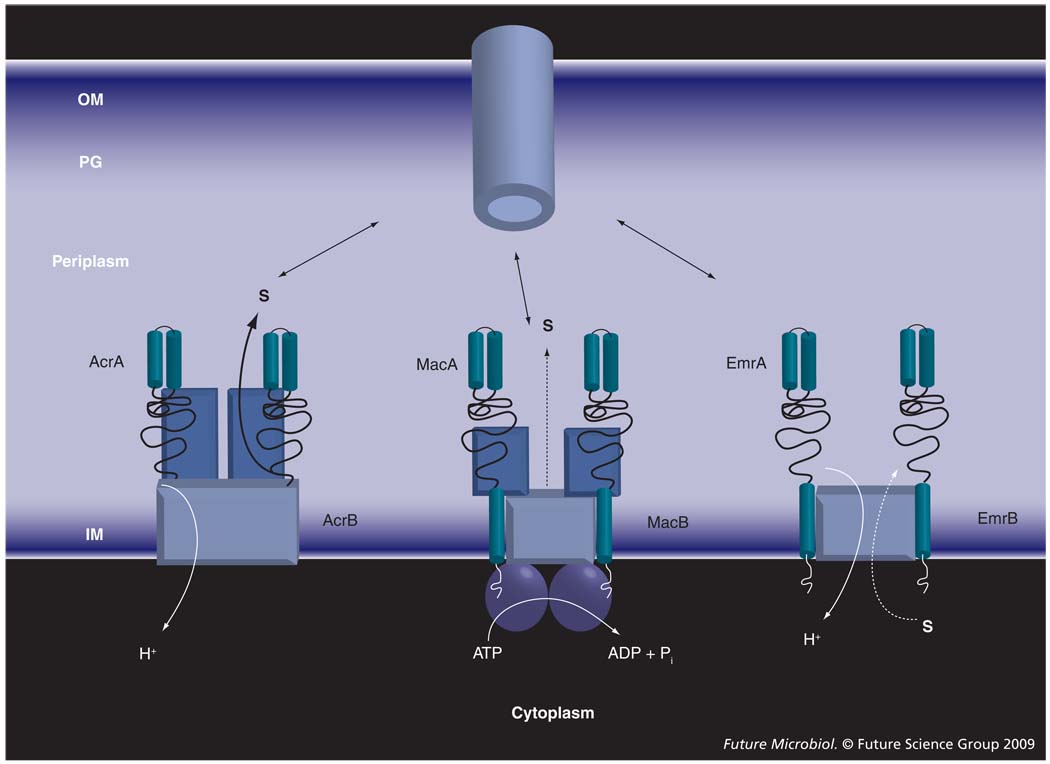
Drug transporters that associate with MFPs and OM channels can belong to any of the three major superfamilies of proteins: resistance–nodulation–cell division (RND), major facilitator superfamily (MFS) and ATP-binding cassette (ABC) [15–18]. These transporters are structurally and mechanistically very diverse (Figure 1). ABC transporters are driven by ATP hydrolysis, whereas drug efflux by RND and MFS pumps is coupled to transport of protons. MFS transporters are thought to function as monomers, whereas ABC and RND transporters are dimers and trimers, respectively. All three types of transporter are broadly represented in Gram-negative bacteria. However, it is the activity of RND-type efflux pumps that is largely responsible for intrinsic resistance in Gram-negative bacteria [19–21].
Figure 1. Membrane fusion protein-dependent transporters are structurally and functionally diverse.
The ATP-binding cassette-type MacB is driven by ATP hydrolysis and functions as a dimer. AcrB and EmrB are secondary transporters driven by proton-motive force. However, EmrB, as a typical member of the major facilitator family, is expected to function as a monomer and transport substrates from the cytoplasm to the periplasm. By contrast, AcrB is a functionally rotating trimer that binds its substrates in the periplasm and transfers them across the OM.
IM: Inner membrane; OM: Outer membrane; PG: Peptidoglycan; S: Substrate.
In this, by no means comprehensive, review we will examine Gram-negative multidrug efflux complexes, and the features of their structure and transport mechanism that make them particularly efficient in blocking the action of antibiotics. We will focus on the recent insights into the structure and function of accessory proteins, the architecture of trans-envelope complexes and how accessory proteins are integrated into drug efflux processes.
RND-type multidrug efflux complexes
AcrAB–TolC from E. coli is the best-characterized multidrug efflux complex capable of trans-envelope transport [22]. Here, AcrA is the periplasmic MFP, AcrB is the RND drug efflux transporter and TolC is the OM channel. AcrAB–TolC and its close homologs in other Gram-negative bacteria, such as MexAB–OprM from P. aeruginosa or AdeABC from Acinetobacter species, are constitutively expressed and protect cells from hundreds of structurally unrelated compounds. Among their substrates are antibiotics, detergents, organic solvents and steroid hormones. Given their low similarity to mammalian RND proteins and high impact on bacterial pathogenesis and infection, the bacterial RND pumps are regarded as very ‘attractive’ targets for drug development [6]. It is now clear that inhibitors of RND pumps potentiate the activity of clinically important antibiotics and could become effective antibacterial agents.
High-resolution structures of all three proteins of the AcrAB–TolC complex are available [23–25]. More recently, the crystal structure of MexB completed structural analyses of components of the P. aeruginosa MexAB–OprM complex [26–28]. These data significantly facilitated functional studies and provided the backbone to develop low-resolution models of these complexes. However, many mechanistic questions about trans-envelope transport by AcrAB–TolC and its homologs remain unanswered.
Structure & mechanism of AcrB
Structural data suggested that AcrB functions as a trimer (Supplementary Figure 1) (see online www.futuremedicine.com/toc/fmb/4/7). This homotrimeric structure consists of two large multidomain subunits, both transmembrane and periplasmic; the latter protrudes approximately 70 Å from the IM. At least three alternative drug binding sites have been observed [29–31]. Importantly, all these binding sites are located in the periplasmic domain of AcrB. These and other studies suggested that AcrB and possibly other RND transporters bind their substrates in the periplasm and transport them across the OM by the ‘alternate occupancy’ mechanism [29,32]. It should be mentioned that another E. coli RND-type transporter, AcrD, was reported to transfer aminoglycoside antibiotics from both the periplasm and cytoplasm [10]. It remains unclear whether substrate binding in the cytoplasm is part of the same transport mechanism. In this mechanism, each protomer of AcrB cycles through three consecutive conformations in a sequential manner, such that at any given time each one of them exists in a different phase. These three consecutive conformations were named as loose (‘access’), tight (‘binding’) and open (‘extrusion’) [29,32]. The result of this cycling is the alternate opening of the drug-binding pocket either into the periplasm or into the funnel connected to TolC. The detailed analysis of this mechanism can be found in a number of excellent recent review articles [33–35].
Site-directed mutagenesis showed that mutations trapping the transporter in one of the three conformations inactivate AcrB. In two independent studies, pairs of cysteine residues were introduced into the periplasmic domain of AcrB in such a way that when one of the protomers adopts the extrusion conformation, these cysteine residues are at a distance of approximately 4.4 Å and can form a disulfide bond [36,37]. By contrast, in two other conformations, these cysteines are too far away from each other to form a bond. AcrB carrying such cysteine residues was deficient in drug transport but could be revived under reducing conditions, which allow breakage of the disulfide bond and release of the trapped protomer. However, in these studies, all three protomers contained the same mutations, leaving it unclear as to which of the multiple possible events leads to the loss of function. Alternatively, AcrB can be trapped in the extrusion conformation by mutating the critical residues involved in proton translocation [38]. Such mutations irreversibly inactivate AcrB. Furthermore, such AcrB mutants have a negative-dominant phenotype when over-produced in wild-type E. coli cells carrying a chromosomal copy of the native gene [39]. This result suggests that AcrB trimers containing both native and mutant subunits are transport deficient. However, alternative explanations for the negative-dominant phenotype exist.
Recently, Takatsuka and Nikaido constructed a covalently linked AcrB trimer and tested the idea that all three protomers function as a single unit [40]. They introduced double-cysteine substitutions in each of the fused protomers and showed that such mutations inactivate the transporter, irrespective of their positions. Similar results were obtained with mutations in the proton translocation pathways.
Kinetic studies with these proteins are complicated because the transport reaction takes place across the OM. In the reconstituted system, the two-membrane envelope was mimicked by reconstitution of two populations of lipid vesicles [12]. AcrB was reconstituted into vesicles containing 1% fluorescently labeled lipids, constituting a fluorescence resonance energy transfer pair (donor vesicles). The second population of vesicles served as a trap to capture fluorescent lipids expelled by AcrB (acceptor vesicles). The transport reaction was initiated by AcrA, which stimulated AcrB and also brought donor and acceptor vesicles into close proximity. Using this reconstituted system it was possible to demonstrate that AcrAB-dependent transport of labeled lipids is ΔpH-dependent and that affinities of substrates to AcrB are in the low micromolar (10–20 µM) range.
The first kinetic study of drug transport by AcrAB–TolC was recently accomplished [41]. β-lactam antibiotics that target synthesis of peptidoglycan in the periplasm were used to characterize the kinetic behavior of this transporter in whole E. coli cells producing the periplasmic β-lactamase AmpC. To increase the permeability of the OM for bulky β-lactams, Nagano and Nikaido used a strain of E. coli mutant that produces a wider porin channel. In this study, the AcrAB–TolC pump was found to vary significantly in its affinity toward substrates (5 µM for nitrocefin vs ~300 µM for cephaloridine) and turnover numbers (10–1000 s−1 depending on substrate). Interestingly, the alternate occupancy mechanism suggests that extrusion of substrate is unlikely to occur unless the neighboring protomer binds the second ligand. Thus, positive cooperative kinetics is anticipated for AcrAB–TolC. Indeed, such kinetics was demonstrated for most of the β-lactams, with the exception of nitrocefin, which is a high-affinity substrate. Taken together with mutagenesis, these results provide a convincing proof that the functional unit of the AcrB transporter is a trimer and that protomers are likely to alternate their conformations as suggested by structural studies [29,32].
Structure & mechanism of TolC
The 3D structures of three OM channels, TolC, OprM and VceC from E. coli, P. aeruginosa and Vibrio cholerae, respectively, have been solved [23,26,42]. Despite very little sequence similarity, they are structurally conserved. Like AcrB, TolC forms stable trimers organized into two barrel structures (Supplementary Figure 1). A 12-stranded, 40 Å-long β-barrel inserts into the OM to form an open pore 30 Å in diameter. The unusual α-helical barrel, 100 Å in length, protrudes deep into the periplasm, where it reaches the TolC docking domain of AcrB. The lower half of this barrel is bound by an equatorial domain of mixed α/β-structure. The tip of the periplasmic end of the channel is closed in an iris-like manner by interacting loops of α-helices. This closed periplasmic entrance must be opened for substrates to gain access to the channel.
How TolC transitions from the closed to open conformation remains unclear. Structural and functional studies showed that disruption of salt bridges, which maintain the iris-like structure of the periplasmic end of TolC, leads to a dilation of the channel [43,44]. In vivo, interactions with the other two components of the complex are suggested to produce the same result. Structural analysis of the possible extent of the interaction between the crown domain of AcrB and the periplasmic part of TolC indicates that such interaction is quite shallow and hence unlikely to be very stable [44]. These interactions are likely restricted to the loops connecting helices H3/H4 and H7/H8 of TolC and the β-hairpins of the TolC-binding domain in AcrB. Thus, AcrA is believed to be the key to pump assembly, which stabilizes the weak AcrB–TolC contact and induces transition to the TolC open state.
After opening of the periplasmic entrance, drugs are believed to move along the TolC channel and are then ultimately released into the aqueous environment. One of the striking features of TolC is that its channel is hydrophilic and electronegative [23]. Most AcrB substrates are charged, amphiphilic molecules with large hydrophobic domains. Furthermore, some of the best substrates, such as sodium dodecyl sulfate and novobiocin, are negatively charged. How this significant barrier is overcome during transport remains unclear.
Structure & mechanism of AcrA
Biochemical and genetic data demonstrate that AcrA interacts with both the RND pump and the OM channel and stabilizes the trans-envelope assembly (Figure 1) [39,45]. On the IM, AcrA seems to actively contribute to transport by stimulating transport activities of AcrB and its close homolog AcrD [10,12]. Similarly, MacA, a close homolog of AcrA, is absolutely required for the ATPase activity of its cognate macrolide transporter MacB (see also below) [11]. Additional data demonstrate that some MFPs directly bind substrates and might initiate transport [46–48]. However, it is unclear how common this feature is among structurally diverse MFPs. On the OM, it is proposed that AcrA recruits the TolC channel in a substrate-independent manner [39]. Once engaged into transport complexes, TolC is thought to open at the periplasmic end, and conformational transitions in AcrA are likely to be involved in this process [24,43]. Thus, AcrA appears to not only stabilize interactions between IM and OM components, but also to actively contribute to transport events that occur at both membranes.
AcrA is a typical MFP [49]. The central domain of these proteins possesses twofold symmetry of coiled–coil and lipoyl-binding motifs. In the recently solved structures of AcrA and its homolog MexA, these domains fold back on themselves at the gap between helical regions, forming an α-helical hairpin (Supplementary Figure 2) (see online www.futuremedicine.com/toc/fmb/4/7) [24,50]. A lipoyl domain connects the α-helical hairpin with the six-stranded β-barrel domain containing a short α-helix. The reported AcrA structure lacks the distal N- and C-terminal domains. However, in the complete structure of MexA, the N- and C-termini together form a compact β-roll domain, which is the most proximal IM domain of these proteins (Supplementary Figure 2).
Biochemical and genetic studies implicate the α-helical hairpin of AcrA in complex formation with TolC, whereas all three of the lipoyl, the α–β-barrel and the membrane proximal β-roll domain form contacts with AcrB [39,45,50–52]. AcrA is conformationally flexible and can undergo large conformational changes [24,53]. This feature of AcrA was used to model the assembled structure of the three-component complex [50]. To satisfy the chemical crosslinking data, AcrA was treated as a series of rigid bodies with interdomain flexibility. As a result, the conformation of AcrA engaged into the modeled tripartite complex differs significantly from that in the crystal structure.
There appears to be a good agreement between functional studies of AcrAB–TolC and its modeled structure. Using a chemical mutagenesis approach we identified the S249N substitution in the β-barrel domain of AcrA, which enabled the functional fit between AcrA and the non-cognate P. aeruginosa MexB transporter [51]. The conclusion that this residue is located at the AcrA–transporter interface is in agreement with the finding that the cysteine residue introduced in position E250 (E196 in [50]) can be crosslinked to AcrB. The C-terminal domain of AcrA, which forms the membrane-proximal β-roll, is the most conserved domain in very diverse MFPs [49]. In the purified AcrA and bipartite AcrA–AcrB and AcrA–TolC complexes, this domain is unstructured and proteolytically labile [54]. However, when the functional tripartite complex is assembled this domain is protected from trypsin digest. In the same study, the importance of the C-terminal domain in AcrA function was further confirmed by site-directed mutagenesis. Mutation in the G363 position of this domain significantly impaired the function of the AcrAB–TolC complex. These results support the AcrAB–TolC model assembled by Symmons et al., in which the β-roll domain interacts intimately with AcrB [50].
The IM AcrAB complex is thought to preassemble without TolC. Furthermore, TolC interactions with both AcrB and AcrA are proposed to contribute to final assembly of the functional complex. A strong body of experimental evidence suggests that the α-helical hairpin of AcrA is the site of interaction with TolC (reviewed in [55]). In the most direct approach, Lobedanz et al. carried out a comprehensive analysis of AcrA–TolC interactions through probing reciprocal cysteine-mediated crosslinking [52]. Their data supported and confirmed a model in which the α-helical hairpin domain of AcrA docks at the intraprotomer grooves of TolC that are observed in the crystal structures of the partially open TolC.
The stoichiometry of the AcrA–TolC complex remains unclear. The crosslinking and complex modeling studies favor 1:1 AcrA:TolC stoichiometry [44,50]. By contrast, reconstitution into lipid sponges, coupled with fluorescence recovery after fringe pattern photobleaching studies, suggest that the stoichiometry of the homologous MexA–OprM complex depends on pH and ranges from 2 to 6 [56]. However, it should be noted that the reconstituted bipartite interactions may not reflect the actual stoichiometry of the components in the assembled tripartite complex.
One of the major steps in the assembly of the AcrAB–TolC complex is believed to be TolC transition into the open state. Based on the structural analysis of a TolC mutant, in which the stabilizing ion bridges are removed (‘leaky’ mutant), Bavro et al. proposed that in vivo interaction with the AcrB alone is likely to be sufficient for the unlocking of the periplasmic gates; however, on its own it is not sufficient to fully open the channel [44]. The analysis of the TolC channel cavity reveals the presence of a second selectivity gate, which is formed by a double ring of aspartate residues, and which is not destroyed in the ‘leaky’ mutants, owing to its location further up the channel. Perhaps interaction with AcrA is necessary to open the second gate. However, the MFP-dependent transition of TolC into the open state has not yet been reconstituted and characterized in vitro.
Despite this structural and functional information, the molecular mechanism of AcrAB–TolC remains unclear. In particular, it is unknown how AcrA stimulates the function of AcrB and how interactions with AcrAB trigger opening of the TolC channel.
ABC-type drug efflux complexes
ATP-binding cassette-type drug efflux transporters contributing to single-drug or multidrug resistance are most prevalent in Gram-positive bacteria, where they often provide protection (immunity) against endogenously produced antibiotics and toxic peptides. Examples include SrmB, OleB and DrrAB from Streptomyces species, lantibiotic exporters PepT and SalXY from Streptococcus species, and BcrABC from Bacillus licheniformis. Only a few bacterial ABC transporters were shown to be clinically important. Inactivation of any of four ABC-type multidrug transporters (abc7, abc11, abc16 and abc23) in Enterococcus faecalis resulted in a dramatic hyper-sensitivity of this bacterium to diverse antimicrobial agents [57]. The only characterized example of an ABC-type drug transporter in Gram-negative bacteria is a macrolide transporter, MacB, from E. coli. Interestingly, MacB homologs are broadly represented in both Gram-negative and Gram-positive bacteria [49]. Similar to their E. coli counterpart, these transporters function with MFPs encoded in the same operon (MacA). In addition, Gram-negative MacAB transporters associate with OM channels [9,58] and transport their substrates across the OM (Figure 1).
Structure & mechanism of ABC transporters
ATP-binding cassette transporters consist of four ‘core’ domains (Figure 1). Two transmembrane domains form a pathway across the membrane through which solutes move. These domains consist of multiple transmembrane segments (TMSs; α-helices) and contain the substrate-binding sites. Typical multidrug ABCs contain six to 12 TMSs and the two transmembrane domains can originate from the same polypeptide or as a result of oligomerization (for recent reviews see [59–61]). The other two domains are highly conserved nucleotide-binding domains (NBDs), which are located at the cytoplasmic face of the membrane and couple ATP hydrolysis to substrate translocation. NBDs are usually located at the C-termini of the same or different polypeptides that dimerize to form a fully functional transporter. The structure of E. coli MacB and its homologs is notably different from that of a typical ABC transporter. Cysteine-scanning mutagenesis established that, along with the NBD atypically located on the N-terminus, MacB contains only four transmembrane segments and a large periplasmic loop between TMS1 and TMS2 [62,63]. The unique transmembrane topology established MacB as a member of the new macrolide exporter family within the ABC superfamily of proteins. However, similar to other ABC proteins, MacB forms dimers and the functional transporter is expected to contain the four core domains [11].
Low- and high-resolution structures of ABC drug transporters demonstrate that two transmembrane domains form a large chamber within the membrane [64–66]. This chamber is opened to the extracellular milieu, and may also be accessible from the lipid phase at the interfaces between the two domains [67,68]. This large chamber is proposed to constitute a drug-binding pocket of ABC pumps. Extensive cysteine mutagenesis studies, coupled with thiol labeling and substrate protection, demonstrated that several TMSs from both transmembrane domains contribute amino acid residues to drug binding.
ATP-binding cassette transporters utilize ATP hydrolysis to drive unidirectional transport of diverse substrates across the membrane. The NBDs of all ABC transporters, irrespective of the transport substrate, share extensive amino acid sequence identity. High-resolution x-ray structures of at least 13 NBDs from ABC transporters and three full-length transporters in different conformations are currently available [69]. These vast structural data, combined with genetic and biochemical results, clearly established the existence of two ATP-binding sites at the interface between the two interacting NBDs so that each bound ATP molecule interacts with amino acid residues from both NBDs [70]. Transport is initiated by substrate binding in transmembrane domains, which drives initial conformational changes in NBDs. These conformational changes initiate ATP-dependent dimerization of NBDs and increase the affinity of NBDs for ATP. Dimerization of the NBDs is proposed to be concomitant with closure of the inward-facing substrate-binding site and opening of the outward-facing binding site. Two competing models are proposed to describe subsequent steps in transport. In the model proposed by Higgins and Linton, ‘the ATP switch model’, the binding of a nucleotide provides the power stroke for transport of the substrate, and the subsequent ATP hydrolysis initiates resetting of the transporter to its basal configuration [70]. In the alternative models, such as the ‘alternating catalytic sites model’ initially proposed by Senior et al. based on studies of human P-glycoprotein [71], drug transport is coupled to relaxation of a high-energy catalytic site conformation generated by the ATP-hydrolysis step. ATP hydrolysis and subsequent release of both Pi and ADP causes NBDs to resume the initial ‘open dimer’ conformation, resetting the whole transport cycle. Detailed analysis of the mechanism of ABC transporters can be found in several excellent review articles [59–61,69,72,73].
ABC transporters in complexes with MFPs & OM channels
In Gram-negative bacteria, the three-component ABC-type transporters are best known as type I secretion systems. These complexes are important virulence factors that deliver a variety of toxins and enzymes into the host. Examples include the hemolysin exporter HlyBD–TolC [74], metal protease exporter PrtCDE [75] and the colicin V exporter CvaAB–TolC [76]. The macrolide pump MacAB–TolC is the first and so far the only ABC-type pump from E. coli confirmed to be involved in drug efflux [77]. However, this transporter was also implicated in the export of enterotoxin STb, which could be its physiological substrate [78]. In addition, the MacAB homolog from Pseudomonas putida may be involved in secretion of putisolvins, the cyclic lipopeptides with antimicrobial, cytotoxic and surfactant properties [79].
In E. coli, type I secretion systems and drug transporters function with the same TolC channel. Thus, all these transporters utilize the same mechanism to transfer their substrates across the OM. MFPs are thought to enable the functional fit between TolC and structurally/mechanistically diverse transporters. However, the stoichiometry and architecture of such complexes remains unknown. Secondary structure analyses suggest that MFPs of type I secretion transporters have structures different from those of drug efflux MFPs [80]. In particular, their α-helical coiled–coil domains are significantly larger and lack the characteristic twofold symmetry found in the RND-type MFPs AcrA and MexA. These proteins also contain a single TMS and a cytoplasmic domain, which are shown to be functionally important [11,47,81]. By contrast, AcrA and MexA lack these domains. Instead, their N-termini are lipid-modified and anchored into the IM. Although no crystal structure is available for type I secretion MFPs, Yum and coauthors recently reported the crystal structure of the ABC-type MFP MacA from E. coli and Actinobacillus actinomycetemcomitans [82]. Similar to MFPs of type I secretion exporters, MacA contains a cytosolic domain and a TMS; however, its periplasmic portion is more similar to that of AcrA. In crystals, MacA contains the same domains but their orientation is different from that in AcrA. In addition, MacA forms hexamers that assemble into the funnel-like structure, the feature that further distinguishes this protein from AcrA. It is unclear how the MacA hexamer, with the central channel and a conical mouth, would fit on top of the MacB dimer. Unfortunately, the crystallized MacA lacks the TMS and the cytoplasmic domain, which could potentially affect the oligomeric state and structure of the protein.
We purified and reconstituted MacAB into proteoliposomes [11]. Unexpectedly, MacB alone could hydrolyze ATP very slowly when reconstituted into proteoliposomes. However, when both MacA and MacB were present in the same phospholipid bilayer, both the affinity and the rate of ATP hydrolysis increased dramatically, leading to an approximately 50-fold increase in MacB ATPase efficiency. The stimulatory effect of MacA was only observed in proteoliposomes with little or no stimulation in detergent or mixed detergent–lipid micelles, suggesting that lipid bilayers are essential for proper MacA–MacB interactions. We also found that both the TMS and the C-terminal domain of MacA are important for this activity. The importance of MacA in stimulation of MacB ATPase was later confirmed independently by Lin and coauthors [83]. In this study, the kinetic analysis suggested that MacA promotes and stabilizes the ATP-binding conformation of MacB, which, according to the current views on the mechanism of ABC transporters (see previously) is the periplasm-facing conformation of MacB.
The stimulating effect of MacA is reminiscent of periplasmic ligand-binding proteins. These proteins specifically associate with ABC transporters involved in the uptake of diverse nutrients in Gram-negative bacteria. Examples include the extensively studied maltose-binding protein (MBP; the product of the malE gene) and the histidine-binding (HisJ) protein [84]. In analogy with these proteins, MFPs might accelerate ATP hydrolysis or proton efflux by functioning as transmembrane signaling molecules. For example, MFPs could signal transporters regarding the formation of a tripartite complex that also includes an OM channel. Studies of the hemolysin transport in E. coli suggest that HlyD mediates the assembly of the transient tripartite complex HlyBD–TolC [14]. The cycle of TolC recruitment and disengagement appears to be linked to the process of substrate translocation. Thus, in HlyBD–TolC and similar complexes, MFPs might coordinate ATP hydrolysis with the transport of substrates across the OM. By contrast, drug efflux transporters such as AcrAB and MacAB form stable complexes with the OM channels [11,39,45]. Furthermore, in reconstitution experiments, TolC did not modulate the ATPase activities of MacB or the MacAB complex, suggesting a passive role of TolC during transport [11].
Alternatively, MFPs could signal to transporters about the presence of substrates. As discussed previously, the RND-type transporters, such as AcrB, contain large periplasmic domains, which are the sites of substrate binding [85–89]. In AcrB and MexB, mutations affecting the substrate specificity of these transporters were mapped to regions predicted to be involved in trimerization or interaction with MFPs rather than to regions expected to contribute to substrate recognition [90]. In the assembled AcrAB–TolC structure, the site of AcrA interaction with AcrB is located away from the region that undergoes conformational rotations and closer to the substrate binding sites of AcrB. Thus, MFPs that associate with the RND-type transporters could be involved in substrate recognition on the periplasmic side of the IM. An unusual feature of the ABC-type transporter of MacB is the presence of a large periplasmic domain [62]. This structural feature is more akin to the RND-type transporters. It is possible that MacB also binds its substrates on the periplasmic side of the IM and that the periplasmic MacA protein plays an active role in this process. We did not detect any interactions of MacA with macrolides [11]. However, Lin et al. showed that MacA increases the erythromycin-binding capacity of MacB [83]. In addition, HlyD, the MFP of the hemolysin exporter, interacts with the substrate on both the cytosolic and the periplasmic side of the IM [47].
MFS-type drug efflux complexes
Major facilitator superfamily is the second largest family of proteins containing multidrug efflux pumps. In Gram-positive bacteria, the best-characterized MFS-type multidrug transporters are QacA and QacB from Staphylococcus aureus [91,92], which extrude mono- and divalent cationic compounds such as quaternary ammonium compounds, dyes and the diamidines LmrP from Lactococcus lactis [93] and Bmr from Bacillus subtilis, which have similar specificities [94]. Increased synthesis of a chromosomal S. aureus NorA transporter is particularly important in the emergence of fluoroquinolone-resistant clinical isolates of this species. E. coli MdfA is a well-studied example of an MFS-type multidrug transporter from Gram-negative bacteria [95]. In addition, some MFS transporters from Gram-negative bacteria function with MFP and OMF accessory proteins (e.g., EmrAB–TolC and EmrKY–TolC from E. coli [4]).
Structure & mechanism of MFS transporters
Major facilitator superfamily-type transporters are broadly represented in bacterial cells [16]. They have either 12 or 14 TMSs and rely on the proton-motive force for their function. The available high-resolution structures of MFS transporters show similar overall architecture [96–98]. A total of 12 TMSs form a compact structure within the membrane. The difference between multidrug (EmrD) and substrate-specific MFS transporters (LacY and GlpT) is the internal cavity formed by TMSs in the core of the transporter. Whereas LacY and GlpT have hydrophilic interiors, the internal cavity of EmrD comprises mostly hydrophobic residues, consistent with its function of transporting lipophilic compounds. Several of these residues are bulky and aromatic, and some are conserved in other multidrug MFS transporters.
Similar to ABC transporters, drugs are believed to gain access to the hydrophobic binding pocket of MFS transporters from the inner leaflet of the cytoplasmic membrane. The mechanism of drug recognition appears to be similar to other multidrug transporters and kinetic studies indicate that different substrates can be bound simultaneously to nonoverlapping binding sites within the same binding pocket [99,100]. Recent studies established that MFSs also operate via reorientation of the substrate-binding site from an inward to outward facing conformation and that this reorientation is coupled to the protonation/deprotonation of specific amino acid residues [101,102]. The negatively charged amino acid residues located in TMSs appear to play a critical role in the protonation/deprotonation step during transport and such residues were identified in representatives of all secondary transporters. Analyses of multidrug LmrP and MdfA showed that no single membrane-embedded acidic residue is critical for the transport mechanism [103,104]. This is in contrast to many other secondary transporters, including transporters belonging to the RND and small multidrug resistance (SMR) families, for which negatively charged, membrane-embedded residues were shown to play irreplaceable roles in proton-coupled transport reactions [34,105].
EmrAB–TolC, the three-component multidrug MFS pump
Although EmrAB was the first multidrug efflux transporter identified in E. coli, it remains the least characterized mechanistically [4]. Similar to AcrAB, EmrAB is constitutively expressed in E. coli and contributes to intrinsic resistance against various dyes (rhodamine and methylviologen), ionophores (carbonyl cyanide m-chloro phenyl hydrazone), detergents (bile salts and sodium dodecyl sulfate) and steroid hormones [106]. However, AcrAB dominates in conferring resistance against these compounds, with EmrAB playing only a minor role. Both EmrA and EmrB are typical representatives of their protein families. EmrA contains a single N-terminal TMS and a central coiled–coil domain similar to that of MacA. However, phylogenetic analysis suggests that EmrA and its homologs are very distant from AcrA and MacA proteins and probably diverted early in evolution [49].
EmrB shares a significant degree of similarity with QacA multidrug transporters of Gram-positive bacteria [107]. Although biochemical studies of EmrB are very limited, QacA is well characterized. These MFS transporters contain 14 TMSs with both N- and C-termini located in the cytoplasm. QacA-mediated transport was shown to occur via the electrogenic drug/nH+ (n > 2) antiport, which conforms to classical Michaelis–Menten kinetics [108]. Interactions with structurally dissimilar compounds occur with high affinity, with Km values in the low micromolar range (<20 µM).
Significant efforts were devoted to the analysis of the oligomeric state of MFS transporters. Although early evidence suggested that oligomerization might be important for activity, LacY was shown to be functional as a monomer by an array of biochemical and genetic approaches (reviewed in [109]). The multidrug transporters MdfA and EmrD were also shown to function as monomers [97,110]. Recently, both EmrA and EmrB were purified and reconstituted into proteoliposomes [111]. Electron microscopy after negative staining suggested that the assembled complexes have a calculated molecular mass of 268 kDa, which could correspond to a tetrameric EmrAB complex 2 × (2 × [56 kDa EmrB and 42 kDa EmrA]). In an independent study, purified EmrA was shown to form dimers and trimers [46]. The N-terminal TMS of EmrA contains a leucine-zipper motif, which is known to promote protein oligomerization. However, in the absence of this domain, EmrA could still form oligomers, presumably through association of its periplasmic coiled–coil domains. Interestingly, phylogenetic analysis of EmrA identified its homologs in Gram-positive bacteria [49]. Furthermore, some of the Gram-positive operons contain two or even three genes encoding MFPs and only a single gene encoding an EmrB-like transporter. Thus, these transporters could function as oligomers or, alternatively, hetero-oligomeric MFPs could function with monomeric transporters. The oligomerization of EmrB is reminiscent of the oligomeric structures of RND and ABC transporters and could be characteristic of transporters that function in complexes with MFPs and OM channels.
EmrA is the only multidrug MFP for which interactions with substrates have been demonstrated [46]. Tryptophan fluorescence quenching with potassium iodide was used to detect binding of ionophores and drugs to EmrA. Substrates were bound by the periplasmic portion of the protein lacking the N-terminal TMS with affinities in the low micromolar range. Topology predictions indicate that EmrB is unlikely to have large periplasmic extentions similar to those found in AcrB and MacB. Yet, drug molecules released from EmrB must reach TolC without escaping into the periplasm; perhaps EmrA receives substrates from the transporter and conveys them to TolC. Further biochemical analyses are needed to reconstruct the mechanism of drug transport by EmrAB–TolC.
Conclusion
In Gram-negative bacteria, drug efflux RND, MFS and ABC transporters require MFPs and OM channels for their functions. Structural analyses suggest that the mode of MFP association with transporters varies depending on the structure of the transporter in question. The MFP–RND interface is located in the periplasm and is well characterized for the AcrA–AcrB complex. By contrast, ABC and MFS transporters presumably interact with MFPs within the IM and in the cytoplasm. The interface and stoichiometry of MFP–ABC and MFP–MFS complexes remain unknown. The emerging view is that in various IM complexes, MFPs play an important role in stimulation of transporters’ activities and substrate binding. However, the biochemical mechanism of these activities remains unclear. Ultimately, substrates from IM complexes must be transferred to the OM channels. In the case of RND transporters, the periplasmic domains of AcrB and TolC are large enough to contact each other. By contrast, ABC and MFS transporters do not have large periplasmic extensions and are unlikely to interact with channels directly. It is unknown how substrates expelled by these transporters reach OM channels.
The structural variety of MFP–transporter complexes does not affect association with OM channels. In E. coli, the majority of the MFP-dependent transporters function with the same OM channel, TolC, regardless of their protein families or substrates. Thus, all these proteins contain a conserved TolC-binding domain. Recent studies suggest that the α-helical coiled–coil hairpins of MFPs might play this role. It remains unclear as to whether the stoichiometry and structure of MFP–TolC complexes differ depending on the IM transporter and how various IM complexes induce TolC transition into the open state.
Future perspective
Future studies are likely to involve structural analyses of MacB and EmrB transporters; alone and in complex with the respective MFP components. Since both MacA and EmrA contain N-terminal TMSs and are prone to oligomerization, their complexes with MacB and EmrB, respectively, are expected to be more stable and possibly more amenable to structural analysis. Hopefully these studies will help us understand how the proposed transport cycles of ABC and MFS transporters are integrated with accessory proteins, how reorientation of binding sites from inward- to outward-facing conformations is achieved in the context of oligomeric structures of MFPs, and whether or not MacA and EmrA form periplasmic tunnels that guide substrates towards TolC. The major hurdle here is that the stable association and functional conformation of MFPs might only be achieved when the OM channel is engaged as well, as was shown in the case of AcrAB–TolC [54]. Thus, the in vivo structural analysis, which was successfully used with AcrAB–TolC [50], could be more straightforward and insightful with MacB and EmrB as well.
A significant emphasis will also be placed on biochemical analyses of reconstituted complexes. These studies are expected to re-enact the transport reaction cycles of three-component transporters, and characterize the mechanism of coupling between conformational changes in each component and energy inputs of ATP hydrolysis or proton transfer. The alternative occupancy mechanism of AcrB requires further analysis to understand whether or not AcrA and TolC contribute to conformational asymmetry of this transporter.
Finally, AcrB and its homologs in Gram-negative pathogens are important drug targets [6]. There will be continuous efforts to understand the mechanism of AcrB/MexB inhibition by already-known chemical entities and identify new potent inhibitors. Biochemical analyses are expected to identify ‘bottlenecks’ in the transport reactions that could be targeted for rational drug design. Located in the OM, TolC is also an attractive drug target. However, so far no inhibitors have been found that specifically interact with TolC and block its function. The same is true for inhibitors that target assembly of complexes in the periplasm. The vast structural information on AcrAB–TolC and new screening approaches promise to facilitate these efforts.
Executive summary
Multidrug efflux transporters are responsible for drug resistance in Gram-negative pathogens
Drug resistance is a serious threat to public health.
Multidrug efflux transporters are the major contributors to intrinsic and acquired drug resistance in Gram-negative pathogens.
These transporters are organized into three-component complexes that transverse the inner and outer membrane and the periplasm, and act in a highly coordinated manner.
Resistance–nodulation–cell division-type multidrug efflux complexes
Escherichia coli AcrAB–TolC is the best-characterized resistance–nodulation–cell division-type multidrug efflux transporter and is an important drug target.
The trimeric AcrB transporter is proposed to function by an ‘alternate occupancy’ mechanism, which involves functional rotation of each protomer through three different conformations.
Genetic and kinetic studies showed that the AcrB trimer transports drugs cooperatively.
TolC provides a route for the expelled drugs to cross the outer membrane.
TolC is a stable trimer organized into a two-barrel structure with one barrel inserted into the outer membrane and the other extending deep into the periplasm.
The periplasmic entrance of TolC is closed.
Association with the periplasmic membrane fusion protein (MFP) AcrA is needed to trigger closed-to-open transition in TolC.
AcrA is a typical representative of the MFP family, which has a flexible, modular structure.
AcrA interacts with both AcrB and TolC and mediates functional cooperation between the transporter and the outer membrane channel.
Further studies are needed to understand how AcrA stimulates the activity of AcrB and triggers the closed-to-open transitions in TolC.
ATP-binding cassette-type multidrug efflux complexes
ATP-binding cassette (ABC) transporters consist of four ‘core’ domains: two transmembrane domains and two nucleotide-binding domains.
ABC transporters operate by reorientation of the substrate-binding site from an inward- to outward-facing conformation.
ATP binding triggers reorientation of the substrate-binding site.
The ABC-type MFPs contain a single transmembrane segment and a cytoplasmic domain.
MFPs stimulate ATP hydrolysis, presumably by stabilizing the ATP-binding conformation of the transporter.
MFPs bind substrates and possibly provide a route for substrates to cross the periplasm.
The ABC-type MFPs are oligomers, and might form a sheath-like structure surrounding the periplasmic entrance of the TolC channel.
The functional association between MFPs and ABC transporters and the structure of the assembled complex remain topics of intense discussion.
Major facilitator superfamily-type multidrug efflux complexes
Major facilitator superfamily (MFS) transporters are drug–proton antiporters.
Similar to ABC and resistance–nodulation–cell division transporters, multidrug MFS transporters have hydrophobic substrate-binding cavities.
Protonation/deprotonation of acidic residues located in transmembrane domains drives reorientation of drug binding sites.
The MFS-type MFPs are structurally similar to ABC-type MFPs.
The MFS-type MFPs interact with drugs and form oligomers.
The mechanism of action of MFS-type tripartite complexes is the least understood and requires further structural and biochemical analyses.
Supplementary Material
Acknowledgments
Financial & competing interests disclosure
Studies in Helen I Zgurskaya’s laboratory are supported by the NIH grant 1-RO1-AI052293. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 2004;10:S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM. The need for new antibiotics. Clin. Microbiol. Infect. 2004;10 Suppl. 4:1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- 3.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl Acad. Sci. USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma D, Cook DN, Alberti M, et al. Molecular cloning and characterization of acrA and acre genes of Escherichia coli. J. Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat. Rev. Drug Discov. 2007;6:56–65. doi: 10.1038/nrd2200. ▪▪ Recent review article presenting the current state of understanding of the mechanisms of multidrug transporters and the efforts to develop clinically useful inhibitors.
- 7.Nikaido H. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell Dev. Biol. 2001;12:215–223. doi: 10.1006/scdb.2000.0247. [DOI] [PubMed] [Google Scholar]
- 8.Zgurskaya HI, Krishnamoorthy G, Tikhonova EB, Lau SY, Stratton KL. Mechanism of antibiotic efflux in Gram-negative bacteria. Front. Biosci. 2003;8:s862–s873. doi: 10.2741/1134. [DOI] [PubMed] [Google Scholar]
- 9.Dinh T, Paulsen IT, Saier MH., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of Gram-negative bacteria. J. Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aires JR, Nikaido H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 2005;187:1923–1929. doi: 10.1128/JB.187.6.1923-1929.2005. ▪ Reports that reconstituted AcrA stimulates the proton transfer activity of the aminoglycoside transporter AcrD.
- 11. Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 2007;63:895–910. doi: 10.1111/j.1365-2958.2006.05549.x. ▪ First reconstitution of the ATP-binding cassette-type multidrug efflux complex.
- 12. Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl Acad. Sci. USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. ▪ The first reconstitution of the resistance-nodulation-cell division-type multidrug efflux complex.
- 13.Andersen C, Hughes C, Koronakis V. Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 2001;13:412–416. doi: 10.1016/s0955-0674(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 14.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen IT, Chen J, Nelson KE, Saier MH., Jr Comparative genomics of microbial drug efflux systems. J. Mol. Microbiol. Biotechnol. 2001;3:145–150. [PubMed] [Google Scholar]
- 16.Saier MH, Jr, Beatty JT, Goffeau A, et al. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 17.Tseng TT, Gratwick KS, Kollman J, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 18.Saier MH, Jr, Paulsen IT, Sliwinski MK, et al. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 19.Poole K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004;10:12–26. doi: 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 20.Sobel ML, Hocquet D, Cao L, Plesiat P, Poole K. Mutations in PA3574 (nalD) lead to increased MexAB–OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zgurskaya HI, Nikaido H. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 2000;37:219–225. doi: 10.1046/j.1365-2958.2000.01926.x. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido H, Zgurskaya HI. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001;3:215–218. [PubMed] [Google Scholar]
- 23. Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. ▪▪ Describes the two-barrel structure of TolC.
- 24. Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. ▪ Reports that AcrA adopts multiple conformations.
- 25. Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. ▪▪ Seminal paper reporting the trimeric structure of AcrB.
- 26.Akama H, Kanemaki M, Yoshimura M, et al. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 27.Akama H, Matsuura T, Kashiwagi S, et al. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 28.Sennhauser G, Bukowska MA, Briand C, Grutter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 30.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 32.Seeger MA, Schiefner A, Eicher T, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 33.Murakami S. Multidrug efflux transporter, AcrB – the pumping mechanism. Curr. Opin. Struct. Biol. 2008;18:459–465. doi: 10.1016/j.sbi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. ▪ Excellent review analyzing the ‘alternate occupancy’ mechanism of AcrB.
- 36.Seeger MA, von Ballmoos C, Eicher T, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 37.Takatsuka Y, Nikaido H. Site-directed disulfide crosslinking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J. Bacteriol. 2007;189:8677–8684. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su CC, Li M, Gu R, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006;188:7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 40.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J. Bacteriol. 2009;191:1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl Acad. Sci. USA. 2009;106:5854–5858. doi: 10.1073/pnas.0901695106. ▪ Reports that AcrAB–TolC functions cooperatively and that MICs are not a good measure of the transporter’s activity.
- 42.Federici L, Du D, Walas F, et al. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 Å resolution. J. Biol. Chem. 2005;280:15307–15314. doi: 10.1074/jbc.M500401200. [DOI] [PubMed] [Google Scholar]
- 43.Andersen C, Koronakis E, Bokma E, et al. Transition to the open state of the TolC periplasmic tunnel entrance. Proc. Natl Acad. Sci. USA. 2002;99:11103–11108. doi: 10.1073/pnas.162039399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bavro VN, Pietras Z, Furnham N, et al. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell. 2008;30:114–121. doi: 10.1016/j.molcel.2008.02.015. ▪ Crystal structure of the open mutant of TolC, suggesting that the initial semi-opened transition in TolC could be triggered by AcrB.
- 45.Touze T, Eswaran J, Bokma E, et al. Interactions underlying assembly of the Escherichia coli AcrAB–TolC multidrug efflux system. Mol. Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- 46.Borges-Walmsley MI, Beauchamp J, Kelly SM, et al. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J. Biol. Chem. 2003;278:12903–12912. doi: 10.1074/jbc.M209457200. [DOI] [PubMed] [Google Scholar]
- 47.Pimenta AL, Racher K, Jamieson L, Blight MA, Holland IB. Mutations in HlyD, part of the type 1 translocator for hemolysin secretion, affect the folding of the secreted toxin. J. Bacteriol. 2005;187:7471–7480. doi: 10.1128/JB.187.21.7471-7480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J. Biol. Chem. 2007;282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 49. Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim. Biophys. Acta. 2009;1794:794–807. doi: 10.1016/j.bbapap.2008.10.010. ▪ Detailed analysis of membrane fusion proteins in both Gram-negative and Gram-positive bacteria.
- 50. Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl Acad. Sci. USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. ▪▪ Describes the most advanced model of the AcrAB–TolC complex, which combines data from site-specific crosslinking in vivo and computer modeling.
- 51.Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J. Bacteriol. 2008;190:691–698. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobedanz S, Bokma E, Symmons MF, et al. A periplasmic coiled–coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl Acad. Sci. USA. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ip H, Stratton K, Zgurskaya H, Liu J. pH-induced conformational changes of AcrA, the membrane fusion protein of Escherichia coli multidrug efflux system. J. Biol. Chem. 2003;278:50474–50482. doi: 10.1074/jbc.M305152200. [DOI] [PubMed] [Google Scholar]
- 54.Ge Q, Yamada Y, Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB–TolC. J. Bacteriol. 2009;191:4365–4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misra R, Bavro VN. Assembly and transport mechanism of tripartite drug efflux systems. Biochim. Biophys. Acta. 2009;1794:817–825. doi: 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reffay M, Gambin Y, Benabdelhak H, et al. Tracking membrane protein association in model membranes. PLoS One. 2009;4:e5035. doi: 10.1371/journal.pone.0005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis DR, McAlpine JB, Pazoles CJ, et al. Enterococcus faecalis multi-drug resistance transporters: application for antibiotic discovery. J. Mol. Microbiol. Biotechnol. 2001;3:179–184. [PubMed] [Google Scholar]
- 58.Paulsen IT, Park JH, Choi PS, Saier MH., Jr A family of Gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from Gram-negative bacteria. FEMS Microbiol. Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 59.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seeger MA, van Veen HW. Molecular basis of multidrug transport by ABC transporters. Biochim. Biophys. Acta. 2009;1794:725–737. doi: 10.1016/j.bbapap.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 61.Locher KP. Review. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:239–245. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kobayashi N, Nishino K, Hirata T, Yamaguchi A. Membrane topology of ABC-type macrolide antibiotic exporter MacB in Escherichia coli. FEBS Lett. 2003;546:241–246. doi: 10.1016/s0014-5793(03)00579-9. [DOI] [PubMed] [Google Scholar]
- 63.Xu Y, Sim SH, Nam KH, et al. Crystal structure of the periplasmic region of MacB, a noncanonic ABC transporter. Biochemistry. 2009;48:5218–5225. doi: 10.1021/bi900415t. [DOI] [PubMed] [Google Scholar]
- 64.Aller SG, Yu J, Ward A, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 66.Reyes CL, Chang G. Structure of the ABC transporter MsbA in complex with ADP.vanadate and lipopolysaccharide. Science. 2005;308:1028–1031. doi: 10.1126/science.1107733. [DOI] [PubMed] [Google Scholar]
- 67.Rosenberg MF, Mao Q, Holzenburg A, et al. The structure of the multidrug resistance protein 1 (MRP1/ABCC1) crystallization and single-particle analysis. J. Biol. Chem. 2001;276:16076–16082. doi: 10.1074/jbc.M100176200. [DOI] [PubMed] [Google Scholar]
- 68.Pleban K, Kopp S, Csaszar E, et al. P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces: a combined photoaffinity labeling-protein homology modeling approach. Mol. Pharmacol. 2005;67:365–374. doi: 10.1124/mol.104.006973. [DOI] [PubMed] [Google Scholar]
- 69.Reyes CL, Ward A, Yu J, Chang G. The structures of MsbA: insight into ABC transporter-mediated multidrug efflux. FEBS Lett. 2006;580:1042–1048. doi: 10.1016/j.febslet.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 70.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 71.Senior AE. Catalytic mechanism of P-glycoprotein. Acta Physiol. Scand. Suppl. 1998;643:213–218. [PubMed] [Google Scholar]
- 72.Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 73.Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holland IB, Blight MA, Kenny B. The mechanism of secretion of hemolysin and other polypeptides from Gram-negative bacteria. J. Bioenerg. Biomembr. 1990;22:473–491. doi: 10.1007/BF00763178. [DOI] [PubMed] [Google Scholar]
- 75.Delepelaire P. PrtD, the integral membrane ATP-binding cassette component of the Erwinia chrysanthemi metalloprotease secretion system, exhibits a secretion signal-regulated ATPase activity. J. Biol. Chem. 1994;269:27952–27957. [PubMed] [Google Scholar]
- 76.Gilson L, Mahanty HK, Kolter R. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 1990;9:3875–3894. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 2001;183:5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J. Bacteriol. 2008;190:7693–7698. doi: 10.1128/JB.00853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dubern JF, Coppoolse ER, Stiekema WJ, Bloemberg GV. Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology. 2008;154:2070–2083. doi: 10.1099/mic.0.2008/016444-0. [DOI] [PubMed] [Google Scholar]
- 80.Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J. Mol. Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 81.Balakrishnan L, Hughes C, Koronakis V. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J. Mol. Biol. 2001;313:501–510. doi: 10.1006/jmbi.2001.5038. [DOI] [PubMed] [Google Scholar]
- 82.Yum S, Xu Y, Piao S, et al. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J. Mol. Biol. 2009;387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 83.Lin HT, Bavro VN, Barrera NP, et al. MacB ABC transporter is a dimer whose ATPase activity and macrolide-binding capacity are regulated by the membrane fusion protein MacA. J. Biol. Chem. 2009;284:1145–1154. doi: 10.1074/jbc.M806964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nikaido H, Hall JA. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–20. doi: 10.1016/s0076-6879(98)92003-1. [DOI] [PubMed] [Google Scholar]
- 85.Yu EW, Aires JR, Nikaido H. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 2003;185:5657–5664. doi: 10.1128/JB.185.19.5657-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric analysis of the multicomponent multidrug efflux transporters from Gram-negative bacteria. J. Bacteriol. 2002;184:6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nehme D, Li XZ, Elliot R, Poole K. Assembly of the MexAB–OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J. Bacteriol. 2004;186:2973–2983. doi: 10.1128/JB.186.10.2973-2983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mao W, Warren MS, Black DS, et al. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 2002;46:889–901. doi: 10.1046/j.1365-2958.2002.03223.x. ▪ Elegant study delineating the molecular recognition of multiple drugs in the periplasmic loops of resistance–nodulation–cell division-type transporter.
- 89.Elkins CA, Nikaido H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 2002;184:6490–6498. doi: 10.1128/JB.184.23.6490-6498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Middlemiss JK, Poole K. Differential impact of MexB mutations on substrate selectivity of the MexAB–OprM multidrug efflux pump of Pseudomonas aeruginosa. J. Bacteriol. 2004;186:1258–1269. doi: 10.1128/JB.186.5.1258-1269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paulsen IT, Brown MH, Skurray RA. Characterization of the earliest known Staphylococcus aureus plasmid encoding a multidrug efflux system. J. Bacteriol. 1998;180:3477–3479. doi: 10.1128/jb.180.13.3477-3479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown MH, Skurray RA. Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 2001;3:163–170. [PubMed] [Google Scholar]
- 93.Bolhuis H, Poelarends G, van Veen HW, et al. The lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J. Biol. Chem. 1995;270:26092–26098. doi: 10.1074/jbc.270.44.26092. [DOI] [PubMed] [Google Scholar]
- 94.Neyfakh AA. The multidrug efflux transporter of Bacillus subtilis is a structural and functional homolog of the Staphylococcus NorA protein. Antimicrob. Agents Chemother. 1992;36:484–485. doi: 10.1128/aac.36.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewinson O, Adler J, Poelarends GJ, et al. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl Acad. Sci. USA. 2003;100:1667–1672. doi: 10.1073/pnas.0435544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Abramson J, Smirnova I, Kasho V, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. ▪ Reports the structure of LacY in the context of vast biochemical studies.
- 97.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lemieux MJ, Huang Y, Wang DN. The structural basis of substrate translocation by the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr. Opin. Struct. Biol. 2004;14:405–412. doi: 10.1016/j.sbi.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 99.Fluman N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 100.Saidijam M, Benedetti G, Ren Q, et al. Microbial drug efflux proteins of the major facilitator superfamily. Curr. Drug Targets. 2006;7:793–811. doi: 10.2174/138945006777709575. [DOI] [PubMed] [Google Scholar]
- 101.Smirnova I, Kasho V, Choe JY, et al. Sugar binding induces an outward facing conformation of LacY. Proc. Natl Acad. Sci. USA. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou Y, Guan L, Freites JA, Kaback HR. Opening and closing of the periplasmic gate in lactose permease. Proc. Natl Acad. Sci. USA. 2008;105:3774–3778. doi: 10.1073/pnas.0800825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sigal N, Molshanski-Mor S, Bibi E. No single irreplaceable acidic residues in the Escherichia coli secondary multidrug transporter MdfA. J. Bacteriol. 2006;188:5635–5639. doi: 10.1128/JB.00422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bapna A, Federici L, Venter H, et al. Two proton translocation pathways in a secondary active multidrug transporter. J. Mol. Microbiol. Biotechnol. 2007;12:197–209. doi: 10.1159/000099641. [DOI] [PubMed] [Google Scholar]
- 105.Schuldiner S. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim. Biophys. Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 106.Elkins CA, Mullis LB. Mammalian steroid hormones are substrates for the major RND- and MFS-type tripartite multidrug efflux pumps of Escherichia coli. J. Bacteriol. 2006;188:1191–1195. doi: 10.1128/JB.188.3.1191-1195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brown MH, Skurray RA. Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 2001;3:163–170. [PubMed] [Google Scholar]
- 108.Mitchell BA, Paulsen IT, Brown MH, Skurray RA. Bioenergetics of the staphylococcal multidrug export protein QacA. Identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem. 1999;274:3541–3548. doi: 10.1074/jbc.274.6.3541. [DOI] [PubMed] [Google Scholar]
- 109.Guan L, Kaback HR. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sigal N, Lewinson O, Wolf SG, Bibi E. E. coli multidrug transporter MdfA is a monomer. Biochemistry. 2007;46:5200–5208. doi: 10.1021/bi602405w. [DOI] [PubMed] [Google Scholar]
- 111.Tanabe M, Szakonyi G, Brown KA, et al. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem. Biophys. Res. Commun. 2009;380:338–342. doi: 10.1016/j.bbrc.2009.01.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.