Abstract
Objective
To investigate the mechanism of glycosylphosphatidylinositol (GPI) anchor deficiency in Burkitt lymphoma cell lines.
Methods
We identified a large GPI anchor protein deficient population in three different Burkitt lymphoma cell lines through proaerolysin treatment of the cells and flow cytometry analysis using a proaerolysin variant (FLAER). The mechanism of GPI anchor protein deficiency was studied by DNA gene sequencing, a cell-free assay to investigate the GPI anchor biosynthetic pathway, microarray analysis, and quantitative real-time polymerase chain reaction.
Results
Burkitt lymphoma cell lines harbor large populations of FLAERneg cells, which are resistant to proaerolysin. In all three cell lines, silencing of a gene involved in an early step in GPI-anchor biosynthesis was responsible for the lack of GPI-anchored proteins on the cell surface. Quantitative polymerase chain reaction and microarray analysis demonstrate that the level of mRNA for PIGL and PIGY is lower in the FLAERneg Ramos cells and that mRNA levels of PIGY are reduced in the Akata and Daudi cells. Hypermethylation of these genes was associated with the low levels of mRNA and treatment of the cells with 5-aza-2′ deoxycytidine restored cell surface GPI-anchored proteins to the FLAERneg cells.
Conclusion
GPI-anchored protein deficiency in Burkitt lymphoma cells is not due to a genetic mutation (e.g., PIGA); rather, the lack of GPI-anchored proteins results from transcriptional silencing of PIGL and PIGY.
Covalent linkage to glycosylphosphatidylinositol (GPI) is an important means of anchoring many cell surface glycoproteins to the cell membrane [1–3]. There are more than a dozen different GPI-anchored proteins (GPI-AP) on blood cells. GPI-AP also serve as receptors for proaerolysin, a pore-forming bacterial toxin secreted by Aeromonas hydrophila [4,5]. The anchor is synthesized in a stepwise manner in the endoplasmic reticulum membrane involving 10 reactions and >20 different gene products [3]. The first step in this pathway is the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to phosphatidylinositol (PI) to yield GlcNAc-PI. This step is catalyzed by GlcNAc:PI α1–6 GlcNAc transferase, an enzyme whose subunits are encoded by seven different genes: PIGA [6], PIGC [7], PIGH [8], GPI1 [9,10], PIGY [11], PIGP, and DPM2 [12]. In the second step, GlcNAc-PI is deacetylated by the gene product of PIG-L to form GlcN-PI [13]. GPI anchor assembly continues in the endoplasmic reticulum with acylation of the inositol and stepwise addition of mannosyl and phosphoethanolamine residues. The preassembled GPI is linked to nascent proteins that contain a C-terminal GPI-attachment signal peptide, displacing it in a transamidase reaction [14]. The GPI-AP then transits the secretory pathway to reach its final destination at the plasma membrane. If the GPI anchor is not attached to the protein, the protein is degraded intracellularly [15,16].
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired clonal hematopoietic stem cell disease characterized by a loss of GPI-AP on the affected stem cell and in all of its progeny [17–19]. Because proaerolysin binds specifically and with high affinity to the glycan portion of the GPI anchor, a fluorescent proaerolysin variant (FLAER) serves as a diagnostic test for PNH [20,21]. GPI-AP deficiency in PNH results from a somatic mutation in the PIGA gene [6,22–24]. Human PIGA resides on the X chromosome and contains six exons extending >16 kb [25]. Rare GPI-AP–deficient cells are also found in the blood and bone marrow of healthy controls, at a frequency of roughly 1 in 50,000 [26–29]. Most of these GPI-AP–deficient blood cells harbor PIGA mutations; however, in contrast to PNH, where the PIGA mutations are clonal and arise in hematopoietic stem cells, most PIGA mutations found in healthy controls arise from colony-forming cells rather than self-renewing hematopoietic stem cells [29]. Others have reported GPI-AP–deficient lymphocyte populations that do not harbor PIGA mutations, but the cause of this deficiency has not been clearly delineated [30–32]. High-grade B-cell lymphomas have surprising heterogeneity in expression of the GPI-AP, CD52, with 25% of cases lacking cell-surface CD52 [33]. Here, we describe a novel mechanism to explain the lack of GPI-APs on Burkitt lymphoma cells.
Materials and methods
Cell lines and cell culture
The human cell lines Daudi, Akata and Ramos were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in complete RPMI-1640 media with 10% fetal bovine serum (FBS). Clonogenic growth was obtained by sorting single Ramos FLAERneg and FLAERpos cell in 96-well plates. Colonies were stained with FLAER at 3 weeks. For demethylation studies, cells were treated with 2 μM 5-aza-2′ deoxycytidine (Sigma-Aldrich, St Louis, MO, USA) for 48 hours, then stained with FLAER, anti-CD27 or anti-CD52 (BD Pharmingen, San Diego, CA, USA) for fluorescein-activated cell sorting analysis.
Patients and cell preparation
Heparinized blood samples were obtained from two Burkitt lymphoma samples following informed consent by the Johns Hopkins internal review board. Peripheral blood mononuclear cells were recovered by Ficoll/Hypaque (density <1.077) centrifugation. Cells were washed, stained with FLAER and CD20 (BD Pharmingen) and analyzed on a FACSVantage SE flow cytometer (Becton Dickinson, Mountain View, CA, USA).
Flow cytometric analysis, cell sorting, and cell-cycle analysis
Cell lines were stained with FLAER (Protox Biotech, Victoria, Canada) for 30 minutes at 4°C, washed, and analyzed on a FACS-Vantage SE flow cytometer (Becton Dickinson). Cells were sorted into FLAERpos and FLAERneg fractions by gating on the lowest and highest 5% FLAER expressing cells, respectively. For cell-cycle analysis, cells were resuspended in 75% ethanol for 24 hours at −20°C, incubated with 20 μg/mL RNAse (Sigma) at 37°C for 30 minutes and then incubated with 50 μg/mL PI for 1 hour, and analyzed by flow cytometry. The percentage of cells in each stage of the cell cycle was calculated using the ModFIT software.
Cell synchronization
Ramos cells were cultured in serum-free medium for 24 hours. Cells were synchronized in late G1 and early S phases by incubation for 12 hours in fresh medium supplemented with 10% FBS and 2 μg/mL aphidicolin. To release cells into S phase, cells were washed twice to remove aphidicolin and resuspended in fresh medium supplemented with 10% FBS. Cells were stained with PI or FLAER before and after aphidicolin incubation.
Immunohistochemical staining
Intact cells were washed with ice-cold PBS, incubated with an Alexa 594-conjugated proaerolysin variant (Protox Biotech, Victoria, Canada) for 20 minutes at 4°C, and then washed with PBS. Cytospins prepared from 5000 cells were fixed in methanol and permeabilized with 0.2% Triton-X 100 for 10 minutes. Slides were blocked for 1 hour with 10% normal serum and stained with either FLAER, fluorescein isothiocyanate–conjugated anti-GM130 (BD Transduction Labs), or phycoerythrin-conjugated anti-CD59 (Research Diagnostics, Flanders NJ, USA) for 20 minutes at room temperature. Nucleus was counterstained with 4′6-diamidino-2-phenylindole (DAPI; Vector Labs, Burlingame, CA, USA), and analyzed by fluorescent microscopy.
In vitro assays of GPI biosynthesis
Assays of the first two steps in GPI biosynthesis, the synthesis of GlcNAc-PI and its conversion to GlcN-PI, were based on published protocols [13,34–36]. Cells were incubated with 5 μg/mL tunicamycin at 37°C for 2 hours. After washing, the cell pellet was agitated with 0.1 mM Nα-p-tosyl-L-lysine chloromethyl ketone (TLCK) and 1 μg/mL leupeptin for 5 minutes on ice. An equal volume of 100 mM HEPES containing 50 mM KCl, 10 mM MgCl2, 0.1 mM TLCK, and 1 μg/mL leupeptin was added and membranes were collected by centrifugation. The membrane fraction was resuspended in 200 μL assay buffer (50 mM HEPES, 25 mM KCl, 5 mM MgCl2, 5 mM MnCl2, 0.5 mM dithiothreitol, 1 mM adenosine triphosphate, 1 mM guanosine triphosphate, 0.1 mM TLCK, 1 μg/mL leupeptin, 0.2 μg/mL tunicamycin) and serial dilutions were made. Two μCi UDP-[3H]GlcNAc (American Radiolabeled Chemicals Inc., St Louis, MO, USA) were added to each tube. After 30 minutes at 37°C, lipids were extracted with 10:10:3 chloroform/methanol/water by centrifugation, and then were dried, dissolved in 15 μL 2:1 chloroform/methanol, spotted onto a Silica Gel 60 thin layer chromatography (TLC) plate (Merck, Whitehouse Station, NJ, USA). The chromatogram was developed by the Fuji Image Analyzer (BAS-2500; Fuji Film Medical Systems, Tokyo, Japan).
To characterize the GPI products, dried samples of n-butanol phases were deaminated by HNO2 for 4 hours at 37°C in 100 μL 50 mM sodium acetate (pH 3.5), 250 mM NaNO2, 0.2% Triton X-100. The products were partitioned in n-butanol/water and analyzed by TLC.
Quantitative real-time polymerase chain reaction (PCR) and microarray analysis
Total RNA was isolated with the TRIzol reagent (Life Technologies, Inc., Gaithersburg, MD, USA). One microgram total RNA was used to synthesize first-strand complementary DNA (cDNA) using the Superscript II-reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA). PCR amplification was carried out with primer sets derived from the genes involved in the GPI-anchor biosynthesis pathway (Suppl. Table S1A) by using SYBR (Bio-Rad, Richmond, CA, USA) on a Bio-Rad ICycler real-time PCR instrument. Each reaction was run in triplicate and contained 1 μL cDNA template in a final reaction volume of 25 μL. Melting curves were performed to ensure only a single product was amplified. Relative quantification (RQ) was normalized to β-actin by using the 2−ΔΔCt method. The fold-change ratio was calculated as RQae/RQwild−1. Values represent the mean ± standard error from three experiments.
The method for preparation of Cy-dye labeled cRNA and array hybridization was provided by Agilent Technologies (Santa Clara, CA, USA). Following hybridization, the arrays were scanned and analyzed. For microarray data clustering, expression data was processed to Agilent log-ration data by standard methods.
DNA extraction and methylation-specific PCR
Genomic DNA was extracted by using a DNeasy Kit (Qiagen) and then bisulfite treated as described previously [37]. Methylation-specific PCR primers were designed according to genomic sequences flanking the presumed transcription start sites for human PIGL (Suppl. Table S1B). Each methylation-specific PCR reaction incorporated 100 ng bisulfite-treated DNA as template in a final reaction volume of 25 μL. Methylation-specific PCR products were analyzed by electrophoresis on 2% agarose.
Transfection of siRNA
The following small interfering RNA (siRNA) sequences were used: Human Rras2-specific 21 nucleotide siRNA oligo (Ambion, Austin, TX, USA) GCACGGCAGCUUAAGGUAAtt and UUACCUUA AGCUGCCGUGCta. Cells were harvested by centrifugation and re-suspended at 3 × 107 cells/mL in Nucleofector kit V buffer (Amaxa, Walkersville, MD, USA), and 100 μL was dispensed to electroporation cuvettes containing 5 μg siRNA. Cells were electroporated by the Nucleofector device (Amaxa) and were incubated for 48 hours in growth medium.
Results
Characterization of GPI-AP–deficient cell lines
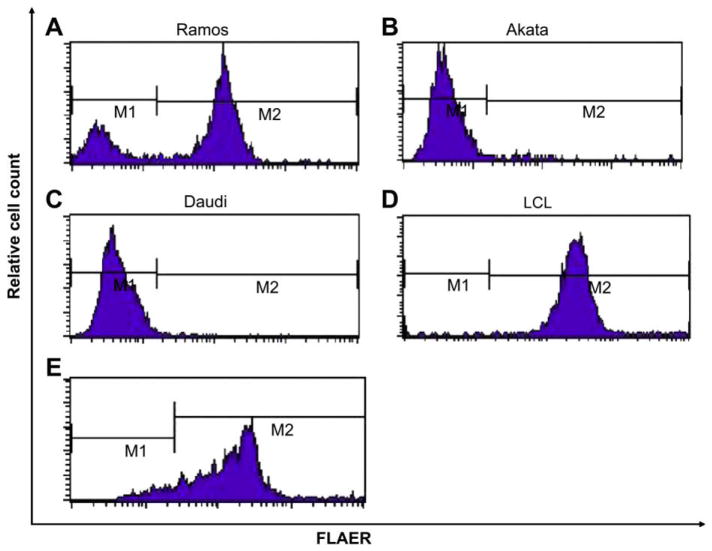
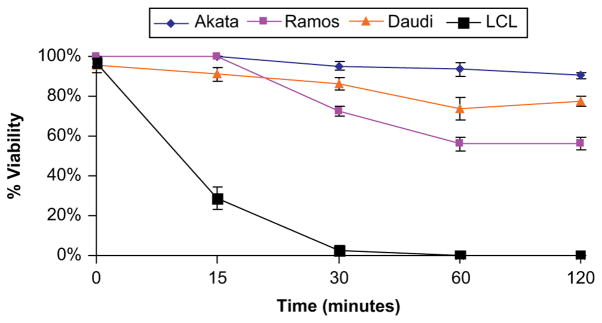
We found that Burkitt lymphoma cell lines harbor large populations of FLAERneg cells. Failure to bind FLAER indicates that these cells lack surface GPI-AP (Fig. 1A, B, and C). The Ramos cell line is an Epstein-Barr virus–negative B-lymphocyte line derived from a patient with Burkitt lymphoma that has been found to contain GPI-AP–deficient cells [32]. The Akata and Daudi are Epstein-Barr virus–positive Burkitt lymphoma cell lines that have also been found to be severely defective in the surface expression of GPI-anchored proteins. We found that 25% to 35% of the cells from the Ramos cell line are FLAERneg by flow cytometry (Fig. 1A) and direct histochemical staining. The Akata and Daudi cells are 99% FLAERneg (Fig. 1B, C). We also studied lymphoblasts from two patients with L3 acute lymphoblastic leukemia and found that one of the samples had a large FLAERlo/neg cell population (Fig. 1E). We expected the population of GPI-anchor–deficient cells to be resistant to proaerolysin compared with the GPI-AP–positive cells based on our previous work. Indeed, Burkitt lymphoma cells are resistant to proaerolysin, compared to normal Epstein-Barr virus–transformed B lymphocytes (Fig. 2).
Figure 1.
Burkitt lymphoma cells lack glycosylphosphatidylinositol-anchored proteins (GPI-AP). Flow cytometry analysis using a proaerolysin variant (FLAER) staining of Burkitt lymphoma cell lines. (A) Ramos, (B) Akata, (C) Daudi, and (D) FLAER staining of Epstein-Barr virus (EBV)–transformed B lymphocytes, and (E) FLAER staining of CD20-positive Burkitt lymphoma cells from a patient.
Figure 2.
Glycosylphosphatidylinositol-anchored proteins (GPI-AP)–deficient cells are resistant to proaerolysin. Viability of Burkitt lymphoma cells and Epstein-Barr virus (EBV)–transformed B lymphocytes after exposure to 1 nM aerolysin at 37°C. Cell viability was determined in triplicate by trypan blue. Error bars represent the standard deviations.
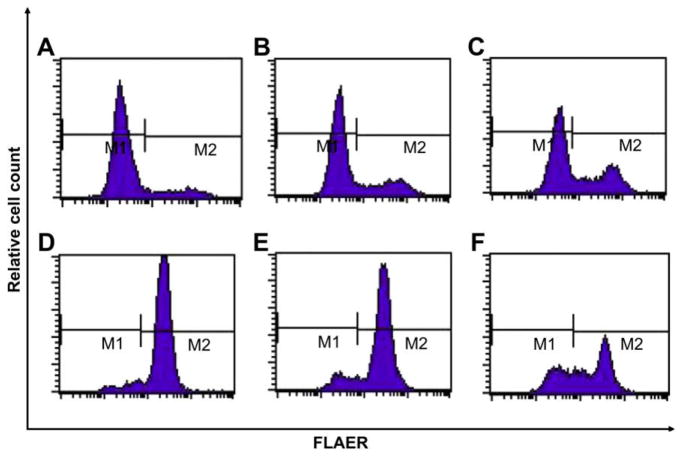
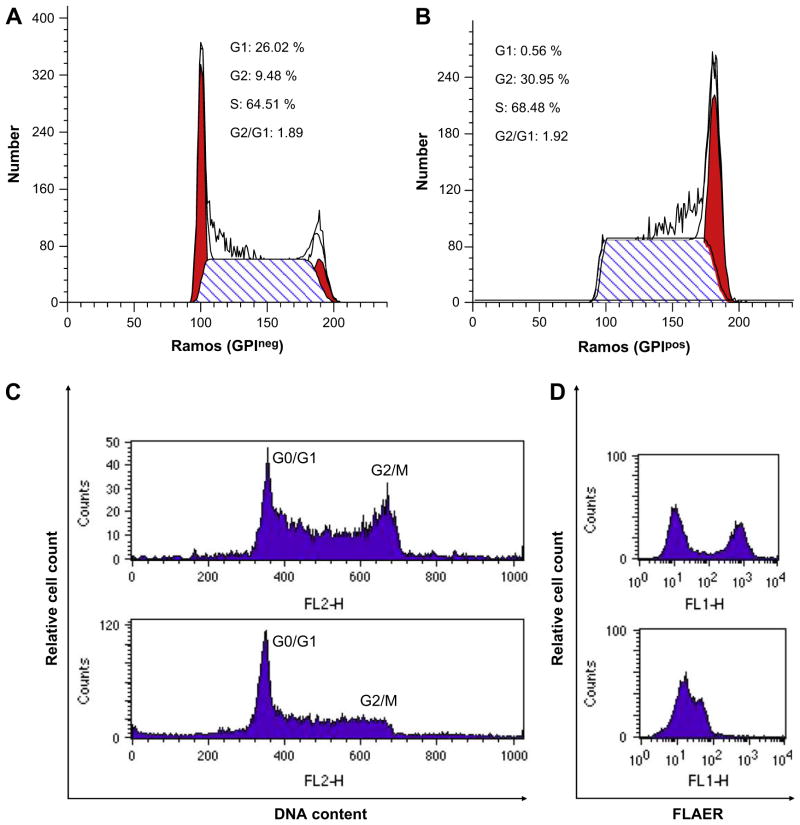
Next, we isolated FLAERneg and FLAERpos Ramos populations by cell sorting. Single FLAERneg or FLAERpos Ramos cells were expanded in 96-well plates. After 3 weeks, the colonies were harvested and assayed for GPI-AP expression using FLAER. Interestingly, the FLAERneg Ramos cells reverted to a mixture of FLAERneg and FLAERpos cells (Table 1 and Fig. 3A, B, and C). Most progeny of the FLAERneg cells remained FLAERneg, but all of the clones acquired a detectable FLAERpos population; in 25% of the clones the FLAERpos cells accounted for >20% of the cells. The progeny of single-cell FLAERpos cells also demonstrated the ability to revert to FLAERneg cells (Table 1 and Fig. 3D, E, and F). Taken together, these data suggest that the GPI-AP deficiency is not due to a fixed mutation because FLAERpos Ramos cells can revert to FLAERneg and vice versa. Indeed, DNA sequencing of the PIGA gene from the proaerolysin-resistant FLAERneg cells did not reveal a PIGA mutation (data not shown). Cell-cycle analysis was performed on sorted Ramos FLAERpos and FLAERneg cells. The FLAERneg population had a greater percentage of cells in G0/G1; suggesting this population is relatively quiescent compared to the FLAERpos cells (Fig. 4A, B). Aphidocolin-induced synchronization of the Ramos cells in G1/S resulted in a marked decrease of the FLAERpos population, further suggesting that the absence of GPI-AP is associated with a more quiescent cell population (Fig. 4C and D).
Table 1.
Single Ramos FLAERneg cells or FLAERpos cells were cultured in growth medium
| Shifting | FLAERneg | FLAERpos |
|---|---|---|
| <10% | 14/24 | 7/24 |
| 10–20% | 4/24 | 15/24 |
| >20% | 6/24 | 2/24 |
There are 24 colonies in each group after 3 weeks. These data are representative of individual colonies which have <10%, 10–20%, >20% shifting.
Figure 3.
Glycosylphosphatidylinositol-anchored proteins (GPI-AP) deficiency of Ramos cells is not fixed. Ramos cells were sorted by flow cytometry into FLAERneg and FLAERpos cells. Single FLAERneg cells (A, B, C) or FLAERpos cells (D, E, F) was expanded in growth medium in 96-well plate. After 3 weeks, cells were analyzed for GPI-AP expression using FLAER. These data are representative of individual colonies that have <10%, 10–20%, and >20% shifting.
Figure 4.
Cell-cycle analysis of Ramos cells. Ramos cells were sorted by flow cytometry into FLAERneg and FLAERpos cells. Cell-cycle analysis was performed on Ramos FLAERneg cells (A) and FLAERpos cells (B). The percentage of cells in each stage of the cell cycle was calculated using the ModFIT software. Left red peak G0/G1, right red peak G2/M. (C) Cell-cycle analysis of Ramos cells before (upper panel) and after (lower panel) synchronization into G1/S with aphidicolin. (D) FLAER staining of Ramos cells before (upper panel) and after (lower panel) aphidicolin incubation.
Mechanism of GPI-AP deficiency
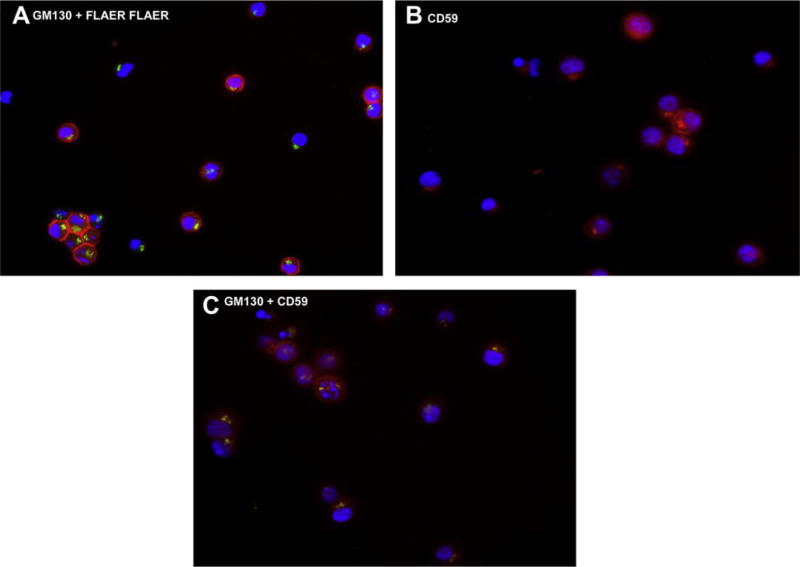
We performed histochemical staining of the Ramos cells using FLAER, anti-GM130 (Golgi-specific monoclonal antibody), and anti-CD59 (Fig. 5). Dual staining with FLAER and anti-GM130 demonstrated that FLAER stained the cell surface of most cells; however, no intracellular or cell surface FLAER staining was seen in 25% of the cells (Fig. 5A). Single staining with anti-CD59 (Fig. 5B) showed fluorescent uptake in all cells; roughly 75% showed staining of the cell surface and the Golgi and the remaining cells demonstrated only intracellular staining. Dual staining with anti-GM130 and anti-CD59 (Fig. 5C) confirmed that CD59 was intracellular, probably localized in the Golgi. Because FLAER does not bind a GPI-AP until the anchor and protein are attached, our immunofluorescence data suggested that a mature GPI precursor was not being synthesized.
Figure 5.
Immunohistochemical staining of Ramos cells. (A) Cells stained with fluorescein isothiocyanate (FITC)–conjugated anti-GM130 and FLAER (Texas Red). (B) Cells stained with phycoerythrin (PE)-conjugated anti-CD59 demonstrating both cell-surface and intracellular staining. (C) Dual staining with FITC-conjugated anti-GM130 and PE-conjugated anti-CD59.
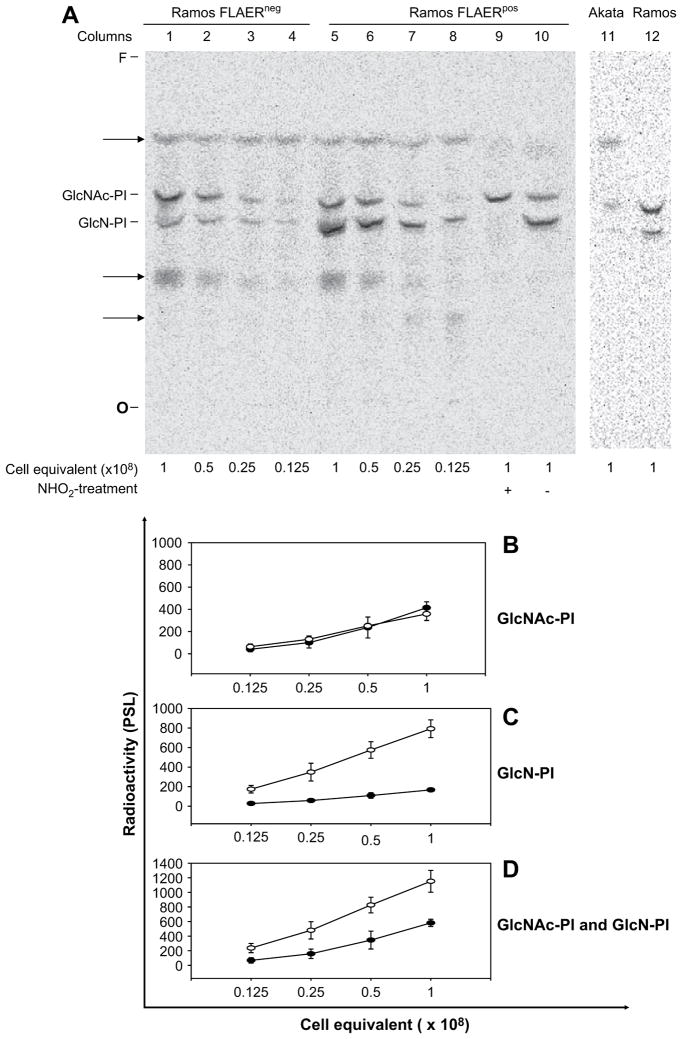
To test whether the deficiency of GPI-AP in FLAERneg cells is due to a defect in synthesis of the GPI precursor, we used a cell-free assay to investigate the first two steps in the biosynthetic pathway. The products of these steps, labeled with UDP-[3H]GlcNAc, are GlcNAc-PI and GlcN-PI, can be resolved by TLC (Fig. 6A). We confirmed the identity of these species by demonstrating the sensitivity of GlcN-PI, but not GlcNAc-PI, to deamination by HNO2 [34–36]. Furthermore, these species migrate similarly on TLC to the corresponding GPIs produced in the well-characterized Trypanosoma brucei cell free system (data not shown) [34]. Comparison of the levels of synthesis by the two cell types (quantitated in Fig. 6B–D) revealed a 5.5-fold decline in production of GlcN-PI in FLAERneg cells relative to FLAERpos cells (Fig. 6C). Because of the level of its precursor, GlcNAc-PI, is virtually identical in both cell types (Fig. 6B), this decline indicates a markedly reduced, but not absent, activity of GlcNAc-PI de-N-acetylase in FLAERneg cells. Measurement of the sum of GPIs produced by the two cell types reveals a 2.5-fold reduction in FLAERneg cells (Fig. 6D). Because the sum of the GPIs reflects the total amount of GlcNAc-PI synthesized, this reduction raises the possibility that there is also a partial decrease in activity in the first enzyme in the pathway, GPI-GlcNAc transferase, in FLAERneg cells. We did not attempt to assay subsequent steps in GPI assembly because the level of GlcN-PI in FLAERneg cells would be too low to allow significant further propagation of the pathway. Additional cell-free assay shown that the first product GlcNAc-PI is not made in the Akata cells (Fig. 6A).
Figure 6.
In vitro glycosylphosphatidylinositol (GPI) synthesis. (A) Phosphorimage of products labeled with UDP-[3H]GlcNAc using membranes from Ramos FLAERneg and FLAERpos cells. Protein measurements on membranes (~1.5 mg per 2 × 108 cell equivalents) confirmed equal loading of samples. Products were characterized by treating with HNO2 (+) or they were mock-treated (−). O, origin; F, front. Lane 1–4 Ramos FLAERneg cells; lane 5–10 Ramos FLAERpos cells; lane 11 Akata cells; lane 12 wild type Ramos cells. (B) GlcNAc-PI. (C) GlcN-PI. (D) Summary of GlcNAc-PI and GlcN-PI. (B–D). Quantitation of GPI products, as represented by PSL value (the intensity of photostimulated luminescence per unit area). Arbritary values of radioactivity were corrected for local background. Diamonds represent radioactivity from FLAERneg Ramos cells; squares represent radioactivity from FLAERpos Ramos cells. Graphs show results of three independent experiments as the mean radioactivity ± standard error.
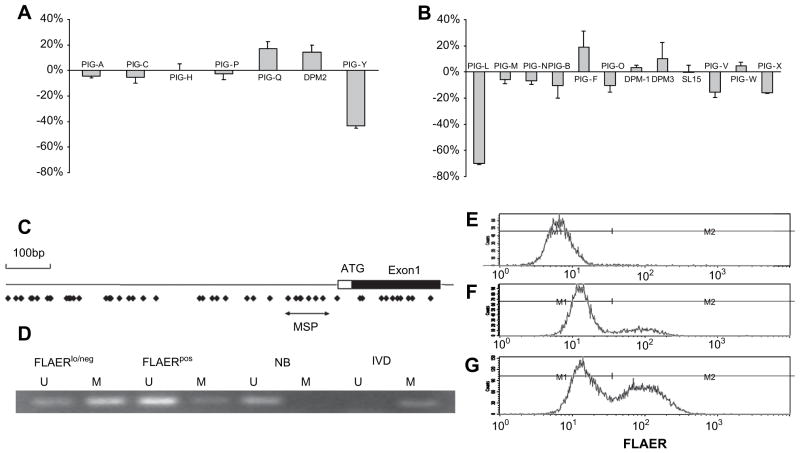
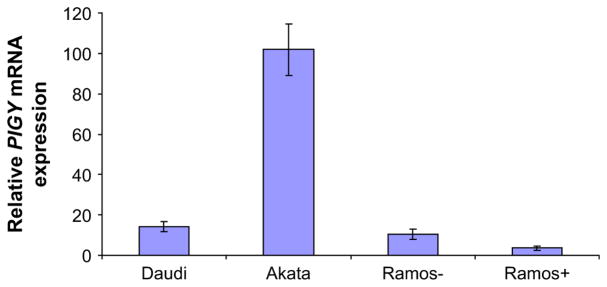
We next performed quantitative real-time PCR on the 24 known genes involved in GPI anchor biosynthesis. Figure 7 demonstrates that the level of mRNA for PIGL, and to a lesser extent PIGY, is lower in the FLAERneg Ramos cells. None of the other genes involved in GPI anchor biosynthesis were suppressed by >18%. Use of methylation specific probes demonstrated that PIGL was hypermethylated in the FLAERneg Ramos cells compared to the FLAERpos Ramos cells (Fig. 7D). Hypermethylation has been associated with silencing of genes [37]. To test whether the demethylating agent 5-aza-2′ deoxycytidine could increase expression of GPI-AP on the FLAERneg Ramos cells, we cultured these cells in normal growth medium or growth medium supplemented with 2 μM 5-aza-2′ deoxycytidine for 48 hours and stained with FLAER to assess GPI-AP expression. Figure 7E through G demonstrates a greater than twofold increase of the FLAERneg Ramos cells displaying GPI-AP after treatment with 5-aza-2′ deoxycytidine.
Figure 7.
PIGL and PIGY are transcriptionally silenced in the FLAERneg Ramos cells. (A) Relative mRNA expression of human genes required for the first step in glycosylphosphatidylinositol (GPI) anchor biosynthesis; (B) remaining genes required for synthesis of mature GPI anchor. Genes required for transamidase reaction not shown. Relative quantification of mRNA extracted from FLAERneg and FLAERpos Ramos cells were normalized to β-actin by using the 2−ΔΔCt method. Values represent the mean ± standard error from three experiments. (C) Schematic representations of 5′ regions of PIGL. Box indicates exon1, including coding (black) and noncoding (white) regions. Dots indicate CpG sites. Black arrows below the CpG sites indicate the region analyzed by methylation-specific polymerase chain reaction. (D) PIGL gene methylation levels in Ramos cells using MSP. M = amplification of methylated alleles; U = amplification of unmethylated alleles. In vitro methylated DNA (IVD) and normal human peripheral lymphocytes (NL) served as the positive and negative methylation controls, respectively. MW = molecular weight. (E), (F), and (G) 5-aza-2′ deoxycytidine leads to increased expression of GPI-anchored protein (GPI-AP). (E) FLAERneg Ramos cells. (F) FLAERneg Ramos cells after 48-hour incubation in growth medium. (G) FLAERneg Ramos cells after 48-hour incubation in growth medium supplemented with 2 μM 5-aza-2′ deoxycytidine.
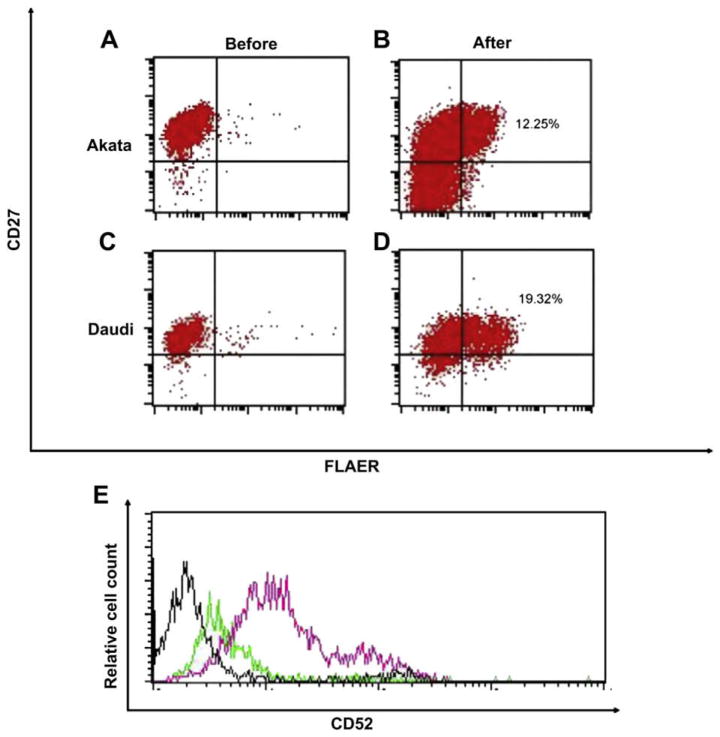
A previous study found that PIGY was responsible for the GPI-AP deficiency in the Daudi cells [11], we too found that the Daudi and Akata cells are defective in PIGY (data not shown). We next examined whether silencing of PIGY is responsible for GPI-AP deficiency. Similar to the Ramos cells, 5-aza-2′ deoxycytidine induced a marked increase in GPI-AP expression in the Akata and Daudi cells. After 5-aza-2′ deoxycytidine treatment, the Akata and Daudi cells became 12.25% and 19.32% FLAERpos, respectively (Fig. 8B and D). Furthermore 5-aza-2′ deoxycytidine induced a marked increase cell surface CD52 on Daudi cells (Fig. 8E). Real-time PCR analysis of PIGY on the Akata and Daudi cells showed that PIGY expression was significantly increased after 5-aza-2′ deoxycytidine treatment (Fig. 9).
Figure 8.
5-aza-2′ deoxycytidine induces glycosylphosphatidylinositol-anchored protein (GPI-AP) expression in Burkitt lymphoma cell lines. (A, C) Akata and Daudi cells in growth medium. (B, D) Akata and Daudi cells after 48 h incubation in growth medium supplemented with 2μM 5-aza-2′ deoxycytidine. (E) Daudi cells after 48 h incubation in growth medium supplemented with 2μM 5-aza-2′ deoxycytidine. Black line histogram represents the isotype control. Cells were stained with CD52 before (green line) and after 5-aza-2′ deoxycytidine (pink line). Relative mRNA expression of PIGY after 48 hour incubation in growth medium supplemented with 2μM 5-Aza-2′ deoxycytidine.
Figure 9.
5-Aza-2′ deoxycytidine increase PIGY expression in Burkitt lymphoma cell lines. Relative mRNA expression of PIGY after 48 h incubation in growth medium supplemented with 2μM 5-Aza-2′ deoxycytidine.
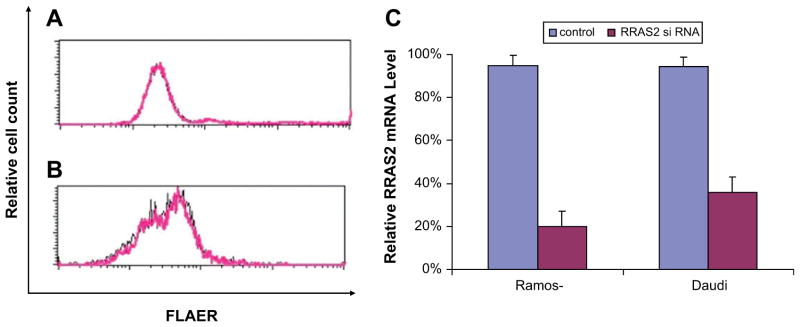
To determine why GPI biosynthesis is suppressed in Burkitt cells, we performed microarray analysis on the Ramos FLAERpos and FLAERneg cells. We carried out t-test and p value <0.05 was used as a cutoff. Other than genes involved with GPI anchor biosynthesis (e.g., PIGY), there were only seven genes that showed a major difference in expression (Table 2). Because Ras2p overexpression has been shown to significantly inhibit GPI-GnT activity in yeast [38] by decreasing expression of Eri1p, the yeast homologue to human PIGY, we examined whether overexpression of human Rras2 in the Ramos FLAERneg cells could be responsible of the decrease in GPI anchor biosynthesis. We transfected Ramos FLAERneg cells and Daudi cells with Rras2 siRNA. In spite of knocking down Rras2 expression by 80% (Fig. 10C), there was no associated increase in GPI-AP expression (Fig. 10A and B), suggesting that Rras2 overexpression does not regulate GPI-GnT activity in mammalian cells.
Table 2.
Genes with the most consistent expression changes in microarray and real-time polymerase chain reaction studies
| Upregulated genes | Fold-change | Validation | Description |
|---|---|---|---|
| Gene name | |||
| ASZ1 | 16.28 | RT-PCR | Ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| PDZRN4 | 12.53 | RT-PCR | PDZ domain containing RING finger 4 |
| RUNX1T1 | 8.82 | RT-PCR | Acute myelogenous leukemia 1 translocation 1 protein |
| RRAS2 | 7.67 | RT-PCR | Related RAS viral (r-ras) oncogene homolog 2 |
| TOX | 6.85 | RT-PCR | Thymus high mobility group box protein TOX |
| LHFP | 6.45 | RT-PCR | Lipoma HMGIC fusion partner |
| Downregulated genes | |||
| FAM3B | −9.98 | RT-PCR | Family with sequence similarity 3, member B |
| PIGY | −14.46 | RT-PCR | Phosphatidylinositol glycan anchor biosynthesis, class Y |
RT-PCR = real-time polymerase chain reaction.
Figure 10.
Knockdown of RRAS2 does not induce glycosylphosphatidylinositol-anchored protein (GPI-AP) expression. (A, B) Ramos FLAERneg cells (A) and Daudi cells (B) were transfected with Rras2 or control small interfering RNA (siRNA) duplex. Cells were stained with FLAER 48 hours after transfection. Black lines represent cells transfected with control siRNA duplex. Pink lines represent cells transfected with Rras2 siRNA. (C). Relative mRNA expression of Rras2 after transfected with Rras2 or control siRNA duplex.
Discussion
GPI-AP are distributed on all normal hematopoietic cells and in a variety of other tissues, but they are absent on hematopoietic cells from patients with PNH. In PNH, the GPI-AP defect is due to a mutation in PIGA. Here, we found that reduced surface expression of GPI-AP is a common feature Burkitt lymphoma cell lines. In contrast to the fixed GPI-AP deficiency in PNH patients, the GPI-APneg phenotype in these Burkitt lymphoma cell lines is reversible. Furthermore, our data implicate that silencing of the genes required for early steps in GPI-anchor biosynthesis, most notably PIGL and PIGY, are responsible for the paucity of GPI-AP on the these cells.
Our previous studies using isolated human CD34+ cells from healthy controls demonstrated that proaerolysin selection can detect a 1 in 100,000 PIGA mutant colony-forming cell [29]. Thus, we were initially surprised to discover that Burkitt cell lines possess large populations of “PNH like” cells. Our data is consistent with previous reports documenting the existence of GPI-AP–deficient lymphoid cell lines. Hatanaka and colleagues [32] also found that a subset of Ramos cells do not display cell surface GPI-AP, even though they express CD55 and CD59 transcripts. These authors hypothesized that the defect was caused by a PIGA mutation, because transient transfection experiments using PIGA cDNA resulted in partial restoration of cell surface GPI-AP; however, they did not sequence the PIGA gene. We found that the PIGA gene is not mutated in the FLAERneg Ramos cells and that simply culturing these cells in growth media leads to partial restoration of cell surface GPI-AP (Fig. 3A, B, and C); thus, the GPI-AP expression following transfection may occur with or without transfection of the PIGA gene. Others have reported that subclones of the Ramos cell line are GPI-AP–deficient due to defect in PIGM, a mannosyl-transferase required for GPI-anchor biosynthesis [39]. This raises the possibility that under selective pressure, GPI-AP–deficient subclones may arise through silencing of more than one of the genes involved in GPI-anchor biosynthesis.
Given that the FLAERneg cells can differentiate into cells that display GPI-AP, it is unlikely that their phenotype is due to a fixed mutation. This finding is in agreement with the work of Taylor et al. [30], who found that alemtuzumab-selected, GPI-AP–deficient lymphocytes had a “reversible” defect and that removal of the cells from selective medium resulted in restoration of the “normal” GPI-APpos phenotype. Rowan et al. [31], also found that when GPI-APneg B cells lines are separated and returned to culture, the GPI-APneg cells revert to their original GPI-AP distribution.
We initially chose the Ramos cell line to investigate the mechanism of GPI-AP deficiency because they have a relatively large population of GPI-AP–deficient and GPI-AP–replete cells. Biochemical analysis of the first two steps in GPI biosynthesis in a cell-free system revealed that FLAERneg cells have a markedly reduced ability to produce the second intermediate, GlcN-PI; there also may be a lesser effect on production of the first intermediate, GlcNAc-PI. In agreement with this finding, expression analysis of genes involved in GPI biosynthesis revealed reduced mRNA levels of PIGL, which encodes the deN-acetylase responsible for synthesis of GlcN-PI and, PIGY, which encodes a subunit of GPI-N-acetylglucosami-nyltransferase, responsible for synthesis of GlcNAc-PI (Fig. 7). Our study did not directly address the mechanism responsible for the reduced levels of PIGL and PIGY mRNA; however, methylation specific primers demonstrated that PIGL was hypermethylated in the FLAERneg Ramos cells. This finding suggests that these genes may be transcriptionally silenced. Furthermore, culturing the FLAERneg Ramos cells in 5-aza-2′ deoxycytidine greatly increased the percentage of cells displaying surface GPI-AP, suggesting that demethylating PIGL and perhaps PIGY may restore surface expression of GPI-AP. Indeed, sodium butyrate has also been reported to cause reexpression of GPI-AP in several GPI-AP–deficient cell lines [40–42].
A previous report demonstrated that Eri1p, the PIGY homolog of S. cerevisiae, was associated with the active form of Ras2p in a cyclic adenosine monophosphate–dependent manner and inhibited downstream signaling. In addition, Ras2p overexpression significantly inhibited GPI-GnT activity in yeast, suggesting that Ras2p negatively regulates GPI biosynthesis [38]. We found that Ras2 was highly overexpressed in the FLAERneg Ramos cells compared to the FLAERpos population; however, in agreement with Murakami et al. [11], our Ras2 knockdown experiment shows that in mammalian cells there is no functional association of the Ras signaling complex and PIGY.
Daudi cells have also been shown to have defective GPI biosynthesis due to a deficiency of the PIGY gene product [11]. Transfection of the PIGY cDNA, but not any of the other six known components of GPI-GnT, restored surface expression of GPI-AP in Daudi cells [11]. These authors did not sequence the PIGY gene; thus, it is unclear whether the GPI-AP defect was due to a PIGY mutation or due to silencing of the PIGY gene. Our data suggests this is due to silencing of the PIGY gene. Real-time PCR the Daudi and Akata cells before and after 5-aza-2′ deoxycytidine treatment showed that PIGY was significantly increased (Fig. 9) and surface expression of GPI-AP were restored (Fig. 8C and D) after 5-aza-2′ deoxycytidine treatment.
Increased expression of genes involved in GPI-anchor biosynthesis following histone deacetylation has been shown to be important in human disease. Recently, an autosomal recessive disease characterized by venous thrombosis, seizures and GPI-AP deficiency has been described [42]. In two unrelated kindred, the disease was shown to be the consequence of a point mutation that disrupts the binding of the transcription factor Sp1 to the promoter of PIGM. Administration of sodium butyrate to the patient increased PIGM transcription, surface GPI-AP expression, and resulted in dramatic clinical improvement [43]. Thus, epigenetic silencing of genes involved in GPI-anchor biosynthesis may be important in human disease, including lymphomas.
It is unclear what, if any, advantage the paucity of GPI-AP confers to Burkitt lymphoma cells. Given the high proliferative potential of Burkitt lymphoma cells, the cell-cycle profile of the FLAERneg Ramos and the relative overexpression of Runx-1 and Ras in the FLAERneg Ramos cells it is possible that these cells represent a cancer stem cell population. Alternatively, because GPI-AP are present on eukaryotes but not prokaryotes, it is possible that the GPI-APneg phenotype may have evolved as a mechanism to protect cells from ubiquitous bacterial toxins that use GPI for its receptor, such as those generated by Aeromonas hydrophila [4,5] and Clostridium septicum [44]. Regardless, the ability to restore cell surface GPI-AP on Burkitt cells may have potential clinical applications. Alemtuzumab is a highly effective monoclonal antibody that is used to treat CD52-positive lymphoproliferative disorders. The ability to raise the cell-surface levels of GPI-AP, including CD52, could make certain aggressive B-cell lymphomas more sensitive to alemtuzumab.
In summary, transcriptional silencing of genes required for GPI-AP biosynthesis is a mechanism to explain reduced cell surface expression of GPI-AP on lymphocytes. This mechanism of GPI-AP deficiency is distinct from that seen in the human disease PNH, but similar to that of a rare autosomal recessive disease resulting in partial GPI-AP deficiency. Treatment with demethylating agents, such as 5-aza-2′ deoxycytidine, can restore cell surface GPI-AP in Burkitt cell lines. The biologic importance of GPI-AP deficiency on these cells is unclear, but seems to be associated with cells of high proliferative potential; furthermore, the lack of complement regulatory proteins on these cells could render these GPI-AP–deficient cells more sensitive to complement-mediated cell toxicity through use of rituximab [45].
Supplementary Material
Acknowledgments
We thank Dr. Carolyn Machamer for assistance and helpful discussions with immunohistochemical staining and Dr. William Matsui for helpful discussions. We thank Dr. Steve Baylin for helpful discussions with methylation studies. This work was generously supported by National Institutes of Health grants CA70970, P50CA096888-01, HL54330 and AI21334.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.exphem.2009.01.003.
References
- 1.Low MG, Saltiel AR. Structural and functional roles of glycosyl-phosphatidylinositol in membranes. Science. 1988;239:268–275. doi: 10.1126/science.3276003. [DOI] [PubMed] [Google Scholar]
- 2.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita T, Inoue N. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr Opin Chem Biol. 2000;4:632–638. doi: 10.1016/s1367-5931(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 4.Diep DB, Nelson KL, Raja SM, Pleshak EN, Buckley JT. Glycosylphosphatidylinositol anchors of membrane glycoproteins are binding determinants for the channel-forming toxin aerolysin. J Biol Chem. 1998;273:2355–2360. doi: 10.1074/jbc.273.4.2355. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky RA, Mukhina GL, Nelson KL, Lawrence TS, Jones RJ, Buckley JT. Resistance of paroxysmal nocturnal hemoglobinuria cells to the glycosylphosphatidylinositol-binding toxin aerolysin. Blood. 1999;93:1749–1756. [PubMed] [Google Scholar]
- 6.Miyata T, Takeda J, Iida Y, et al. The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science. 1993;259:1318–1320. doi: 10.1126/science.7680492. [DOI] [PubMed] [Google Scholar]
- 7.Inoue N, Watanabe R, Takeda J, Kinoshita T. PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2. Biochem Biophys Res Commun. 1996;226:193–199. doi: 10.1006/bbrc.1996.1332. [DOI] [PubMed] [Google Scholar]
- 8.Kamitani T, Chang HM, Rollins C, Waneck GL, Yeh ET. Correction of the class H defect in glycosylphosphatidylinositol anchor biosynthesis in Ltk- cells by a human cDNA clone. J Biol Chem. 1993;268:20733–20736. [PubMed] [Google Scholar]
- 9.Tiede A, Schubert J, Nischan C, et al. Human and mouse Gpi1p homologues restore glycosylphosphatidylinositol membrane anchor biosynthesis in yeast mutants. Biochem J. 1998;15(334 Pt 3):609–616. doi: 10.1042/bj3340609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe R, Inoue N, Westfall B, et al. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 1998;17:877–885. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami Y, Siripanyaphinyo U, Hong Y, Tashima Y, Maeda Y, Kinoshita T. The initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-Y, a seventh component. Mol Biol Cell. 2005;16:5236–5246. doi: 10.1091/mbc.E05-08-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe R, Murakami Y, Marmor MD, et al. Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 2000;19:4402–4411. doi: 10.1093/emboj/19.16.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe R, Ohishi K, Maeda Y, Nakamura N, Kinoshita T. Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphos-phatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem J. 1999;339(Pt 1):185–192. [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma DK, Vidugiriene J, Bangs JD, Menon AK. A cell-free assay for glycosylphosphatidylinositol anchoring in African trypanosomes. Demonstration of a transamidation reaction mechanism. J Biol Chem. 1999;274:16479–16486. doi: 10.1074/jbc.274.23.16479. [DOI] [PubMed] [Google Scholar]
- 15.Tartakoff AM, Singh N. How to make a glycoinositol phospholipid anchor. Trends Biochem Sci. 1992;17:470–473. doi: 10.1016/0968-0004(92)90491-q. [DOI] [PubMed] [Google Scholar]
- 16.Singh N, Singleton D, Tartakoff AM. Anchoring and degradation of glycolipid-anchored membrane proteins by L929 versus by LM-TK-mouse fibroblasts: implications for anchor biosynthesis. Mol Cell Biol. 1991;11:2362–2374. doi: 10.1128/mcb.11.5.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodsky RA. Paroxysmal nocturnal hemoglobinuria. In: Hoffman R, Benz EJ Jr, Shattil SJ, et al., editors. Hematology: Basic Principles and Practice. 4. Philadelphia: Elsevier Churchill Livingstone; 2005. pp. 419–427. [Google Scholar]
- 18.Moyo VM, Mukhina GL, Garrett ES, Brodsky RA. Natural history of paroxysmal nocturnal hemoglobinuria using modern diagnostic assays. Br J Haematol. 2004;126:133–138. doi: 10.1111/j.1365-2141.2004.04992.x. [DOI] [PubMed] [Google Scholar]
- 19.Brodsky RA. Narrative review: paroxysmal nocturnal hemoglobinuria: the physiology of complement-related hemolytic anemia. Ann Intern Med. 2008;148:587–595. doi: 10.7326/0003-4819-148-8-200804150-00003. [DOI] [PubMed] [Google Scholar]
- 20.Brodsky RA, Mukhina GL, Li S, et al. Improved detection and characterization of paroxysmal nocturnal hemoglobinuria using fluorescent aerolysin. Am J Clin Pathol. 2000;114:459–466. doi: 10.1093/ajcp/114.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhina GL, Buckley JT, Barber JP, Jones RJ, Brodsky RA. Multilineage glycosylphosphatidylinositol anchor deficient hematopoiesis in untreated aplastic anemia. Br J Haematol. 2001;115:476–482. doi: 10.1046/j.1365-2141.2001.03127.x. [DOI] [PubMed] [Google Scholar]
- 22.Takahaski M, Takeda J, Hirose S, et al. Deficient biosynthesis of N-acetylglucosaminyl-phosphatidylinositol, the first intermediate of glycosyl phosphatidylinositol anchor biosynthesis, in cell lines established from patients with paroxysmal nocturnal hemoglobinuria. J Exp Med. 1993;177:517–521. doi: 10.1084/jem.177.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessler M, Mason PJ, Hillmen P, et al. Paroxysmal nocturnal haemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene. EMBO J. 1994;13:110–117. doi: 10.1002/j.1460-2075.1994.tb06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagarajan S, Brodsky R, Young NS, Medof ME. Genetic defects underlying paroxysmal nocturnal hemoglobinuria that arises out of aplastic anemia. Blood. 1995;86:4656–4661. [PubMed] [Google Scholar]
- 25.Bessler M, Hillmen P, Longo L, Luzzatto L, Mason PJ. Genomic organization of the X-linked gene (PIG-A) that is mutated in paroxysmal nocturnal haemoglobinuria and of a related autosomal pseudogene mapped to 12q21. Hum Mol Genet. 1994;43:751–757. doi: 10.1093/hmg/3.5.751. [DOI] [PubMed] [Google Scholar]
- 26.Rawstron AC, Rollinson SJ, Richards S, et al. The PNH phenotype cells that emerge in most patients after CAMPATH-1H therapy are present prior to treatment. Br J Haematol. 1999;107:148–153. doi: 10.1046/j.1365-2141.1999.01676.x. [DOI] [PubMed] [Google Scholar]
- 27.Araten DJ, Nafa K, Pakdeesuwan K, Luzzatto L. Clonal populations of hematopoietic cells with paroxysmal nocturnal hemoglobinuria genotype and phenotype are present in normal individuals. Proc Natl Acad Sci U S A. 1999;96:5209–5214. doi: 10.1073/pnas.96.9.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ware RE, Pickens CV, DeCastro CM, Howard TA. Circulating PIG-A mutant T lymphocytes in healthy adults and patients with bone marrow failure syndromes. Exp Hematol. 2001;29:1403–1409. doi: 10.1016/s0301-472x(01)00746-9. [DOI] [PubMed] [Google Scholar]
- 29.Hu R, Mukhina GL, Piantadosi S, Barber JP, Jones RJ, Brodsky RA. PIG-A mutations in normal hematopoiesis. Blood. 2005;105:3848–3854. doi: 10.1182/blood-2004-04-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor VC, Sims M, Brett S, Field MC. Antibody selection against CD52 produces a paroxysmal nocturnal haemoglobinuria phenotype in human lymphocytes by a novel mechanism. Biochem J. 1997;322(Pt 3):919–925. doi: 10.1042/bj3220919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowan WC, Hale G, Tite JP, Brett SJ. Cross-linking of the CAM-PATH-1 antigen (CD52) triggers activation of normal human T lymphocytes. Int Immunol. 1995;7:69–77. doi: 10.1093/intimm/7.1.69. [DOI] [PubMed] [Google Scholar]
- 32.Hatanaka M, Seya T, Matsumoto M, et al. Mechanisms by which the surface expression of the glycosyl-phosphatidylinositol-anchored complement regulatory proteins decay-accelerating factor (CD55) and CD59 is lost in human leukaemia cell lines. Biochem J. 1996;314(Pt 3):969–976. doi: 10.1042/bj3140969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodig SJ, Abramson JS, Pinkus GS, et al. Heterogeneous CD52 expression among hematologic neoplasms: implications for the use of alemtuzumab (CAMPATH-1H) Clin Cancer Res. 2006;12:7174–7179. doi: 10.1158/1078-0432.CCR-06-1275. [DOI] [PubMed] [Google Scholar]
- 34.Doering TL, Masterson WJ, Englund PT, Hart GW. Biosynthesis of the glycosyl phosphatidylinositol membrane anchor of the trypanosome variant surface glycoprotein. Origin of the non-acetylated glucosamine. J Biol Chem. 1989;264:11168–11173. [PubMed] [Google Scholar]
- 35.Stevens VL. Regulation of glycosylphosphatidylinositol biosynthesis by GTP. Stimulation of N-acetylglucosamine-phosphatidylinositol deacetylation. J Biol Chem. 1993;268:9718–9724. [PubMed] [Google Scholar]
- 36.Hirose S, Ravi L, Hazra SV, Medof ME. Assembly and deacetylation of N-acetylglucosaminyl-plasmanylinositol in normal and affected paroxysmal nocturnal hemoglobinuria cells. Proc Natl Acad Sci U S A. 1991;88:3762–3766. doi: 10.1073/pnas.88.9.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 38.Sobering AK, Watanabe R, Romeo MJ, et al. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell. 2004;117:637–648. doi: 10.1016/j.cell.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Maeda Y, Watanabe R, Harris CL, et al. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 2001;20:250–261. doi: 10.1093/emboj/20.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tisdale EJ, Schimenti JC, Tartakoff AM. Sodium butyrate causes reexpression of three membrane proteins on glycolipid-anchoring mutants. Somat Cell Mol Genet. 1991;17:349–357. doi: 10.1007/BF01233060. [DOI] [PubMed] [Google Scholar]
- 41.Ohannesian DW, Lotan D, Thomas P, et al. Carcinoembryonic antigen and other glycoconjugates act as ligands for galectin-3 in human colon carcinoma cells. Cancer Res. 1995;55:2191–2199. [PubMed] [Google Scholar]
- 42.Almeida AM, Murakami Y, Layton DM, et al. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat Med. 2006;12:846–851. doi: 10.1038/nm1410. [DOI] [PubMed] [Google Scholar]
- 43.Almeida AM, Murakami Y, Baker A, et al. Targeted therapy for inherited GPI deficiency. N Engl J Med. 2007;356:1641–1647. doi: 10.1056/NEJMoa063369. [DOI] [PubMed] [Google Scholar]
- 44.Gordon VM, Nelson KL, Buckley JT, et al. Clostridium septicum alpha toxin uses glycosylphosphatidylinositol-anchored protein receptors. J Biol Chem. 1999;17(274):27274–27280. doi: 10.1074/jbc.274.38.27274. [DOI] [PubMed] [Google Scholar]
- 45.Nagajothi N, Matsui WH, Mukhina GL, Brodsky RA. Enhanced cyto-toxicity of rituximab following genetic and biochemical disruption of glycosylphosphatidylinositol anchored proteins. Leuk Lymphoma. 2004;45:795–799. doi: 10.1080/10428190310001625700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.