Abstract
Calbindin-D28k is a calcium binding protein with six EF hand domains. Calbindin-D28k is unique in that it functions as both a calcium buffer and sensor protein. It is found in many tissues including the brain, pancreas, kidney and intestine, playing important roles in each. Calbindin-D28k is known to bind four calcium ions and upon calcium binding undergoes a conformational change. The structure of apo- calbindin-D28k is in an ordered state, transitioning into a disordered state as calcium is bound. Once fully loaded with four calcium ions, it again takes on an ordered state. The solution structure of disulfide-reduced holo- calbindin-D28k has been solved by NMR, while the structure of apo- calbindin-D28k has yet to be solved. Differential surface modification of lysine and histidine residues analyzed by mass spectrometry have been used in this study to identify, for the first time, the specific regions of calbindin-D28k undergoing conformational changes between the holo- and apo- states. Using differential surface modification in combination with mass spectrometry, EF hands 1 and 4 as well as the linkers before EF hand 1 and the linkers between EF hands 4–5 and 5–6 were identified as regions of conformational change between the apo- and holo- calbindin-D28k. Under the experimental conditions employed, EF hands 2 and 6, which are known to not bind calcium were unaffected in either form. EF hand 2 is highly accessible; however, EF hand 6 was determined to not be surface accessible in either form. Previous research has identified a disulfide bond between cysteines 94 and 100 in the holo- state. Until now it was unknown whether this bond also exists in the apo- form. Our data confirm the presence of the disulfide bond between cysteines 94 and 100 in the holo- form, and indicate that there is predominantly no disulfide bond between these residues in the apo- protein.
The calcium binding protein, calbindin-D28k plays a unique role in eukaryotic cells, acting as both a calcium buffer and sensor (1–4). It is found in several tissue types and serves many functions. For example, it is responsible for the selective reabsorption of calcium in the kidney and the intestine, as well as regulating the release of insulin in pancreatic islet cells (5, 6). Calbindin-D28k is very abundant in the brain, making up 0.1–1.5% of the total soluble protein. It is essential in neural functioning, altering synaptic interactions in the hippocampus, modulating calcium channel activity and neuronal firing (6–8). In addition to this, calbindin-D28k has been found to modulate the activity of proteins involved in the development of neurodegenerative disorders including Alzheimer’s and Huntington’s disease, as well as bipolar disorder (9–14). Deciphering how calbindin-D28k functions is essential for further understanding of the pathogenesis of these neurodegenerative disorders.
Structurally, calbindin-D28k is made up of six EF hand domains, four of which bind calcium (2, 15). The EF hand domain is a helix-loop-helix calcium binding domain. The loop consists of twelve conserved residues responsible for coordinating calcium binding. EF hand calcium binding proteins are subdivided into two general groups, calcium buffers and calcium sensors (16–18). As stated above, calbindin-D28k is unique in that it functions as both. EF hand proteins in the sensor category are characterized by undergoing a conformational change that occurs upon calcium binding (19, 20). Mass spectrometry (MS) and nuclear magnetic resonance (NMR) experiments have shown that calbindin-D28k binds four calcium ions, in EF hands 1, 3, 4 and 5 and that the calcium ions are sequentially bound with EF hand 1 first, followed by 4 and 5 and finally EF hand 3 (15). NMR titration experiments have also demonstrated that the structure of calbindin-D28k is ordered in the apo- state (15). As calcium ions are bound, calbindin-D28k transitions into a disordered state and once fully-loaded with four calcium ions it returns to an ordered state. The effect that the conformational change has on the surface hydrophobicity of calbindin-D28k has been investigated and the studies showed that the surface hydrophobicity of rat brain calbindin-D28k is lowest in the holo- form (2). Similar experiments for human calbindin-D28k also showed a lower surface hydrophobicity for the holo- form versus the apo- form (1). Circular dichroism (CD) spectroscopy has shown that the secondary structure remains unperturbed upon calcium binding but the tertiary structure is much more sensitive to calcium binding (21). These experiments have provided some insight into the effect calcium binding has on the structure of calbindin-D28k. They do not, however, define the specific areas of the protein affected by the conformational change. The high resolution structure of the disulfide-reduced holo- rat brain calbindin-D28k structure has been solved using NMR (2), however, the structure of apo- calbindin-D28k remains to be solved. In the absence of a high resolution structure of apo- calbindin-D28k it has not been possible to compare the three dimensional structures of the apo- and holo- conformational states of calbindin-D28k.
This study uses differential surface modification analyzed by MS to identify, for the first time, the specific regions of calbindin-D28k affected by the conformational changes between the apo- and holo- forms. MS analysis of differential surface modifications has been used for several purposes including mapping protein surfaces, studying protein-protein complexes and to determine ligand induced conformational changes by differentially modifying specific amino acid residue side chains (22–32). These side chain modifications reflect both the reactivity and surface accessibility of a specific residue. Side chain reactivity is affected by its surface accessibility as well as its surrounding microenvironment, including the presence of electrostatic interactions, such as the formation of a salt bridge (22–24, 33). Several studies have established that the relative reactivity data from differential surface modification experiments of both lysine and histidine residues correlate with their surface accessibility (22–24, 34). This allows the differential surface modification patterns to be used to identify regions of structural change that occur as a result of a specific perturbation, such as the binding of calcium (33, 35). Lysine acetylation and histidine modification analyzed by mass spectrometry have been used in this study to identify, for the first time, the specific regions of calbindin-D28k undergoing a conformational change upon calcium binding. We have investigated the status of the potential disulfide bond in the apo- form and have found the apo- form to be predominantly reduced. We have also modeled the conformational changes occurring as a result of disulfide bond formation based on the reduced holo- calbindin-D28k NMR structure, and have interpreted our differential reactivity results in terms of these structural changes.
Experimental Procedures
Materials
GST resin was purchased from Novagen (Gibbstown, NJ). Sequencing grade trypsin was obtained from Promega (Madison, WI). Sequencing grade GluC and chymotrypsin were purchased from Roche Applied Sciences (Indianapolis, IN). Acetonitrile was purchased from Caledon Laboratory Chemicals (Georgetown, ON). Formic acid (FA), (96%), acetic anhydride and diethylpyrocarbonate (DEPC) were supplied by Sigma Aldrich (St. Louis, MO).
Expression and Purification of Calbindin-D28k
The cloning, expression and purification protocol for rat calbindin-D28k has previously been described (36). Briefly, calbindin-D28k is expressed as a GST fusion protein. A glutathione S-transferase (GST) resin column is used to isolate the calbindin-D28k-GST fusion protein. The fractions containing the calbindin-D28k-GST fusion protein were combined and the N-terminal GST-tag was removed by thrombin cleavage. A second GST column is used to isolate the calbindin-D28k from the GST-tag. Purity was confirmed by running the sample on a denaturing SDS-PAGE gel (Figure S1). The holo- calbindin-D28k sample was dialyzed into 20 mM HEPES, 6 mM CaCl2, pH 7.0. The apo- calbindin-D28k sample was dialyzed into 20 mM HEPES, 2 mM EDTA, pH 7.0
NMR Spectroscopy
NMR samples contained 0.5 mM of apo- or holo- calbindin-D28k in their appropriate buffers. A 1H-15N-HSQC-TROSY experiment was collected at 25 °C on a Varian Inova 600 MHz spectrometer using a 1H/13C/15N triple- resonance Z-gradient probe. Data were processed using NMRPIPE (37) and analyzed using NMRVIEW (38).
Differential Surface Modification Experiments
All surface reaction experiments were performed in triplicate or duplicate and analyzed individually as described below.
Lysine Acetylation
Both the apo- and holo- calbindin-D28k were acetylated in parallel, in their respective buffers as described above, using 1000-fold molar excess acetic anhydride with respect to amines. The pH was maintained at 7.0. The pH was monitored with a pH probe and 5 M NaOH was titrated in as necessary immediately after the addition of the acetic anhydride. Reactions proceeded at room temperature for 30 minutes and were then quenched by adding an equal volume of 1 M Tris, pH 8.0 in their respective buffers.
Histidine Modification
Both apo- and holo- calbindin-D28k were diethyl pyrocarbonate (DEPC)-modified in parallel, using 5x molar excess DEPC to histidines. Reactions proceeded at room temperature for 30 minutes in their respective buffers and then quenched by the addition of 10 mM histidine.
Circular Dichroism
CD spectra were measured using a Jasco J-600 spectropolarimeter (Jasco Inc. Easton, MD) using a 1 cm (near-UV) Hellma cuvette (Hellma Corp). A concentration of 40 uM for each protein sample was used. Spectra were recorded five times and averaged at 25 ± 1.0 °C from 250 to 320 nm. Acetylated samples were prepared as described above. Spectra were recorded in 20 mM HEPES 6 mM CaCl2, pH 7.0 for the holo-calbindin-D28k and 20 mM HEPES, 2 mM EDTA, pH 7.0 for apo- calbindin-D28k. Measurements were corrected for the appropriate buffer signal.
Enzymatic Digestion
Following each chemical modification reaction, the proteins were digested with trypsin, chymotrypsin, or GluC, at a protein:enzyme ratio of 20:1, at pH 8.0 in their respective buffers as described. Trypsin digests proceeded for 2 hours at 37°C. The chymotrypsin and GluC digests proceeded overnight at 25°C.
Mass Spectrometry
A Waters Q-Tof Ultima Premier mass spectrometer equipped with a nanoAquity UPLC system and NanoLockspray source (Waters, Milford, MA) was used for the acquisition of the LC/ESI/MS and LC/ESI/MS/MS data. Separations were performed using a 3 µm nanoAquity Atlantis column dC18 100 µm×100 µm (Waters) at a flow rate of 300 nL/min. A nanoAquity trapping column 5 µm C18 180 µm×200 mm (Waters) was positioned in-line with the analytical column. The digested samples were diluted to 0.5 pmol/µL just prior to analysis with their respective buffers as described above and a 2 µL aliquot was injected. Peptides were eluted using a linear gradient of 98% solvent A (water/0.1% formic acid (v/v)) and 2% B (acetonitrile/0.1% formic acid (v/v)) to 95% B over 60 minutes. Mass spectrometer settings for MS analysis were a capillary voltage of 3.5 kV, cone voltage of 30 V, and a collision energy of 8.0 V. The mass spectra were acquired over a scan range of 200–2000 Da. MS/MS data were acquired using a data dependent acquisition method, selecting collision energies based on mass and charge state of the candidate ions. Data analyses were performed using MassLynx 4.0 software (Waters, Milford, MA). Extracted ion chromatograms were used to calculate ratios of the abundances of modified peptides to unmodified peptides. In calculating the percent modification, only results for those ions that could be verified by MS/MS are reported.
A Waters Q-Tof Ultima Global hybrid tandem mass spectrometer (Waters, Milford, MA) was also used for the acquisition of ESI/MS and ESI/MS/MS data for the native apo- and holo- proteins and peptide digests. This instrument is equipped with a nanoflow ESI source and consists of a quadrupole mass filter and an orthogonal acceleration time-of-flight mass spectrometer. The needle voltage was ~3500 V and the collision energy was 10 eV for the MS analyses and 20–40 eV for the MS/MS analyses. Samples were diluted 1:1 just prior to analysis with a solution of 50:50 ACN:H2O (0.1% FA) and infused at ~300 nL/min using a pressure injection vessel.
Structural Model
Using molecular dynamics (MD) simulations, conformational changes in specific areas of the solution structures of apo- calbindin-D28k and non-reduced holo- calbindin-D28k were determined. The initial structures were based on the disulfide-reduced, holo- NMR structure [model 1 of the PDB entry: 2G9B] (2). For the apo- calbindin-D28k simulation, the coordinates of the NMR-determined structure were directly used for the initial model as the NMR structure more closely resembles the holo- structure than the apo- form. This apo- calbindin-D28k structure was solvated in a box of 16124 water molecules. For the holo- calbindin-D28k simulation, four calcium ions were introduced to EF hands 1, 3, 4 and 5 according to the information given in references 2 and 19. Also, for this case a disulfide bond was introduced between the sulfur atoms of cysteine residues 94 and 100. This holo- calbindin-D28k system was solvated in a box of 16127 water molecules.
Prior to structural equilibration, both systems were subjected to several stages of energy minimizations and relaxations under constant volume. As the first step, 100 ps belly dynamics runs on water molecules relaxed their initial positions while the protein was kept frozen. This was followed by an energy minimization step followed by a low temperature NPT (constant temperature/constant volume) step with frozen protein to obtain a reasonable starting density (around 1g/cc). After a complete energy minimization (10,000 conjugate gradient steps), and a step-wise heating procedure at constant volume (NVT, 200 ps), constant volume equilibration runs were carried out for one nanosecond at NVT at 300K for each system. The final trajectories were calculated at 300K under NPT for about 15 ns. During the production runs, no constraints were used in dynamics. All final MD runs were carried out with time steps of 1.0 fs and particle mesh Ewald method (39) was used to treat long range electrostatics in all simulations. The PMEMD module of the Amber10 molecular dynamics package was used for all energy minimizations and MD trajectory calculations. The force field parameters for all amino acid residues and the calcium ions were taken from the ff03 force field (40) included in the AMBER10 package (41). It should be noted that the 15 ns simulation is insufficiently long to observe large conformational changes but is sufficient to capture less drastic conformational changes occurring within specific regions of the molecule.
Results
Rat brain calbindin-D28k is a ~30 kDa protein comprised of 261 residues (Figure 1). Chemical surface modification of both lysine and histidine residues have been used to identify the regions of calbindin-D28k undergoing a conformational change upon calcium binding. The purity of the samples was confirmed using SDS-PAGE (Figure S1). In addition 1H/15N-HSQC-TROSY spectra confirmed the protein samples were pure and in the correct conformation as seen in previously published data (Figure S2) (2, 15). A 3D HNCACB spectrum (Figure S3) confirms that, under these conditions, no deamidation of the protein occurs at Asn203/Gly204. If deamidation was present, a negative crosspeak in the HNCACB spectrum would be seen at the 1HN, 15N shifts of Gly204 and the 13Cα chemical shift of Asn203(42). As is evident in Figure S3, such a crosspeak is not observed.
Figure 1. Amino acid sequence of rat brain calbindin-D28k.
Individual EF hand domains are designated on the left of the sequence. The amino acid sequence of each EF hand is subdivided into the individual structural components of the domain. Lysine and histidine residues are highlighted in bold. The line between cysteine 94 and cysteine 100 indicates the site of possible disulfide bond formation. Amino acid residues that were observed in the digests of the modification reactions are underlined; the solid line applies to the apo- form and the dashed line applies to the holo- form
Of the 261 residues, there are twenty-five lysine residues and four histidine residues. Lysines were modified by acetylation and histidines were modified to their N-ethoxycarbonyl derivatives. CD spectra of the near-UV region were acquired to confirm the modification reactions did not affect the conformation of the proteins (Figure S4). No gross conformational change is observed between the modified and non-modified proteins. However, local structural changes that may be caused by the modifications may have been missed. As expected, a large conformational change between the apo- and holo- calbindin-D28k proteins was observed. The reaction products were enzymatically digested and then analyzed by LC/ESI/MS/MS and sequence coverage of greater than 95% in the apo- state and greater than 90% for the holo- state was achieved after combining all enzymatic digestion results (Figure 1). In addition, 23 of the 25 lysine residues were observed in the apo- state and 22 of the 25 lysine residues were observed for the holo- state. Samples of both apo- and holo- calbindin-D28k that had not undergone surface modification were also analyzed and greater than 99% sequence coverage was achieved, with all lysine and histidine residues being observed. Additional acetylation products in which free cysteine thiol groups modified are also observed. No additional N-ethoxycarbonyl derivatized amino acid residues were observed. All sites of modification were confirmed by tandem MS.
Modification of lysine residues
Surface accessible lysine residues of calbindin-D28k in both the apo- and holo- states were acetylated under neutral pH conditions using excess acetic anhydride. The modified proteins were then subjected to proteolytic digestion by either trypsin, GluC, or chymotrypsin. Tryptic digestion results in the hydrolysis of the peptide backbone after either a lysine or arginine residue. Trypsin, however, will not cleave after an acetylated lysine, thus, resulting in a “missed-cleavage”. Independent of tryptic digestion, endoproteinase GluC which cleaves after glutamate and aspartate residues was also used in order to increase sequence coverage. The resulting peptides were analyzed using LC/ESI/MS and LC/ESI/MS/MS. Acetylated lysine residues were identified by a +42 Da mass shift for the [M+H]1+ charge state, a +21 Da mass shift for a [M+2H]2+ charge state and a +14 Da mass shift for a [M+3H]3+ charge state. The relative quantitation of peptides observed in both the acetylated and non-acetylated state were calculated from three independent surface modification data sets using extracted ion chromatograms (EIC’s). The absolute ion counts from the EIC’s of each form were compared and tabulated as the percent modification of the residue. Table 1 summarizes the calculated percentages for each lysine residue observed and ratios of the percent modification of the apo- to the holo- form. Several residues showed no difference in the extent of acetylation between the apo- and holo- forms of calbindin-D28k. For example, lysine residues 235, 236 and 246 were observed as more than 95% non-acetylated in both forms. Conversely, lysine residues 48, 49, 59, 72, 98, 105, 109, 133, 136, 142, 193, and 216 were observed to be greater than 95% acetylated in both the apo- and holo- calbindin-D28k. This indicates similar surface accessibility and/or reactivity in or around these residues in both forms. In both the apo- and holo- forms of calbindin-D28k, ions corresponding in mass to proteolytic peptides containing lysine residue 189 (EF hand 5) were not observed.
Table 1. Comparison of modification states between apo- and holo- calbindin-D28k.
The percent modification for each residue in each state were calculated from three different modification experiments (except as noted) and the mean determined. In addition, the ratio of the percent modification observed for each residue in the apo- and holo- calbindin-D28k were calculated. These ratios represent the amount of change that occurs between the two forms.
| Residuea,b | Peptidec,d,e | Apo-percent modified±SDf |
Holo-percent modified±SD |
Ratio of Apo- to Holo- |
|---|---|---|---|---|
| K34g | A25-35 | 100 | <0.1 | 100 |
| K48 | T48-68 | 96.2±3.8 | 98.1±1.9 | 0.98 |
| K49 | T48-68 | 96.2±3.8 | 98.1±1.9 | 0.98 |
| K59 | T48-68 | 96.2±3.8 | 98.1±1.9 | 0.98 |
| K72 | A58-77 | 95.0±3.6 | >100 | 0.95 |
| K98 | T94-108 | 100 | 100 | 1 |
| K105 | T94-108 | 100 | 100 | 1 |
| K109 | Apo:T109-128; Holo:f,gY108-117 |
100 | 100 | 1 |
| K124 | Apo:T109-128; Holo:* |
100 | hNC | NC |
| K128 | Apo:T109-128; Holo:* |
<0.1 | NC | NC |
| K133 | A133-145 | 100 | 100 | 1 |
| K136 | A133-145 | 100 | 100 | 1 |
| K142 | A133-145 | 100 | 100 | 1 |
| K152 | gA149-163 | 100 | <0.10 | 100 |
| K161 | gA149-163 | 100 | <0.10 | 100 |
| K180 | T170-180 | 14.0±3.6 | 31.4±4.6 | 0.45 |
| K185 | T170-180 | 14.0±3.6 | 31.4±4.6 | 0.45 |
| K189 | * | NC | NC | NC |
| K193 | A191-196 | 100 | 100 | 1 |
| K216 | A211-220 | 96.3±2.1 | 97.0±2.6 | |
| K221 | Apo*; Holo: T217-221 |
*NC | <0.1 | NC |
| K223 | T222-236 | 44.0±8.5 | 28.7±3.2 | 1.5 |
| K235 | T222-236 | <0.10 | <0.10 | 1 |
| K236 | T222-236 | <0.10 | <0.10 | 1 |
| K246 | T237-246 | <0.10 | <0.10 | 1 |
| H5 | A4-18 | 64.0±1.2 | 22.0±8.1 | 2.9 |
| H22 | gA19-24 | 51±0.5 | 32.0±5.8 | 1.6 |
| H80 | A78-86 | 60.0±3.0 | 60.0±9.9 | 1 |
K = lysine residue
H = histidine residue
A refers to GluC digest peptide
T refers to trypsin digest peptide
Y refers to chymotrypsin digest peptide
SD=standard deviation, no SD given when one form not observed
average of two analyses
NC= unable to be calculated
=residue unobserved
Many other lysine residues, however, showed differences in the extent of acetylation between the two forms of the protein. For example, lysine 34 (EF hand 1) in the apo- form was identified as acetylated in the GluC peptide corresponding to amino acid residues 25–35 (ADGSGYLEGKE). The theoretical [M+2H]2+ for this peptide is m/z 563.262+. However, an ion of m/z 584.302+ was observed which corresponds in mass to this peptide plus one acetyl group. From the MS/MS data of this ion, lysine 34 was identified as the site of acetylation. Conversely in the holo- form, this peptide was observed as unmodified based on the observation of an ion of m/z 563.292+ and its corresponding MS/MS spectrum. These data identify lysine 34 as being acetylated in the apo- form and non-acetylated in the holo- form of calbindin-D28k.
Lysine residues 48, 49 (linker region between EF hands 1 and 2), and 59 (EF hand 2) are all contained within one tryptic peptide corresponding to amino acid residues 48–68. In both the apo- and holo- forms an ion of m/z 851.103+ was observed which corresponds to an m/z increase of 42 which translates to a mass increase of 126 Da over the expected ion of m/z 809.093+ for this amino acid sequence. For the [M+3H]3+ charge state this mass increase correlates to the addition of three acetylation modifications for both the apo- and holo- forms. MS/MS analysis confirmed the acetylation of lysine residues 48, 49 and 59 (Figure S5). This peptide was also observed, albeit to a much lower extent, with no acetylated residues as indicated by the observation of two peptides corresponding to ions with no modifications in both the apo- and holo- forms of the protein. One of these tryptic peptides corresponds to residues 48–59 and was observed in both the apo- and holo- forms of the protein. The other tryptic peptide corresponds to residues 50–59. This peptide is formed by cleavage after lysine 49 which indicates that lysine 49 was not modified, as well as lysine 59. The abundances of the peptides arising from or containing an unmodified lysine are less than 2% of the abundance of the triply acetylated peptide. To calculate the extent to which each peptide was modified, extracted ion chromatograms (EIC’s) were generated and the abundances of the triply-acetylated peptide 48–68 and the unmodified peptides were compared. For the tryptic peptide corresponding to residues 48–68, this percentage was 98.1±2.0 for the holo- form and 96.2±3.9 for the apo- form (Table 1).
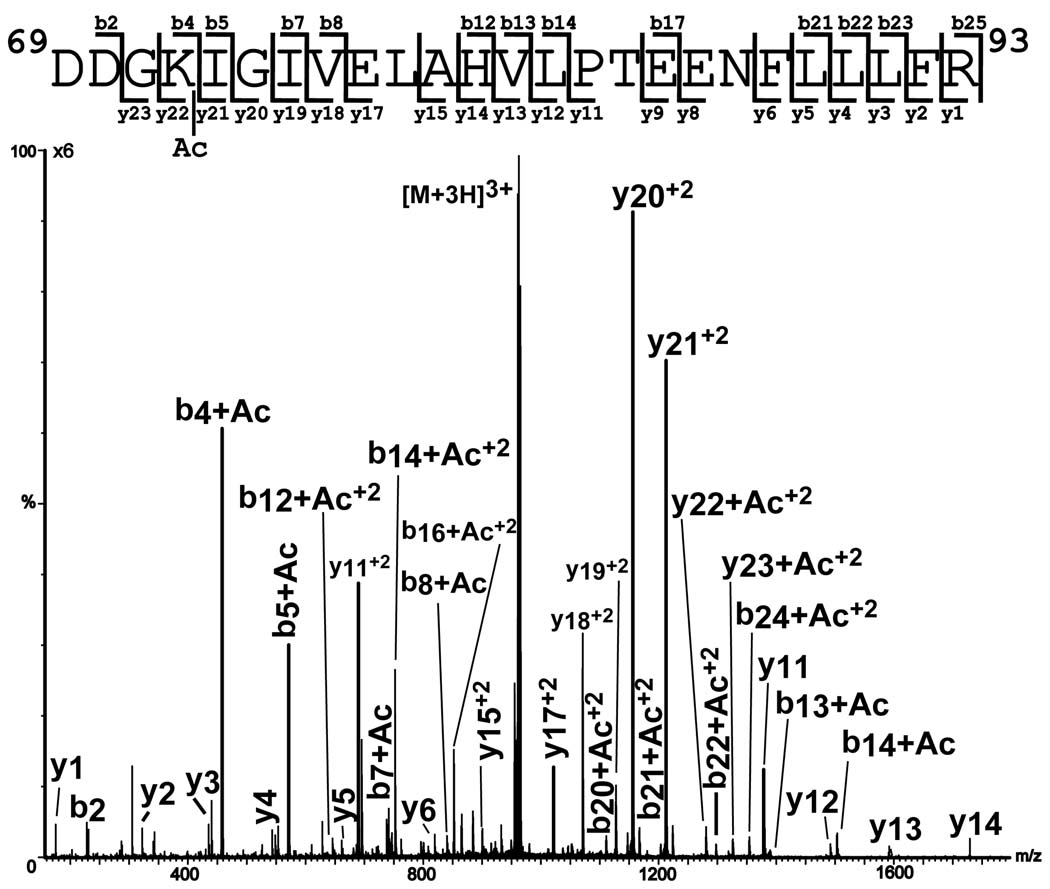
Ions corresponding in mass to the unmodified and mono-acetylated tryptic peptide comprising residues 69–93, which contains lysine residue 72 of EF hand 2, were observed at m/z 946.903+ and m/z 960.963+, respectively, in the apo- form. In the holo- form, only an ion corresponding to a mono- acetylated peptide was observed. The MS/MS data of the ion of m/z 960.963+ from the apo- form is shown in Figure 2. A nearly complete series of both b and y ions are observed which correspond to cleavages along the peptide backbone (43, 44). The y series ions result from C-terminal peptide backbone cleavages and the b series ions result from N-terminal backbone cleavages. Many of these ions correspond to the backbone cleavage plus an acetyl group; thereby providing the necessary data to definitively assign the location of the acetyl modification to lysine 72. To compare the extent of acetylation in the apo- form EIC’s were generated. When calculating the percent of modified peptide, EIC’s for the GluC peptide 58–77 were used instead of the tryptic peptide 69–93 because the GluC digests yielded peptides with identical amino acid sequences (with or without acetylation) while the trypsin digest yielded multiple peptides arising from cleavages of unmodified lysines as well as peptides with ‘missed-cleavages’ arising from modified lysines. Comparison of the counts showed that this peptide was modified 95% ± 5 in the apo- form. These data indicate that lysine residue 72 is found to be in a mainly acetylated state in both the apo- and holo- states. Lysine 72 is located in the loop of EF hand 2, which does not bind calcium. This similarity between the two forms indicates this region does not undergo a significant change upon binding calcium.
Figure 2. MS/MS spectrum of the tryptic peptide corresponding to residues 69–93 plus one acetyl group in apo- calbindin-D28k.
MS/MS spectrum of the [M+3H]3+ ion of m/z 960.96 from apo- calbindin-D28k, corresponding in mass to tryptic peptide 69–93 plus one acetyl group (Ac) on lysine 72. A multiplication factor of six was applied to the MS/MS spectrum.
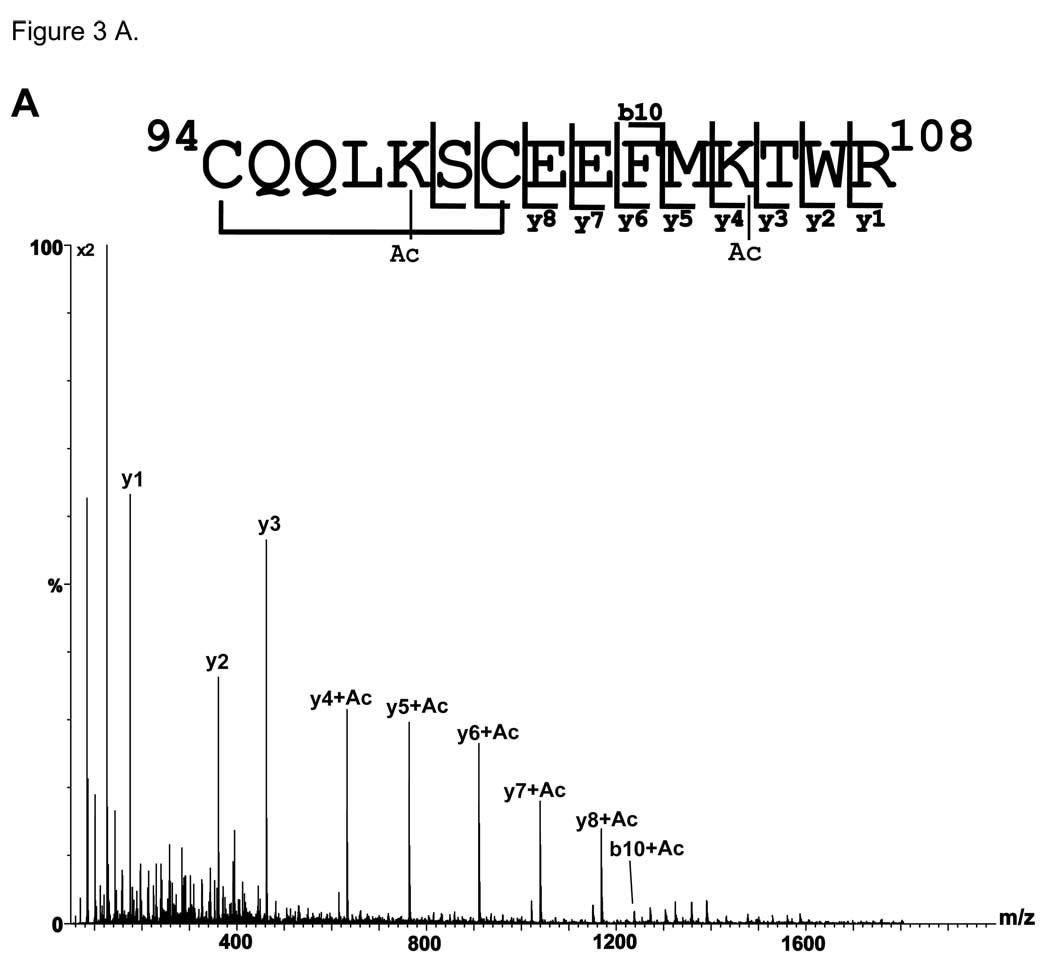
Tryptic peptide 94–108 has two lysine residues, 98 and 105 (EF hand 3). The theoretical m/z value of the [M+2H]2+ of this peptide is m/z 958.942+. In the tryptic digests of the apo- and holo- calbindin-D28k, doubly-charged ions of neither m/z 958.94 (corresponding to no acetylation) nor m/z 979.932+, corresponding to one acetylation, were observed. However, an ion of m/z 1000.002+ was observed in both the apo- and holo- form. This is 1 Da lower than the expected m/z ion corresponding to the doubly charged tryptic peptide 94–108 plus two acetyl groups (theoretical m/z=1000.962+). This 1 Da decrease in the [M+2H]2+ ion corresponds to a 2 Da mass shift in the [M+H]1+ ion; thereby implicating the presence of a disulfide bond between cysteine residues 94 and 100 within this peptide. To verify the nature of this ion, the MS/MS data were acquired and are shown in Figure 3A (holo) and Figure S6A (apo). The mass difference between the y3 and y4 ions (i.e. 170 Da) corresponds to an acetylated lysine residue at lysine 105. The observed m/z values corresponding to the y1–y8 cleavages are the same as the predicted m/z values for these fragment ions (i.e. no 1 or 2 Da mass shift observed for these ions). The mass of the b10 ion is also that of the predicted m/z value for the ion containing one acetylation and a disulfide bond. Thus, the presence of a disulfide bond between cysteine 94 and cysteine 100 as well as the sites of acetyl modification at lysine 105 can be determined from these data for the doubly acetylated form. Low abundance ions are also observed above mass 1200, possibly due to rearrangements and fragmentations within the disulfide ring.
Figure 3. MS/MS spectra of tryptic peptides corresponding to residues 94–108 in calbindin-D28k.
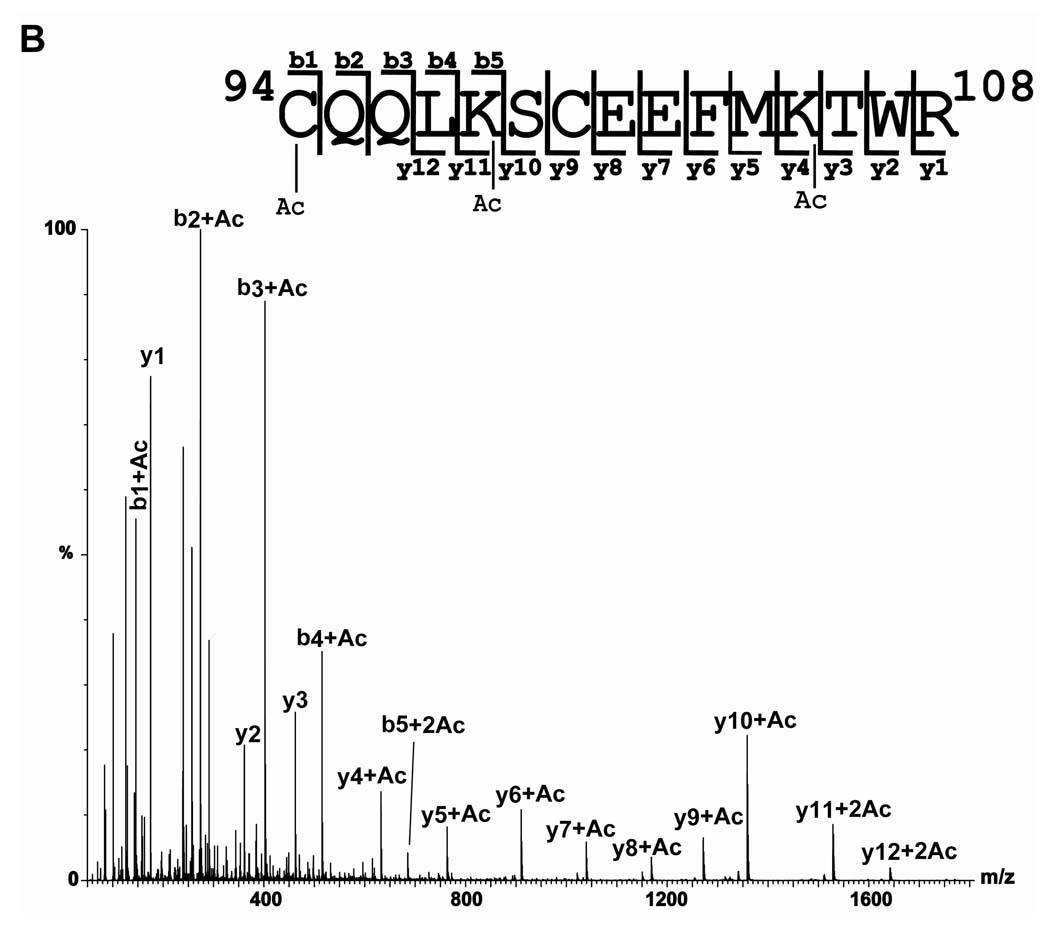
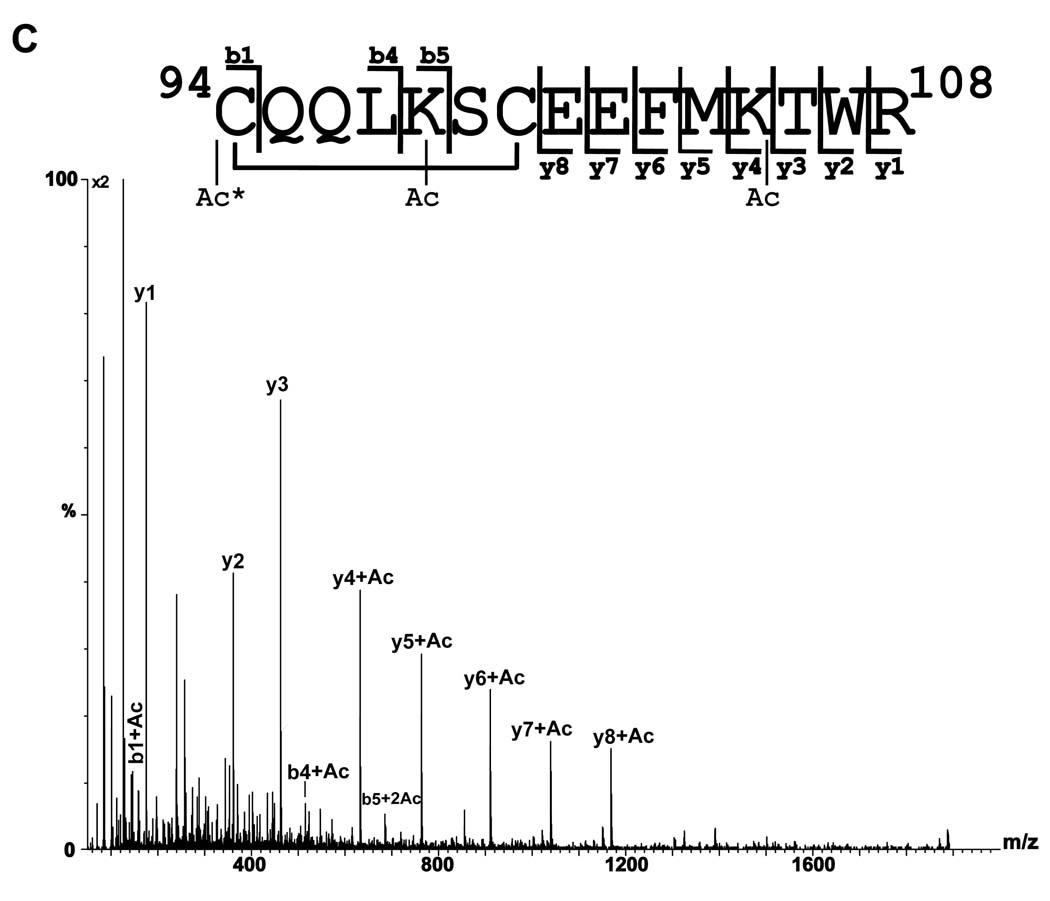
A) MS/MS spectrum of the [M+2H]2+ ion of m/z 1000.00 from the holo- form corresponding in mass to tryptic peptide amino acids 94–108 plus two acetyl groups (Ac). The Ac groups were determined to be located at Lys98 and Lys105. A disulfide bond between Cys94 and 100 was observed and is designated by a black line connecting the two residues. A multiplication factor of two was applied to the MS/MS spectrum. B) MS/MS spectrum of the [M+2H]2+ ion of m/z 1021.90 from the apo- form corresponding in mass to tryptic peptide amino acids 94–108 plus three acetyl groups (Ac). C) MS/MS spectrum of the [M+2H]2+ ion of m/z 1020.90 from the holo-form corresponding in mass to tryptic peptide amino acids 94–108 plus three acetyl groups (Ac). Ac* indicates N-terminal acetylation. A multiplication factor of two was applied to the MS/MS spectrum.
Ions corresponding to the addition of three and four acetylations were also observed in tryptic peptide 94–108. These ions have a theoretical m/z of 1021.962+ and 1042.962+ respectively. In the apo- form, an ion with an m/z of 1021.912+ was observed, corresponding to the addition of three acetyl groups. The MS/MS spectrum contains abundant acetylated b ions (b1 through b4) indicating acetylation of cysteine 94, a diacetylated b5 ion, and a y series indicating acetylation at lysine 98 and lysine 105 (Figure 3B). In the holo- form an ion with an m/z of 1020.922+ was observed which is that expected if the disulfide bond is intact. In the MS/MS analysis of the triacetylated holo-form there are no abundant acetylated b ions as observed in the MS/MS spectrum of the triacetylated apo-form. Instead the MS/MS spectrum is dominated by y-ions through y8, which correspond to cleavage being initiated at the disulfide bond cyclic system. This indicates that the second and third acetylation occur in this ring, The b1 ion indicates acetylation of cys-94 and the mass difference between the b4 and b5 ion locates the second acetylation in the disulfide ring as being at lysine 98 (Figure 3C).
In the peptide corresponding to the addition of four acetyl groups in the apo- form, MS/MS analysis confirmed diacetylation of cysteine 94 and acetylation of both lysines 98 and 105 (Figure S6B). There are, however, several relatively low abundance ions indicating some acetylation is also occurring at cysteine 100, the (y10 + 2Ac) and (y9 + 2Ac) ions. Thus, acetylation predominantly occurs at the lysines followed by acetylation of the N-terminal cysteine. The latter acetylation probably occurs after digestion while still in the presence of excess acetic anhydride. The MS/MS spectrum of the quadruply acetylated peptide from the holo- form (Figure S6C) is similar to the quadruply acetylated apo-peptide (Figure S6B).
Because the MS/MS of the doubly acetylated apo- form indicated the presence of a disulfide bond (Figure S6B) while the mass of the triply acetylated ion was consistent with a reduced disulfide, we used EIC’s to determine the extent of disulfide-bond formation as well as the extent of modification of this peptide in both the holo- and apo- forms. For the apo- form, the relative abundance of the disulfide bonded diacetylated peptide corresponds to 16% of the summed relative abundances of the plus 2–4 acetylated peptides while, in the holo- state, the relative abundance of peptides including a disulfide bond correspond to 76% of the summed relative abundances of the plus 2–4 acetylated peptides. From these calculations it is clear that the holo- form exists predominantly with a disulfide bond while the apo- form exists predominantly in a reduced state.
Lysine residues 109, 124 and 128 (EF hand 3) are located in tryptic peptide 109–128. An [M+3H]3+ ion of m/z 805.403+ was expected for the unmodified peptide, however, in the apo- form an m/z of 833.503+ was observed. This mass difference corresponds to the addition of two acetyl groups to this peptide. MS/MS analysis identified and confirmed lysine residues 109 and 124 as acetylated and lysine 128 as non-acetylated (Figure S7). Of note, however, this peptide was not observed in the holo- form. In the chymotrypsin digest of the holo-form, however, a peptide corresponding to residues 108–117 was observed with an m/z of 634.212+, a 21 Da increase over the expected m/z of 613.282+. MS/MS analysis identified lysine 109 as acetylated in this peptide generated from the holo- form of the protein. No peptides containing lysine residues 124 and 128 were observed in the tryptic digest of the holo- protein, therefore, the modification state of these lysine residues was unable to be determined. We speculate, however, that these two residues might both be acetylated in the holo- form. This would result in a missed cleavage by trypsin and, therefore, a large peptide (aa 109–152) with a theoretical mass of at least 5165.831+ would be expected due to the acetylation pattern of lysine residues 109, 124, 128, 133, 136, and 142. No ions corresponding to multiply charged forms of possible peptides resulting from modification of these residues were observed in this range in the MS spectra with an m/z of 2000 or less. There were also no corresponding peptides observed in the GluC digestion.
Lysine 152 is located in helix 1 of EF hand 4. It was observed as 100% modified in the apo- form as a GluC peptide 149–163. No peptide containing lysine 152 was observed in the holo- form. We did, however, observe the tryptic peptide 153–161, indicating that lysine 152 was unmodified to some extent. Lysine 161 is located in the loop of EF hand 4 and was observed as 100% modified in the apo- form, while the holo- form was observed to be 100% unmodified.
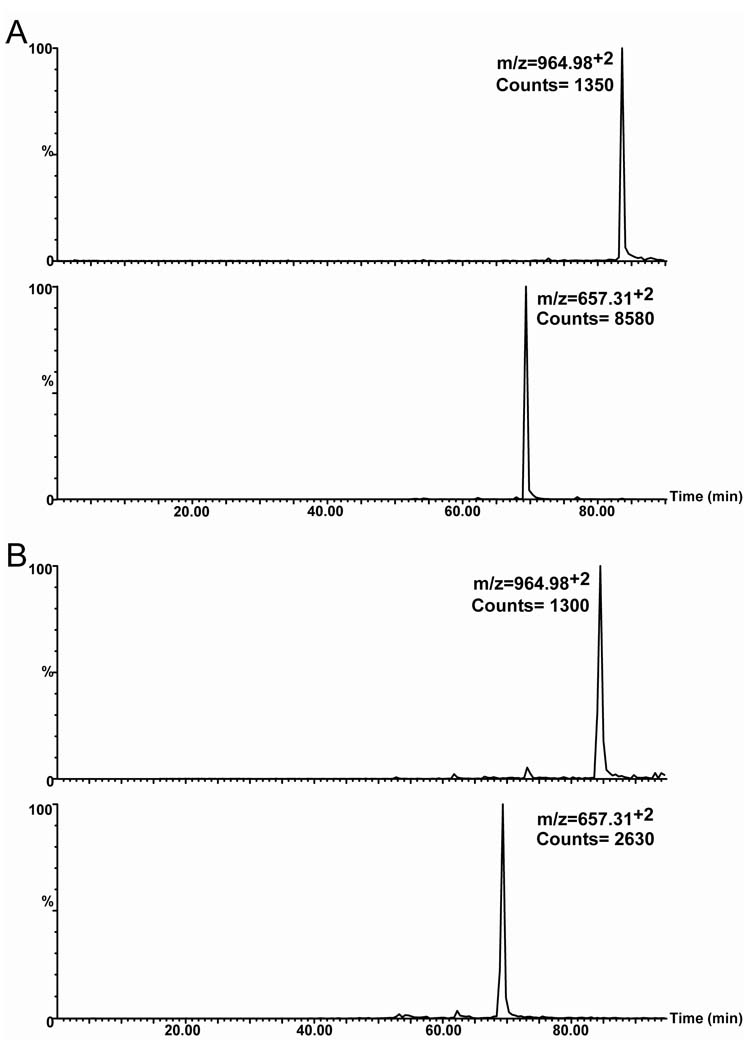
Lysines 180, 185 and 189 are located in the linker region between EF hands 4 and 5. No ions were detected for peptides containing lysine 189. However, lysines 180 and 185 underwent a significantly greater extent of modification in the holo- form (31.4%± 4.62) than in the apo- form (14.0% ± 3.6). These percentages are based on the extracted ion chromatograms (EIC’s) of the ions corresponding to peptides from this linker region.. In this case, we compared the EIC for m/z 964.982+ of the tryptic peptide corresponding to residues 170–185, where lysine 180 is in a modified state and the EIC of tryptic peptide 170–180, m/z of 657.312+, where lysine 180 is in an unmodified state (Figure 4). No corrections were made for differences in ionization efficiency of the two peptides.
Figure 4. Extracted ion chromatograms of the acetylated and non- acetylated peptides corresponding to lysine 180.
A) EIC’s of the apo- calbindin-D28k tryptic peptides corresponding to the modification state of K180. Top EIC corresponds to the mono- acetylated tryptic peptide (amino acids 170–185), m/z of 964.982+. The bottom EIC corresponds to the unmodified K180 (amino acids 170–180), m/z of 657.312+. B) EIC’s of the holo- calbindin-D28k tryptic peptides corresponding to the modification state of K180. Top EIC corresponds to the mono- acetylated tryptic peptide (amino acids 170–185), m/z of 964.982+. The bottom EIC corresponds to the unmodified K180 (amino acids 170–180), m/z of 657.312+.
Lysines 193 and 216 are located in EF hand 5, in helix 1 and helix 2, respectively. Lysine 193 was observed as 100% modified in both the apo- and holo- forms. The GluC peptide corresponding to residues 211–220 (LDALLKDLCE) contains lysine residue 216 and cysteine 219. Peptides corresponding in mass to an unmodified peptide, a mono-acetylated peptide and a di-acetylated peptide were observed in both the apo- and holo- forms of the protein. MS/MS verified that only lysine 216 was modified in the mono-acetylated peptide (Figure S8). The acetylation of cysteine 219 was only observed in the diacetylated peptide. Lysine 216 was found to be mostly modified in both the apo- and holo- forms. To calculate the modification percentages, the mono- and di-acetylated peptide EIC counts were added together. A modification percentage of 96.3% ± 2.1 was observed in the apo- form and 97.0% ± 2.6 in the holo- form. This indicates that this region in EF hand 5 is not affected by the binding of calcium. EF hand 5 also contains four additional lysine residues (K185, K189, K221, and K223); all of which are found in the linker regions. As mentioned previously, no ions were observed corresponding to any proteolytic peptides containing K189 in either the apo- or holo- calbindin-D28k digests. Ions corresponding in mass to the non-acetylated K221-containing tryptic peptide were observed in the digest of the holo- protein, but these same ions were not observed in the digest of the apo- form. What was observed, however, was the ion corresponding to tryptic peptide 222–236. The observation of this ion implies that lysine 221 is not acetylated because cleavage after lysine 221 is required for the formation of the tryptic peptide 222–236. If K221 were acetylated, tryptic peptide 222–236 would not be observed.
Modification of histidine residues
The surface accessibility of histidine residues in calbindin-D28k was investigated using DEPC modification to the N-ethoxycarbonyl derivative. Following DEPC-modification and enzymatic digestion, three of the four histidine residues in the sequence were observed in the MS and MS/MS analyses, histidine residues 5, 22, and 80. No proteolytic peptide ions that could be confirmed as containing histidine 114, modified or unmodified, were observed in any of the analyses of the modification reactions. For histidine 5 in EF hand 1, [M+2H]2+ ions of m/z 847.902+ and of m/z 883.982+ were observed in both the apo- and holo- form GluC digests. An expected theoretical mass for an [M+2H]2+ ion for an unmodified peptide containing residues 4–18 would be 847.922+. Therefore, the observed ions would correspond to a non-modified peptide and a singly modified peptide, respectively. MS/MS data verified that this peptide is present as both DEPC-modified (Figure S9) and unmodified in the apo- as well as the holo- form of the protein under the experimental conditions used. In the apo- form, the extent of DEPC-modification was 63.6% ± 1.2 (calculated from the EIC’s for the GluC peptides containing histidine 5). In the holo- form, the percent DEPC-modification was 22.0% ± 8.1. These data imply that histidine 5 becomes less surface accessible upon calcium binding. Similarly, for histidine 22 (EF hand 1), ions corresponding to the GluC peptides were observed for both DEPC-modified and unmodified peptides in both states of the protein. Relative quantitation using ion counts from the extracted ion chromatograms show the extent of DEPC-modification to be of 31.5% ± 5.8 for the holo- form versus 50.5%±0.5 for the apo- form (Table 1).
Ions corresponding to both DEPC-modified and unmodified GluC peptides were observed for histidine 80 (EF hand 2) in both forms of the protein. MS/MS analysis of the [M+2H]2+ ion of m/z 540.77, corresponding to the mono- N-ethoxycarbonyl derivative, confirmed histidine 80 as the site of modification. This residue was observed to be modified to the same extent (60%) in both the apo- and holo- proteins (Table 1). These data indicate no change in accessibility of histidine 80 upon calcium binding. For both forms, two distinct chromatographic peaks were observed in the LC/MS extracted ion chromatogram, m/z 540.772+, for the DEPC-modified GluC peptide (Figure S10). To verify the identity of these ions, the corresponding MS/MS data were acquired from each of the chromatographic peaks (Figure S11). These MS/MS spectra were essentially identical and confirm the amino acid sequence as LAHVLPTEE with the histidine residue identified as DEPC-modified. No evidence for threonine acetylation was observed in either tandem MS (Figure S11). It has been shown that DEPC is able to modify both tautomeric states of histidine (45). If histidine 80 exists in both tautomeric states, then two structurally distinct DEPC products could form; thereby giving rise to two distinct chromatographic peaks. To calculate the percent modification of His 80, ion counts from the extracted ion chromatograms of each peak were added together and compared to the extracted ion chromatogram for the unmodified peptide.
Molecular Dynamics
Molecular dynamics simulations yielded stable solution configurations after 10 ns. Root mean square deviations (RMSD) calculated for the backbone atoms, with respect to the NMR structure of holo- disulfide-reduced calbindin-D28K as the reference configuration, can be used to gauge the achievement of stability (Figure S12 and S13). For the apo- structure, the observed overall RMSD is slightly larger than the corresponding RMSD of the holo- structure, an average of 3.7 Å and 2.9 Å, respectively. Most individual EF hand domains maintain relatively stable folds in both the apo- and holo- systems (Figure S14). From the RMSD of the molecular dynamics simulations, the helices stay relatively close to their orientations with respect to those observed in the NMR structure. The exceptions are EF hands 3, 5 and 6 of the apo- structure (Figure S12A, blue, brown and orange lines, respectively) and EF hands 4 and 6 of the holo- structure (Figure S13A, yellow and orange, respectively) (showing an average RMSD over 2 Å). For peptides connecting adjacent EF hands, the region between EF hands 4 and 5 (Figure S12B, blue line) in the apo- and the regions between EF hands 2 and 3, and EF hands 4 and 5 (Figure S13B, blue and brown lines, respectively) in the holo- display the largest deviations compared with corresponding segments of the NMR structure. In the case of the peptide between EF hands 2 and 3 in the holo- system, the disulfide bond formation may lead to the observed differences.
Discussion
Calbindin-D28k is a unique calcium binding protein that functions as both a calcium sensor and buffer in eukaryotic cells. Although an NMR structure of disulfide-reduced, holo- calbindin-D28k has been published (2), no structure for apo- calbindin-D28k or for holo- calbindin-D28k with an intact disulfide bond has been reported. This study used differential surface modification in combination with mass spectrometry to identify the regions of calbindin-D28k affected by the conformational change that occurs upon calcium binding. As mentioned in the introduction, NMR titration experiments indicate that calbindin-D28k passes through a disordered state upon transitioning from the apo- form to the holo- form. Based on this, we expect disulfide-bridged holo- calbindin-D28k to be more structurally similar to the disulfide-reduced holo- structure than to the apo- structure, and we will discuss structural differences between the apo- and holo- forms in these terms. We will also discuss specific structural changes between disulfide-reduced and disulfide-bridged holo- forms based on and consistent with specific differential chemical reactivities.
Disulfide Bond
Rat brain calbindin-D28k contains four cysteine residues, 94, 100, 186 and 219. It has been shown in the homologous human brain calbindin-D28k that the two N-terminal cysteine residues 94 and 100 are essential to the function of calbindin-D28k (46). Previous research has shown that a disulfide bond between cysteines 94 and 100 is present in rat brain holo- calbindin-D28k (47). It is unclear from this report, however, whether this disulfide bond forms in both the apo- and holo- calbindin-D28k. Our results indicate that upon calcium-binding, calbindin-D28k exists predominantly with a disulfide bond between cysteines 94 and 100. The presence of a disulfide bond was also detected for the apo- form to a limited extent. However, it is possible that the tryptic digestion conditions (pH 8) oxidized cysteines 94 and 100 in the apo- form (48). In the apo- form, cysteine 94 is modified 84% and cysteine 100 52% of the time. In the holo- form, however, modification of cysteine 94 only, occurred at a rate of 24%. These results indicate that the formation of a disulfide bond is calcium dependent. This confirms previous speculation that the formation of a disulfide bond in the calcium bound form is involved in fine tuning the surface hydrophobicity, which could play a role in the ability of calbindin-D28k to interact with different target proteins or the affinity with which it binds these different targets (2).
EF Hand 1
The first linker region (residues 1–15) of EF hand 1 contains a histidine residue at position 5. The ratio of the percents of DEPC-modification of the histidine 5 peptide in the apo- to holo- forms is 2.9 indicating a significant increase in reactivity of this residue in the apo- form, and is thus predicted to be in a more flexible, less protected conformation. The helix-loop-helix of EF hand 1 extends from amino acid 16 through amino acid 44. Within this region, the reactivity of two different residues was evaluated, histidine 22 and lysine 34. Histidine 22 is located in the initial helix of the EF hand and our modification results indicate a somewhat increased reactivity/accessibility for this residue (percent ratio = 1.6) in the apo- form. The loop region of this EF hand contains lysine residue 34. The differential modification experiments indicate a significant difference in reactivity of this residue between the holo- and apo- forms with the apo- form only detected as modified while the holo- form was only detected as unmodified. This indicates that this region undergoes a significant conformational change leading to a more surface exposed lysine residue in the apo- form. The linker region between EF hands 1 and 2 contains two lysines (K48 and K49). These residues were observed as part of a relatively long peptide that also contained lysine 59 of EF hand 2. This peptide showed only a minor to no difference in reactivity between the holo- and apo- forms, indicating that there may be little structural change in this linker between the two forms.
EF hand 2
EF hand 2 contains lysine 59 (helix 1), lysine 72 (loop) and histidine 80 (helix 2). As noted above, there was little or no change in the reactivity of lysine 59 between the holo- and apo- forms, and lysine 72 (located in the middle of the loop in EF hand 2) also showed little change in reactivity. As this hand does not bind calcium, the similar reactivities shown by K72 between the apo-and holo- forms is not unexpected. Histidine 80 is located in helix 2 and is three residues C-terminal to the EF hand 2 loop. Histidine 80 was also observed to be unchanged in the extent of modification, again, indicating that the tertiary structure in this region does not differ significantly on calcium binding.
Linker region between EF hands 2 and 3
Lysine 98 is located in the linker region between EF hand 2 and EF hand 3, and is located between cysteines 94 and 100. From these analyses, lysine 98 was observed solely as acetylated in both apo- and holo-calbindin-D28k. These data indicate that the absence or presence of calcium with or without the presence of the disulfide bond does not significantly affect the conformation of this region. A recent study used NMR chemical shift perturbation experiments to identify the potential binding interface of holo- calbindin-D28k with peptides of previously identified protein binding partners (49). This type of experiment identified select residues that show significant chemical shift changes, line broadening or both upon peptide binding, giving site specific interaction information. It was determined that the region including part of the second helix of EF hand 2 and the linker region between EF hand 2 and EF hand 3 (residues 82–100) as a dominant site of peptide binding in the holo- form. Our data show that this region is equally accessible in both forms, suggesting that this region is available for interaction in the apo- form as well. This discovery makes this region of great interest in understanding how calbindin-D28k functions as both a buffer and sensor protein as EF hand 2 is not involved in calcium binding.
EF hand 3
Several lysine residues and a histidine residue are found in the amino acid sequence comprising EF hand 3. No histidine 114-containing proteolytic peptide ions, however, were observed from the histidine modification reactions. Lysines 105 and 109 are located in helix 1 of this hand and do not exhibit any change in reactivity between the two forms under our experimental conditions. Lysines 124 and 128 are located in helix 2. In the apo- protein, lysine 124 is found acetylated and lysine 128 is found non-acetylated. No peptide containing either of these residues was detected in the holo- protein digest. Interestingly, these residues were observed in both tryptic and GluC peptides in a digest of a holo- control protein that had not undergone any chemical modification reactions, suggesting that, in the acetylated holo- protein, both of these lysines are modified (in addition to the confirmed sites of modification at lysines 109, 133, 136 and 142) resulting in six missed cleavages and a high mass peptide which may result in reduced sensitivity in the mass spectrometer. Thus, we hypothesize that both lysines are fully modified in the holo- form (as are lysines 133 and 136 that would be part of that tryptic peptide, see below), while only lysine 124 and not lysine 128 is modified in the apo- form. These data are consistent with a major conformational change occurring in this region. The linker between EF hands 3 and 4 contains lysines 133, 136 and 142. Because all three lysines were observed to be fully modified in both the holo- and apo- forms, this indicates an open conformation in this region in either state.
EF Hand 4
This EF hand contains one lysine residue in the first helix (K152) and one lysine residue in the loop (K161). Both of these lysines are observed to be modified in the apo- form, indicating an open conformation. No peptide containing lysine 152 was detected in the holo- form. However, the tryptic peptide 153–161 was observed indicating that lysine 152 did exist to an indeterminate extent in the unmodified state in the holo-form. This is consistent with a decrease in accessibility in the holo- state. Lysine 161 is found 100% modified in the apo- form, but no modified lysine 161 was observed in the holo- form. Thus, the lysine 161 results are also consistent with a decrease in accessibility in this region in the holo- form. The linker between EF hand 4 and EF hand 5 contains lysines 180, 185 and 189. Both lysines 180 and 185 are more readily modified in the holo- form (31.3%±4.62) than in the apo- form (14%±3.6) indicating that these lysines in the holo- form are less constrained than in the apo- form. Proteolytic peptides containing lysine 189 were not observed in our experiments.
EF Hand 5
The amino acid sequence of the helix-loop-helix of EF hand 5 contains two lysine residues, K193 and K216. Lysine 193 is located in helix 1 and was observed only modified in both forms of the protein, indicating an open conformation for this helix. Lysine 216 was also observed greater than 95% modified in both forms. These results indicate that there is little change in protection of these residues. The linker between EF hand 5 and 6 contains lysines 221 and 223. Peptides containing lysine 221 were not observed in the digest of the apo- form although a lysine 221-containing peptide was observed in digests of the holo- form as being unmodified. These data are more consistent with a conformational change in this region to a more open state in the apo- form in that one would expect to see a K221 containing peptide in the apo- form if it were unmodified.
EF Hand 6
The sixth EF hand of calbindin-D28k contains lysines 235, 236, and 246. All three residues were observed only acetylated in both the holo- and apo- forms. Therefore, no difference in reactivity between the holo- and apo- forms was observed, indicating little or no conformational change.
Structural Consequences
Major differences in reactivity were observed in several regions of the protein: the initial linker; helix 1 of EF hand 1; the loop of EF hand 1; the first helix and the loop of EF hand 4; the linker between EF hands 4 and 5; and the first part of the linker to EF hand 6. The very significant differences in reactivities observed in EF hand 1, EF hand 4 and the linker regions on both sides of EF hand 5 are consistent with the significant conformational changes between these two forms observed by CD [Figure S4 and (21)]. The results presented here provide specific information about the regions of the molecule undergoing significant changes in conformation.
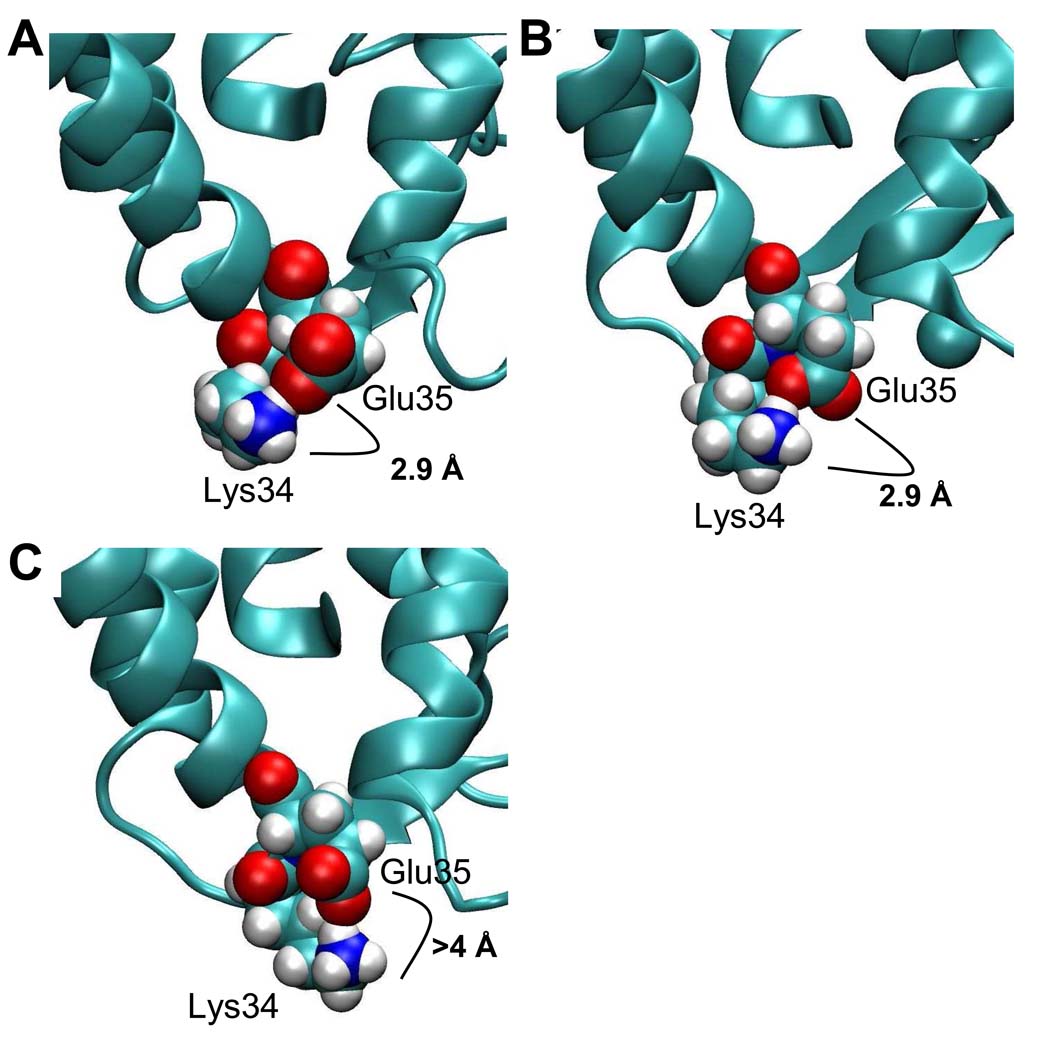
Using molecular modeling, structural explanations for differences in the reactivity for several specific residues are proposed. Molecular dynamics simulations over 15 ns were then used to obtain a model of conformational changes in specific regions of the solution structure of apo- calbindin-D28k as well as the non-reduced holo- calbindin-D28k. It should be noted that the 15 ns dynamic window may not be long enough to capture extensive overall conformational changes in its entirety. The initial coordinates for these simulations were based on the disulfide-reduced, holo- NMR structure (PDB: 2G9B). The chemical modification data for lysine 34 of the EF hand 1 loop indicate this region as being involved in a conformational change between the apo- and holo- forms. A comparison of the NMR structure of the reduced holo-, our non-reduced holo- model and our apo- model (Figure 5 A–C) shows that lysine 34 is more solvent exposed and away from any potential salt bridge-forming partners (>4 Å) in the apo- form whereas in the disulfide-containing holo- form model, lysine 34 is able to form a salt bridge with glutamic acid 35 (~2.9 Å). Salt bridges are thought to play an important role in the structure and function of a protein. Salt bridges can form between oppositely charged amino acid residues that are within a maximum of 4 Å of each other, but a more typically accepted range is less than 3.5 Å (50, 51).
Figure 5. Comparison of the NMR structure to our structural models around residues lysine 34 and glutamic acid 35.
Images of A) NMR structure with a reduced disulfide bond. B) holo- model with a non-reduced disulfide bond and C) apo- model with reduced disulfide bond. Glu35 and Lys34 are shown in space-filled form. Atoms are represented by blue=N, red=O, white=H, and yellow=C. Also, in panels A, B and C the averaged NZ to OE1 or OE2 distances are shown.
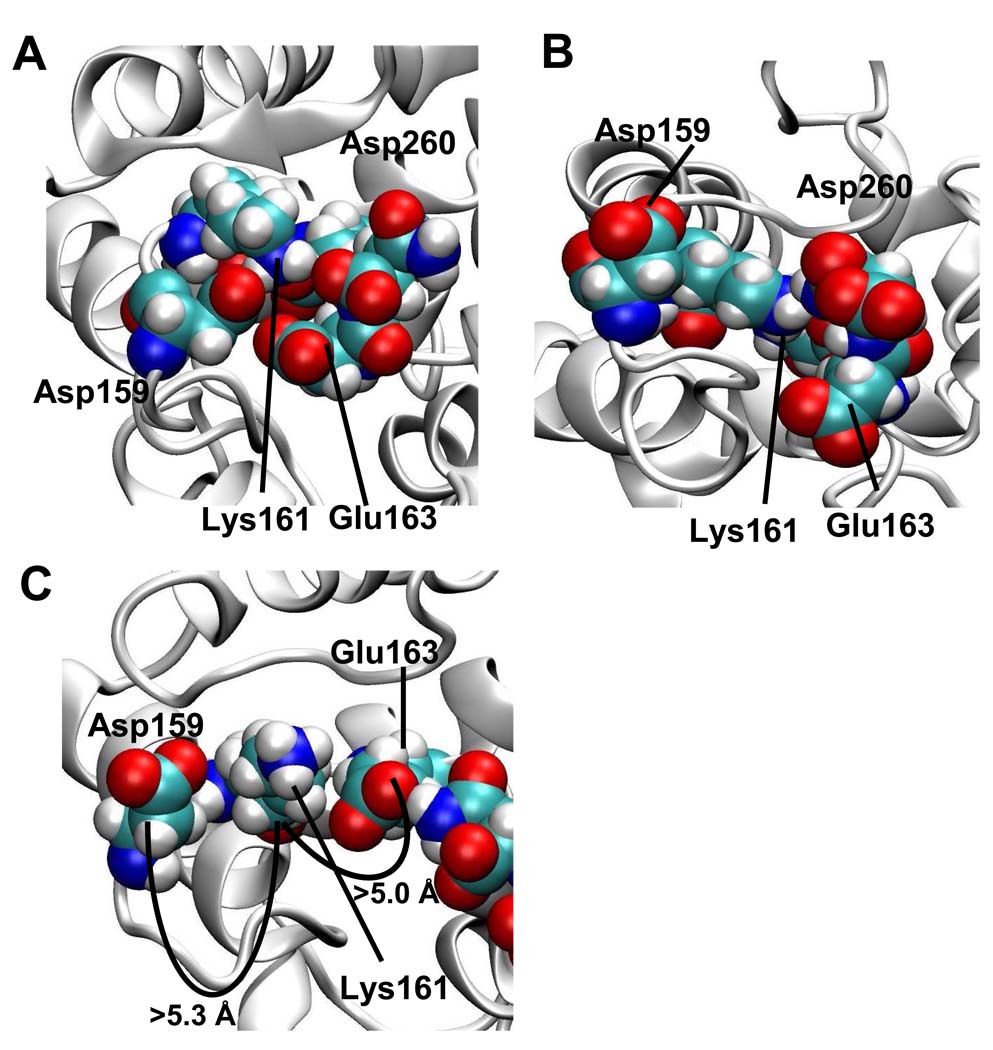
The reactivity of lysine 161 was also significantly affected upon calcium binding, going from a completely modified state in the apo- to a fully unmodified state in the holo- calbindin-D28k. Comparison of the apo- model to both the NMR structure of the reduced holo-, and our non- reduced holo- form showed a significant structural difference in the residues surrounding lysine 161 (Figure 6). In both holo- forms (Figures 6A and 6B) lysine 161 is completely surrounded by acidic residues giving it many opportunities to form salt bridges, resulting in our observation of lysine 161 as unmodified in the holo- form. However, the opposite is true in the apo- model, displaying lysine 161 as available for modification in the non- calcium bound state. Lysine 161 is located in the middle of the calcium binding loop of EF hand 4, identifying EF hand 4 as a site of significant change upon calcium binding.
Figure 6. Comparison of the NMR structure to our structural models of lysine 161 and surrounding residues.
Images of A) NMR structure with a reduced disulfide bond. B) holo-model with a non-reduced disulfide bond and C) apo- model with reduced disulfide bond. Asp159, Lys161, Glu163 and Asp260 are shown in space-filled form. As seen in panels A and B for the holo- structures lysine 161 is completely surrounded by acidic residues, whereas in panel C (apo- model) lysine 161 is no longer surrounded by this pocket of acidic residues. Atoms are represented by blue=N, red=O, white=H, and yellow=C.
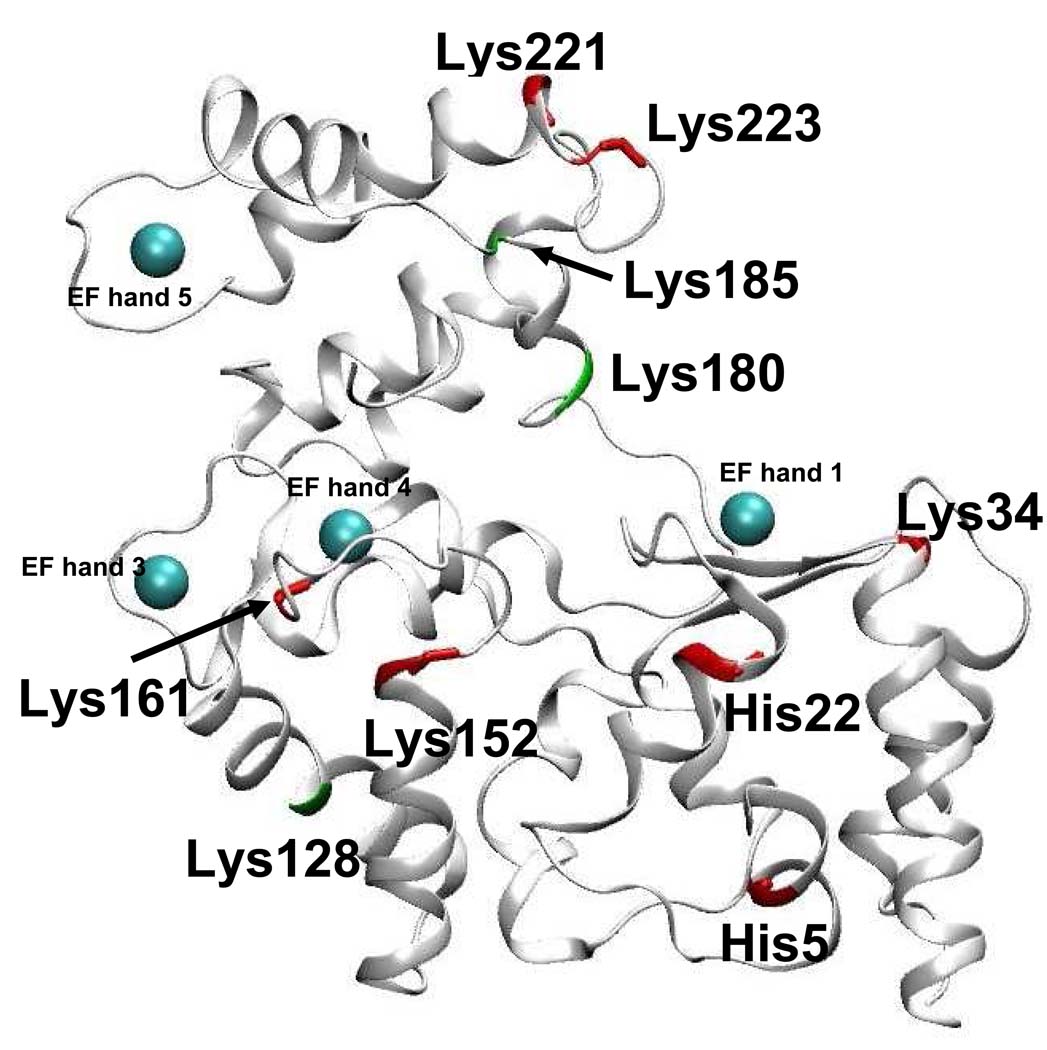
Our computational model of holo-calbindin-D28k with a disulfide bond is shown in Figure 7. In this figure the residues that we observed to undergo significant change in surface reactivity between the holo- and apo- forms are highlighted. The locations of these residues may be indicative of the areas of the protein undergoing extensive conformational change upon calcium binding.
Figure 7. Computational model of disulfide-bonded calbindin-D28K with residues showing significant differences in surface reactivities between the apo- and holo- forms.
Residues showing a decrease in reactivity upon binding calcium are highlighted in red. Residues showing an increase in reactivity upon binding calcium are highlighted in green. Calcium binding EF hands are designated, with calcium ions shown as cyan spheres.
In summary, changes in local and global electrostatic and hydrophobic properties allow a protein to fine tune the surface it presents, which is essential to its function. These changes result in a different surface being presented to potential binding partners. The conformational change characterized here using differential surface modification analyzed by mass spectrometry identifies the regions affected by the conformational change that occurs between the holo- and apo- states of calbindin-D28k; thereby allowing, for the first time, for structural differences between apo- and holo- calbindin-D28k to be determined.
Supplementary Material
Acknowledgement
The authors would like to thank Dr. Ronald A. Venters for providing the deamidation NMR spectra. The authors would also like to thank Dr. James G. Smedley III and Dr. Mark Rance for critical review of the manuscript and Dr. Ron Ventors, Duke University for the 3D HNCACB data.
Abbreviations
- NMR
nuclear magnetic resonance
- MS
mass spectrometry
- CD
circular dichroism
- GST
glutathione S-transferase
- DEPC
diethyl pyrocarbonate
- MD
molecular dynamics
- LC/ESI/MS
liquid chromatography electrospray ionization mass spectrometry
- LC/ESI/MS/MS
liquid chromatography electrospray ionization tandem mass spectrometry
- EIC
extracted ion chromatogram
- PDB
Protein Data Bank
Footnotes
This research was supported by a grant from the Kenan Institute for Engineering, Technology and Science and by the Intramural Research Program of the National Institute of Environmental Health Sciences /National Institutes of Health (projects ES050127/ES04301023).
Supporting Information Available
Figures identified in the text can be found in this section. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Berggard T, Silow M, Thulin E, Linse S. Ca(2+)- and H(+)-dependent conformational changes of calbindin D(28k) Biochemistry. 2000;39:6864–6873. doi: 10.1021/bi992394g. [DOI] [PubMed] [Google Scholar]
- 2.Kojetin DJ, Venters RA, Kordys DR, Thompson RJ, Kumar R, Cavanagh J. Structure, binding interface and hydrophobic transitions of Ca2+-loaded calbindin-D(28K) Nat Struct Mol Biol. 2006;13:641–647. doi: 10.1038/nsmb1112. [DOI] [PubMed] [Google Scholar]
- 3.Lutz W, Frank EM, Craig TA, Thompson R, Venters RA, Kojetin D, Cavanagh J, Kumar R. Calbindin D28K interacts with Ran-binding protein M: identification of interacting domains by NMR spectroscopy. Biochem Biophys Res Commun. 2003;303:1186–1192. doi: 10.1016/s0006-291x(03)00499-6. [DOI] [PubMed] [Google Scholar]
- 4.Berggard T, Miron S, Onnerfjord P, Thulin E, Akerfeldt KS, Enghild JJ, Akke M, Linse S. Calbindin D28k exhibits properties characteristic of a Ca2+ sensor. J Biol Chem. 2002;277:16662–16672. doi: 10.1074/jbc.M200415200. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JA, Grande JP, Roche PC, Kumar R. Immunohistochemical localization of the 1,25(OH)2D3 receptor and calbindin D28k in human and rat pancreas. Am J Physiol. 1994;267:E356–E360. doi: 10.1152/ajpendo.1994.267.3.E356. [DOI] [PubMed] [Google Scholar]
- 6.Oberholtzer JC, Buettger C, Summers MC, Matschinsky FM. The 28-kDa calbindin-D is a major calcium-binding protein in the basilar papilla of the chick. Proc Natl Acad Sci U S A. 1988;85:3387–3390. doi: 10.1073/pnas.85.10.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohr G, Lambert CE, Mody I. Calbindin-D28K (CaBP) levels and calcium currents in acutely dissociated epileptic neurons. Exp Brain Res. 1991;85:543–551. doi: 10.1007/BF00231738. [DOI] [PubMed] [Google Scholar]
- 8.Mody I, Reynolds JN, Salter MW, Carlen PL, MacDonald JF. Kindling-induced epilepsy alters calcium currents in granule cells of rat hippocampal slices. Brain Res. 1990;531:88–94. doi: 10.1016/0006-8993(90)90761-y. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt H, Schwaller B, Eilers J. Calbindin D28k targets myo-inositol monophosphatase in spines and dendrites of cerebellar Purkinje neurons. Proc Natl Acad Sci U S A. 2005;102:5850–5855. doi: 10.1073/pnas.0407855102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berggard T, Szczepankiewicz O, Thulin E, Linse S. Myo-inositol monophosphatase is an activated target of calbindin D28k. J Biol Chem. 2002;277:41954–41959. doi: 10.1074/jbc.M203492200. [DOI] [PubMed] [Google Scholar]
- 11.Su JH, Zhao M, Anderson AJ, Srinivasan A, Cotman CW. Activated caspase-3 expression in Alzheimer's and aged control brain: correlation with Alzheimer pathology. Brain Res. 2001;898:350–357. doi: 10.1016/s0006-8993(01)02018-2. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Porta A, Peng X, Gengaro K, Cunningham EB, Li H, Dominguez LA, Bellido T, Christakos S. Prevention of glucocorticoid-induced apoptosis in osteocytes and osteoblasts by calbindin-D28k. J Bone Miner Res. 2004;19:479–490. doi: 10.1359/JBMR.0301242. [DOI] [PubMed] [Google Scholar]
- 13.Christakos S, Liu Y. Biological actions and mechanism of action of calbindin in the process of apoptosis. J Steroid Biochem Mol Biol. 2004;89–90:401–404. doi: 10.1016/j.jsbmb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Shamir A, Elhadad N, Belmaker RH, Agam G. Interaction of calbindin D28k and inositol monophosphatase in human postmortem cortex: possible implications for bipolar disorder. Bipolar Disord. 2005;7:42–48. doi: 10.1111/j.1399-5618.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 15.Venters RA, Benson LM, Craig TA, Bagu J, Paul KH, Kordys DR, Thompson R, Naylor S, Kumar R, Cavanagh J. The effects of Ca(2+) binding on the conformation of calbindin D(28K): a nuclear magnetic resonance and microelectrospray mass spectrometry study. Anal Biochem. 2003;317:59–66. doi: 10.1016/s0003-2697(03)00084-8. [DOI] [PubMed] [Google Scholar]
- 16.Nelson MR, Chazin WJ. Structures of EF-hand Ca(2+)-binding proteins: diversity in the organization, packing and response to Ca2+ binding. Biometals. 1998;11:297–318. doi: 10.1023/a:1009253808876. [DOI] [PubMed] [Google Scholar]
- 17.Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. [PubMed] [Google Scholar]
- 18.Yap KL, Ames JB, Swindells MB, Ikura M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins. 1999;37:499–507. doi: 10.1002/(sici)1097-0134(19991115)37:3<499::aid-prot17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 19.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 20.Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359:509–525. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 21.Venyaminov SY, Klimtchuk ES, Bajzer Z, Craig TA. Changes in structure and stability of calbindin-D(28K) upon calcium binding. Anal Biochem. 2004;334:97–105. doi: 10.1016/j.ab.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 22.Glocker MO, Borchers C, Fiedler W, Suckau D, Przybylski M. Molecular characterization of surface topology in protein tertiary structures by amino-acylation and mass spectrometric peptide mapping. Bioconjug Chem. 1994;5:583–590. doi: 10.1021/bc00030a014. [DOI] [PubMed] [Google Scholar]
- 23.Suckau D, Mak M, Przybylski M. Protein surface topology-probing by selective chemical modification and mass spectrometric peptide mapping. Proc Natl Acad Sci U S A. 1992;89:5630–5634. doi: 10.1073/pnas.89.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hnizda A, Santrucek J, Sanda M, Strohalm M, Kodicek M. Reactivity of histidine and lysine side-chains with diethylpyrocarbonate -- a method to identify surface exposed residues in proteins. J Biochem Biophys Methods. 2008;70:1091–1097. doi: 10.1016/j.jbbm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Beardsley RL, Running WE, Reilly JP. Probing the structure of the Caulobacter crescentus ribosome with chemical labeling and mass spectrometry. J Proteome Res. 2006;5:2935–2946. doi: 10.1021/pr060170w. [DOI] [PubMed] [Google Scholar]
- 26.Janecki DJ, Beardsley RL, Reilly JP. Probing protein tertiary structure with amidination. Anal Chem. 2005;77:7274–7281. doi: 10.1021/ac050891z. [DOI] [PubMed] [Google Scholar]
- 27.Strohalm M, Santrucek J, Hynek R, Kodicek M. Analysis of tryptophan surface accessibility in proteins by MALDI-TOF mass spectrometry. Biochem Biophys Res Commun. 2004;323:1134–1138. doi: 10.1016/j.bbrc.2004.08.217. [DOI] [PubMed] [Google Scholar]
- 28.Mendoza VL, Vachet RW. Protein surface mapping using diethylpyrocarbonate with mass spectrometric detection. Anal Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochleitner EO, Borchers C, Parker C, Bienstock RJ, Tomer KB. Characterization of a discontinuous epitope of the human immunodeficiency virus (HIV) core protein p24 by epitope excision and differential chemical modification followed by mass spectrometric peptide mapping analysis. Protein Sci. 2000;9:487–496. doi: 10.1110/ps.9.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin K, Yang Y, Mastrangelo P, Westaway D. Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J Biol Chem. 2002;277:1981–1990. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 31.Zappacosta F, Ingallinella P, Scaloni A, Pessi A, Bianchi E, Sollazzo M, Tramontano A, Marino G, Pucci P. Surface topology of Minibody by selective chemical modifications and mass spectrometry. Protein Sci. 1997;6:1901–1909. doi: 10.1002/pro.5560060911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiner RF, Albaugh S, Fenselau C, Murphy C, Vestling M. A mass spectrometry method for mapping the interface topography of interacting proteins, illustrated by the melittin-calmodulin system. Anal Biochem. 1991;196:120–125. doi: 10.1016/0003-2697(91)90127-f. [DOI] [PubMed] [Google Scholar]
- 33.Iacob RE, Keck Z, Olson O, Foung SK, Tomer KB. Structural elucidation of critical residues involved in binding of human monoclonal antibodies to hepatitis C virus E2 envelope glycoprotein. Biochim Biophys Acta. 2008;1784:530–542. doi: 10.1016/j.bbapap.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalkum M, Przybylski M, Glocker MO. Structure characterization of functional histidine residues and carbethoxylated derivatives in peptides and proteins by mass spectrometry. Bioconjug Chem. 1998;9:226–235. doi: 10.1021/bc970162t. [DOI] [PubMed] [Google Scholar]
- 35.Carven GJ, Stern LJ. Probing the ligand-induced conformational change in HLA-DR1 by selective chemical modification and mass spectrometric mapping. Biochemistry. 2005;44:13625–13637. doi: 10.1021/bi050972p. [DOI] [PubMed] [Google Scholar]
- 36.Gross MD, Kumar R, Hunziker W. Expression in Escherichia coli of full-length and mutant rat brain calbindin D28. Comparison with the purified native protein. J Biol Chem. 1988;263:14426–14432. [PubMed] [Google Scholar]
- 37.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 39.Essman U, Perera L, Berkowitz ML, Darden HLT, Pederson LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995b:8577–8592. [Google Scholar]
- 40.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 41.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley MF, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg G, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell S, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. San Francisco: University of California; 2008. [Google Scholar]
- 42.Tugarinov V, Hwang PM, Kay LE. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 43.Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 44.Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom. 1988;16:99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- 45.Altman J, Lipka JJ, Kuntz I, Waskell L. Identification by proton nuclear magnetic resonance of the histidines in cytochrome b5 modified by diethyl pyrocarbonate. Biochemistry. 1989;28:7516–7523. doi: 10.1021/bi00445a004. [DOI] [PubMed] [Google Scholar]
- 46.Vanbelle C, Halgand F, Cedervall T, Thulin E, Akerfeldt KS, Laprevote O, Linse S. Deamidation and disulfide bridge formation in human calbindin D28k with effects on calcium binding. Protein Sci. 2005;14:968–979. doi: 10.1110/ps.041157705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson KL, Veenstra TD, Londowski JM, Tomlinson AJ, Kumar R, Naylor S. On-line sample clean-up and chromatography coupled with electrospray ionization mass spectrometry to characterize the primary sequence and disulfide bond content of recombinant calcium binding proteins. Biomed Chromatogr. 1999;13:37–45. doi: 10.1002/(SICI)1099-0801(199902)13:1<37::AID-BMC810>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 48.Wu J. Disulfide bond mapping by cyanylation-induced cleavage and mass spectrometry. Methods Mol Biol. 2008;446:1–20. doi: 10.1007/978-1-60327-084-7_1. [DOI] [PubMed] [Google Scholar]
- 49.Kordys DR, Bobay BG, Thompson RJ, Venters RA, Cavanagh J. Peptide binding proclivities of calcium loaded calbindin-D28k. FEBS Lett. 2007;581:4778–4782. doi: 10.1016/j.febslet.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Nussinov R. Close-range electrostatic interactions in proteins. Chembiochem. 2002;3:604–617. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Nussinov R. Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys J. 2002;83:1595–1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.