Abstract
Stratification by age at onset has been useful for genetic studies across all of medicine. For the past 20 years, the National Institute of Mental Health has been systematically recruiting patients with onset of schizophrenia before age 13 years. Examination of familial transmission of known candidate risk genes was carried out, and a 10% rate of cytogenetic abnormalities was found. Most recently, high-density, array-based scans for submicroscopic rare copy number variations (CNVs) have suggested that this kind of genetic variation occurs more frequently than expected by chance in childhood-onset schizophrenia (COS) and at a higher rate than observed in adult-onset disorder. Several CNVs and cytogenetic abnormalities associated with COS are also seen in autism and mental retardation. Populations with COS may have more salient genetic influence than adult-onset cases. The relationship of rare CNVs to prepsychotic development is being studied further.
Introduction
Schizophrenia is a severe, debilitating brain disorder characterized by hallucinations, delusions, flat and/or inappropriate affect, and cognitive impairment. Worldwide prevalence shows a lifetime risk of approximately 1%. Age at onset typically peaks in late adolescence through early adulthood, with slightly earlier age at onset seen in males. Although schizophrenia is not a childhood disorder per se, onset does occur rarely between 6 and 12 years of age. Clinically, childhood-onset schizophrenia (COS) has a severe, progressive, nonepisodic course with poorer premorbid functioning and higher incidence of prepsychotic developmental abnormalities than the more typical adult-onset form of the disorder (AOS). We have established the phenotypic and neurobiologic continuity between COS and AOS [1–3]. Given the rarity of COS patients and therefore the lack of good population estimates, the true familial risk is not known.
The strategy of identifying a subset of rare, extreme early-onset cases of common, complex disorders is not new to the field of medical genetics. Studies of breast cancer [4], type 2 diabetes [5], and Alzheimer's disease [6] provide clear examples in which ascertainment of early-onset cases has facilitated identification of causal genetic variants. Given the success of stratification by age at onset, the study of this rare form of schizophrenia was planned with the hypothesis that such a sample would be unusually informative for genetic studies. Over the past 20 years, our research team in the child psychiatry branch of the National Institute of Mental Health has screened more than 3000 charts, conducted in-person screenings of more than 300 children and adolescents, and to date has enrolled 105 patients meeting the criteria of onset of schizophrenia by age 12 years, but only after 1 to 3 weeks of careful inpatient, medication-free observation. Furthermore, follow-up approximately every 2 years has ensured the continuity and reliability of the diagnosis.
Traditional linkage and association studies in schizophrenia—and most other psychiatric disorders, for that matter—have been unsuccessful in attempting to identify unequivocal risk variants. In spite of this, although the National Institute of Mental Health COS sample size is small (and therefore relatively underpowered for detection of genetic associations), we were able to replicate associations with several schizophrenia susceptibility genes implicated in adult patient cohorts, including DAOA (previously G72) [7], NRG1 [8], DTNBP1 [9], and GAD1 [10]. These associations provided the first evidence that genetic associations in this population may represent more highly penetrant effects of genes in COS as compared with AOS.
Cytogenetic Abnormalities and Copy Number Variations
The emerging technology of high-density microarrays has provided us with insight into ascertainment of submicroscopic genomic changes that were previously unknown. Now that we have the tools in place, these recent advances have led to an abundance of studies of healthy and diseased populations, leading to the discovery that variation in which some large stretches of DNA are present in fewer or more than the expected two copies (one inherited from each parent) is common in all human populations. Copy number variations (CNVs) thus can represent “normal” genetic variation or can contribute to risk for disease. The deluge of information on the topic that has been published over the past 3 years is overwhelming. One thing that is clear is that we are just scratching the surface of things yet to come. This source of variation clearly contributes a great deal to any number of diseases, as well as to what makes us uniquely human, as demonstrated through evolutionary and comparative genomic studies [11••].
The observation that 10% of our COS sample showed cytogenetic abnormalities (Table 1) motivated the study of additional undiscovered submicroscopic structural alterations. Ascertainment of CNVs was accomplished by running three complementary array platforms: Affymetrix 500K SNP (Santa Clara, CA), Agilent 185/244K array comparative genomic hybridization (CGH) (Santa Clara, CA), and a custom bacterial artificial chromosome array developed to target known regions containing segmental duplications.
Table 1.
Chromosomal abnormalities in 96 patients with childhood-onset schizophrenia
| Chromosomal abnormality | Inherited or de novo | Discovery method |
|---|---|---|
| 3-MB deletion 22q11 | – | FISH |
| 3-MB deletion 22q11 | De novo | FISH |
| 3-MB deletion 22q11 | De novo | FISH |
| 3-MB deletion 22q11 | De novo | FISH |
| 46,X,del(X)(q24-qter) | De novo | High-resolution karyotype |
| 78% XO; 22% idicXq | De novo | High-resolution karyotype |
| Trisomy X | – | High-resolution karyotype |
| 1;7 balanced translocation | Inherited | High-resolution karyotype |
| 5q32-qter uniparental isodisomy | De novo | Affymetrix 500K SNP* |
Santa Clara, CA.
FISH—fluorescent in situ hybridization; MB—megabyte; SNP—single nucleotide polymorphism.
The overarching theme that resulted from our study and others like it is that many rare CNVs observed among patients but not in healthy controls are likely involved in disease etiology. The resolution of current molecular platforms allows for detection of CNVs down to 100 kb in size, which is about 30 times the resolution of standard cytogenetic techniques. Given the high rate of large chromosomal anomalies in this cohort, we hypothesized that we would have a similarly high rate of smaller chromosomal abnormalities. Although discovery of novel de novo events was the initial aim of the study given that the overwhelming majority of our cases appear to be sporadic as opposed to familial, we found that nearly all the CNVs that we observed were inherited. The only exception to this was a 2.5-megabyte deletion on chromosome 2q31.2-31.3, which was not observed in either parent; however, the same deletion was observed in the healthy sibling of the proband, indicating a unique situation likely involving germline mosaicism in one of the parents. Other events were either inherited or of unknown origin (because both parents were unavailable for study). With a few exceptions (four 22q11 deletions, three sex chromosome anomalies, two 16p11.2 duplications, and two duplications in MYT1L), each novel CNV was found in an individual patient. Nonetheless, after excluding patients with previously known de novo chromosomal abnormalities, there was an overall excess of novel CNVs that overlapped or disrupted known genes in patients when compared with the nontransmitted parental “control” chromosomes (28% vs 13%; odds ratio, 2.6; P = 0.03) [12••]. This observation confirmed the findings of an independent research group from Seattle that reported that the overabundance of rare CNVs impacting known genes was even more pronounced among younger-onset cases.
The observation that little to no overlap between CNVs is found in our samples as compared with those found in other samples of schizophrenia patients does not immediately discount the potential role of such variants in the etiology of schizophrenia. We acknowledge that some of these CNVs likely will be benign, which is why further follow-up through resequencing and association of common variants in large cohorts is warranted. Even for the disease-related CNVs, given the extreme genetic complexity and heterogeneity of the disorder, none are likely to be fully penetrant. Nonetheless, as converging evidence from multiple sources mounts, we can begin to put the pieces of the puzzle together.
Highlights of Genomic Abnormalities in COS
22q11.21 deletions and sex chromosome anomalies
We previously observed four patients who harbored the well-documented hemizygous 3-megabit 22q11.21 deletion, which is the most common spontaneous deletion in live births (~ 1 in 4000) and is associated with a highly variable phenotype affecting several tissues and organs, many of which are derived from neural crest cells [13]. The rate seen in the COS cohort is significantly higher than that seen in the general population or in unselected AOS populations [14,15••,16••]. The deleted region contains approximately 40 known genes. Attempts to find associations between any particular gene in this region and schizophrenia were not fruitful [17–19]. Contiguous gene effects are plausible [20,21]. Many variable pheno-types have been associated with this deletion, including autism, mental retardation, cardiac anomalies, and facial dysmorphologies [22•].
We have identified three additional patients who have unusual sex chromosome anomalies, which is a significantly increased rate compared with the general population or samples of AOS [23–25]. Although we hypothesized that these rare, atypical sex chromosome anomalies may merely be indicators of more general “genomic instability,” screening for additional submicroscopic structural variants in these patients did not reveal any additional chromosomal events common to these individuals that could be implicated in the etiology of schizophrenia. However, the one patient who was missing 35 megabits of one Xq arm had a 17-megabit duplication of chromosome 16 that apparently attached to the missing X as a possible case of “chromosomal rescue” [26]. The involvement of these X chromosome anomalies in the risk for schizophrenia remains uncertain.
16p11.2 duplications
A CNV of particular interest was observed in two COS probands who harbor an identical 50 0-kb duplication at 16p11.2, a region recently implicated in autism, with deletions and duplications estimated to account for approximately 1% of cases [27,28]. This region is flanked by segmental duplications (stretches of DNA with near-identical sequence) that are more prone to misalignment and rearrangement during meiosis [29]. Again, the rate of 2% seen in the COS cohort is significantly greater than that seen in large, recently published case-control studies of schizophrenia [15••,16••]. This region contains approximately 24 known genes; thus, sorting out which genes are the most likely culprits in causing illness will be challenging. The first COS proband with the 16p11.2 duplication had a history of pervasive developmental disorder not otherwise specified prior to onset of schizophrenia at age 8 years and inherited the duplication from a father with schizotypal personality disorder. The second proband inherited the duplication from a healthy father but had second-degree paternal relatives who had a history of psychosis, although we did not have their DNA for testing. Similar to the first proband with this duplication, this patient had poor social and motor development from a very young age, before the onset of schizophrenia at age 10 years.
MYT1L
The other novel recurrent CNV that we observed in our cohort was a duplication that disrupts the gene MYT1L on chromosome 2p25.3. Unlike the 22q11 deletions or the 16p11.2 duplications, this region is not flanked by segmental duplications, which may explain why these CNVs do not have the same breakpoints. Vrijenhoek et al. [30] recently found two additional schizophrenia patients with duplications that disrupted or spanned the MYT1L gene. Although not much work has been done on this specific gene, it is a member of the myelin transcription factor 1 family, which regulates proliferation and differentiation of oligodendrocytes by regulating transcription activity in the central nervous system [31,32]. Furthermore, a patient with severe mental retardation and facial dysmorphologies was found to have a de novo deletion that spanned the MYT1L gene on chromosome 20 [33].
NRXN1
We observed a novel, hemizygous 115-kb deletion in a single proband that affects exons 22 to 23 in the α- and exons 4 to 5 of the β- transcript of NRXN1, a locus previously implicated in schizophrenia, autism, and mental retardation. The observed deletion is in a different region of the gene than had been previously reported in the literature in schizophrenia, autism, or mental retardation. Neurexins are polymorphic cell surface proteins that are expressed in neurons. Marshall et al. [34] identified a 79-kb (intronic α transcript) inherited deletion in autism spectrum disorder (ASD). Szatmari et al. [35] identified two female siblings with ASD who had apparently identical 300-kb CNV losses of chromosome 2p16 not detected in either parent. Feng et al. [36] identified two putative missense structural variants in four Caucasian patients with autism (all of whom were also mentally retarded) and not in 535 healthy Caucasian controls. Kim et al. [37] identified two unrelated patients with ASD who had chromosomal disruptions either within or near NRXN1. They also sequenced 57 individuals with ASD and found seven novel variants within coding sequence: two were missense changes; the other five did not change amino acids. One of the missense variants was seen in two independent ASD families. Zahir et al. [38] described a child with developmental delay, unusual autistic-like behaviors, multiple vertebral anomalies, and an unusual facial appearance who was found to have a de novo 321-kb deletion involving the most 5′ portion of the NRXN1 gene. Kirov et al. [39] reported the observation of an inherited 250-kb deletion in affected siblings with schizophrenia. The deletion was transmitted by an asymptomatic mother. Furthermore, a recent report by Rujescu et al. [40•] revealed that only deletions that impact exons in NRXN1 are associated with schizophrenia.
Taken together, these studies provide strong evidence for the involvement of NRXN1 in early neurodevelopmental difficulties, and whether particular genetic “lesions” confer specific risks for specific disorders or more generalized risk remains to be determined.
SRGAP3
Another CNV that is of particular interest because it appears to track disease within one of our multiple affected families (from a bipolar father to the COS proband and one affected sibling) is a 120-kb duplication overlapping two exons in the SRGAP3 gene on chromosome 3p25.3. This gene is predominantly and highly expressed in the adult and fetal brain, with low expression detectable in kidney tissue and very low expression in other tissues. In the adult brain, the SRGAP3 gene is expressed in all tested regions, including the cerebellum, cortex, occipital pole, frontal and temporal lobe, amygdala, hippocampus, substantia nigra, and thalamus. SRGAP3 plays a critical role in neural development and axonal guidance and was reportedly disrupted by a translocation in a patient with severe mental retardation [41].
CNVs and premorbid development
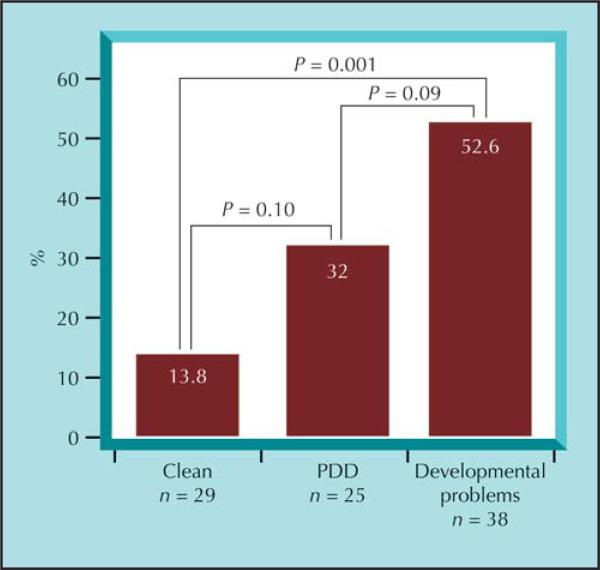
Beyond the anecdotal cases of various rare CNVs that likely play a role in the development of schizophrenia in these patients, we found that overall, the strongest clinical association for those who harbor a karyotypic and/or submicroscopic rare CNV was with a nonspecific developmental delay (social, language, or motor) [42]. Cases with a “clean” premorbid developmental history were least likely to harbor a novel CNV impacting a known gene, whereas those with a history of pervasive developmental disorder were in between (Fig. 1).
Figure 1.
Percentage of patients with childhood-onset schizophrenia (COS) with rare copy number variations and/or chromosomal anomalies by prepsychotic developmental pattern. Patients with COS had varied developmental histories, and we assigned them to one of three basic categories: pervasive developmental disorder (PDD, 23 met criteria for PDD not otherwise specified, one for autism and one for Asperger's syndrome); developmental problems, for anyone who had documented difficulties in the areas of speech and language, motor delays, or social/peer relationship problems; and clean, for those who did not have any developmental delays or problems before onset of schizophrenia.
One question that has arisen more often than not is how specific the association is with a particular psychiatric diagnosis and a particular genetic defect. The recent paper by Mefford and colleagues [43••] provides a great example for assessing this very issue. What is clear is that these genomic syndromes have a wide range of phenotypes associated with them, from autism to schizophrenia to epilepsy to cardiac problems and so forth. Thus, the challenge will lie with the genetic counselors who are faced with the task of informing parents of the implications of having a child with such genomic abnormalities.
Conclusions
The question of whether COS arises from genetic effects that are more penetrant than in AOS remains difficult to answer with certainty, but mounting evidence appears to support this hypothesis. The overabundance of specific genetic abnormalities compared with either AOS cases (Table 2) or the general population is compelling.
Table 2.
Rates of specific genetic abnormalities in adult-onset schizophrenia compared with childhood-onset schizophrenia
| AOS, n | COS, n | Odds ratio (95% CI) | Fisher's exact test | |
|---|---|---|---|---|
| Genetic abnormality | ||||
| 22q11 deletion | 21/7229 (0.3%) [15••,16••] | 4/96 (4.2%) [14] | 15.0 (5.3–42.6) | P < 0.001 |
| 45,X atypical/mosaic | 11/6483 (0.17%) [25] | 2/40 (5.0%) [23] | 31.0 (7.5–126.7) | P = 0.003 |
| 47,XXX | 22/8837 (0.25%) [24] | 1/40 (2.5%) [23] | 10.3 (1.7–61.7) | P = 0.099 |
| Recurrent CNVs | ||||
| 16p11.2 duplication | 5/7229 (0.07%) [15••,16••] | 2/96 (2.1%) | 30.7 (6.8–139.4) | P = 0.003 |
| MYT1L duplication | 2/806 (0.25%) [30] | 2/96 (2.1%) | 8.6 (1.49–49.1) | P = 0.06 |
| NRXN1 deletion | 1/2977 (0.03%) [40•] | 1/96 (1.0%) | 31.3 (3.3–302.3) | P = 0.06 |
AOS—adult-onset schizophrenia; CNV—copy number variation; COS—childhood-onset schizophrenia.
Although this is a relatively new area of study, exploration of CNVs already has revealed clear risk factors for schizophrenia, which, though rare, carry sizeable relative risks. If we allow ourselves to assume that all the novel CNVs that impact genes discovered in the COS cohort to date are pathogenic, then we can explain the etiology of almost 40% of our patients. This is a huge leap from where we stood just 1 or 2 years ago. As whole genome sequencing becomes a feasible reality, many more insights will be gained on the genomic architecture of affected individuals. Still, a great deal of work needs to be done to determine the rates of these variations in unselected populations, as well as in birth cohorts so that we can estimate the penetrance and specificity of each lesion. Nonetheless, in-depth resequencing of novel candidate genes and regions identified through CNV studies to identify other rare sequence variants that may be related to disease is ongoing. Ultimately, identification of syndromic causes of schizophrenia and related disorders will allow for better prenatal screening, as well as early intervention and treatment strategies.
Footnotes
Disclosure
No potential conflicts of interest relevant to this article were reported.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Nicolson R, Rapoport JL. Childhood-onset schizophrenia: rare but worth studying. Biol Psychiatry. 1999;46:1418–1428. doi: 10.1016/s0006-3223(99)00231-0. [DOI] [PubMed] [Google Scholar]
- 2.Asarnow JR, Tompson MC, Goldstein MJ. Childhood-onset schizophrenia: a followup study. Schizophr Bull. 1994;20:599–617. doi: 10.1093/schbul/20.4.599. [DOI] [PubMed] [Google Scholar]
- 3.Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 4.Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 5.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 6.Murrell J, Farlow M, Ghetti B, Benson MD. A mutation in the amyloid precursor protein associated with hereditary Alzheimer's disease. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 7.Addington AM, Gornick M, Sporn AL, et al. Polymorphisms in the 13q33.2 gene G72/G30 are associated with childhood-onset schizophrenia and psychosis not otherwise specified. Biol Psychiatry. 2004;55:976–980. doi: 10.1016/j.biopsych.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Addington AM, Gornick MC, Shaw P, et al. Neuregulin 1 (8p12) and childhood-onset schizophrenia: susceptibility haplotypes for diagnosis and brain developmental trajectories. Mol Psychiatry. 2007;12:195–205. doi: 10.1038/sj.mp.4001906. [DOI] [PubMed] [Google Scholar]
- 9.Gornick MC, Addington AM, Sporn A, et al. Dysbindin (DTNBP1, 6p22.3) is associated with childhood-onset psychosis and endophenotypes measured by the Premorbid Adjustment Scale (PAS). J Autism Dev Disord. 2005;35:831–838. doi: 10.1007/s10803-005-0028-3. [DOI] [PubMed] [Google Scholar]
- 10.Addington AM, Gornick M, Duckworth J, et al. GAD1 (2q31.1), which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol Psychiatry. 2005;10:581–588. doi: 10.1038/sj.mp.4001599. [DOI] [PubMed] [Google Scholar]
- 11••.Perry GH, Yang F, Marques-Bonet T, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [•• Of major importanceThis paper directly compared CNVs in 30 human with 30 chimpanzee genomes using array CGH and found a great deal of overlap in the regions of recurrent CNVs, indicating that there are shared sequence motifs that may predispose certain regions to structural instability. Furthermore, the authors identified specific genes that may have faced exceptional natural selection pressures at the copy number level during human and chimpanzee evolution] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [•• Of major importanceThis was the first study to report an association between CNVs that impact known genes and schizophrenia. The association was even more pronounced among younger-onset patients. In addition, the CNVs observed among patients with schizophrenia were more likely to disrupt genes with neurodevelopmental functions] [DOI] [PubMed] [Google Scholar]
- 13.Prescott K, Ivins S, Hubank M, et al. Microarray analysis of the Df1 mouse model of the 22q11 deletion syndrome. Hum Genet. 2005;116:486–496. doi: 10.1007/s00439-005-1274-3. [DOI] [PubMed] [Google Scholar]
- 14.Sporn A, Addington A, Reiss AL, et al. 22q11 deletion syndrome in childhood onset schizophrenia: an update. Mol Psychiatry. 2004;9:225–226. doi: 10.1038/sj.mp.4001477. [DOI] [PubMed] [Google Scholar]
- 15••.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [•• Of major importanceThe researchers from deCODE genetics in Reykjavik, Iceland, used a strategy of identifying regions prone to deletions or duplications to minimize further testing for association with schizophrenia using a huge population-based case-control design. They identified three novel regions in which deletion carried strong risks for schizophrenia] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [•• Of major importanceThe International Schizophrenia Consortium concurrently identified two of the same regions found by the deCODE group using another very large case-control sample collected by 13 different groups. The clear association of the 1q21 and 15q13 as strong genetic risk factors for schizophrenia is unequivocal] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy KC. Schizophrenia and velo-cardio-facial syndrome. Lancet. 2002;359:426–430. doi: 10.1016/S0140-6736(02)07604-3. [DOI] [PubMed] [Google Scholar]
- 18.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 19.Murphy KC, Owen MJ. Velo-cardio-facial syndrome: a model for understanding the genetics and pathogenesis of schizophrenia. Br J Psychiatry. 2001;179:397–402. doi: 10.1192/bjp.179.5.397. [DOI] [PubMed] [Google Scholar]
- 20.Maynard TM, Haskell GT, Lieberman JA, LaMantia AS. 22q11 DS: genomic mechanisms and gene function in DiGeorge/velocardiofacial syndrome. Int J Dev Neurosci. 2002;20:407–419. doi: 10.1016/s0736-5748(02)00050-3. [DOI] [PubMed] [Google Scholar]
- 21.Maynard TM, Haskell GT, Peters AZ, et al. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc Natl Acad Sci U S A. 2003;100:14433–14438. doi: 10.1073/pnas.2235651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Ousley O, Rockers K, Dell ML, et al. A review of neuro-cognitive and behavioral profiles associated with 22q11 deletion syndrome: implications for clinical evaluation and treatment. Curr Psychiatry Rep. 2007;9:148–158. doi: 10.1007/s11920-007-0085-8. [• Of importanceThis is a very good review of the highly variable phenotypes associated with the hemizygous 22q11 deletion] [DOI] [PubMed] [Google Scholar]
- 23.Eckstrand K, Addington AM, Stromberg T, et al. Sex chromosome anomalies in childhood onset schizophrenia: an update. Mol Psychiatry. 2008;13:910–911. doi: 10.1038/mp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLisi LE, Friedrich U, Wahlstrom J, et al. Schizophrenia and sex chromosome anomalies. Schizophr Bull. 1994;20:495–505. doi: 10.1093/schbul/20.3.495. [DOI] [PubMed] [Google Scholar]
- 25.Prior TI, Chue PS, Tibbo P. Investigation of Turner syndrome in schizophrenia. Am J Med Genet. 2000;96:373–378. doi: 10.1002/1096-8628(20000612)96:3<373::aid-ajmg26>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 26.Kotzot D. Complex and segmental uniparental disomy (UPD): review and lessons from rare chromosomal complements. J Med Genet. 2001;38:497–507. doi: 10.1136/jmg.38.8.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 28.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 29.Bailey JA, Yavor AM, Massa HF, et al. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Mol Cell Neurosci. 2004;25:111–123. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroepfl T, Petek E, Schwarzbraun T, et al. Mental retardation in a girl with a subtelomeric deletion on chromosome 20q and complete deletion of the myelin transcription factor 1 gene (MYT1). Clin Genet. 2008;73:492–495. doi: 10.1111/j.1399-0004.2008.00982.x. [DOI] [PubMed] [Google Scholar]
- 34.Marshall CR, Noor R, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Autism Genome Project Consortium. Szatmari P, Paterson AD, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng J, Schroer R, Yan J, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Kim HG, Kishikawa S, Higgins AW, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahir FR, Baross A, Delaney AD, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 39.Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 40•.Rujescu D, Ingason A, Cichon S, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2008 Oct 22; doi: 10.1093/hmg/ddn351. (Epub ahead of print). [• Of importanceThis recent review of the various deletions that have been reported in the NRXN1 gene in schizophrenia, autism, and mental retardation (as well as controls) revealed that only deletions that impact an exon in the gene were more common among patients with schizophrenia] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endris V, Wogatzky B, Leimer U, et al. The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci U S A. 2002;99:11754–11759. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollis C. Child and adolescent (juvenile onset) schizophrenia. A case control study of premorbid developmental impairments. Br J Psychiatry. 1995;166:489–495. doi: 10.1192/bjp.166.4.489. [DOI] [PubMed] [Google Scholar]
- 43••.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [•• Of major importanceThese researchers have laid the groundwork for trying to identify the vast range of phenotypic outcomes that can be associated with a specific “genomic syndrome.” The implications of these findings for medical geneticists and genetic counselors cannot be underestimated] [DOI] [PMC free article] [PubMed] [Google Scholar]