Abstract
Because the two sexes share a common gene pool while performing many different biological functions, mutations benefiting one sex may not accumulate due to counter selection in the other sex. In these experiments 99% of a haploid genome of Drosophila melanogaster was constrained to segregate like a male-limited Y chromosome for 41 generations, thereby eliminating potential counter selection in females. The synthetic Y chromosomes rapidly accumulated genetic variation that increased male fitness and decreased female fitness. The survival and fertility of females declined when they were mated to males expressing the synthetic Y chromosomes. These results suggests that opposing selection between the sexes may substantially interfere with sex-specific adaptation. They also demonstrate how intersexual evolutionary conflict can lead to perpetual degeneration of the Y via genetic hitchhiking of deleterious mutations.
Ecological studies (1–3) demonstrate that traits such as coloration and behavior can be selected discordantly in the two sexes, yet there is little evidence that such sexually antagonistic selection substantially impedes adaptation of each sex. In prior experiments with Drosophila melanogaster, I forced small chromosomal regions, linked to synthetic sex determining genes, to be passed from mother to daughter for 29 generations (4). When these female-limited regions were reinserted into males, male fitness was substantially reduced. Reduced fitness of the excluded sex is predicted by theory (5–7) if sexually antagonistic (SA) alleles (i.e., those favored in one sex but disfavored in the other) are rare at individual loci but common when summed across the genome. But theory (5–7) also predicts a small benefit/detriment ratio for many SA alleles that accumulate near a new sex determining locus. So experimental power to resolve benefits to the included sex was low.
Experimental power is increased in the experiments reported here by constraining a much larger segment of the genome to be sex-limited. For technical reasons (i.e., the absence of intrachromosomal recombination in males), male-limited gene transmission replaces the previous female-limited protocol. I used an experimental protocol that caused 99% of the haploid set of genes in D. melanogaster to be transmitted from father to son, thereby releasing these giant, synthetic Y chromosomes from counterselection in females. If male adaptation is substantially impeded by counterselection in females, then this male-limited gene pool should lead to the evolution of increased male fitness.
Below I provide evidence that male-limited gene transmission leads to a rapid increase in male fitness and decrease in female fitness. I then discuss the significance of this finding in the context of male-female antagonistic coevolution and the degeneration of the Y sex chromosome.
MATERIALS AND METHODS
Experimental Protocol.
This work extends the analysis of experimental lines from my prior study of coevolution between the sexes (8), which contains details of the experimental protocol and genetic constructs. Briefly, each of the two experimental lines contained 774 males and each male carried a male-limited genomic haplotype (i.e., a synthetic Y chromosome that was composed of chromosomes X, II, and III, but not the dot chromosome IV). Male-limited transmission of the synthetic Y chromosomes was accomplished by mating males to specially constructed clone-generator females that carried a compound X chromosome, a Y chromosome, and a homozygous II-III autosomal translocation. To speed the rate of adaptation (9–13), a small fraction (4%) of the synthetic Y chromosomes were forced to recombine by passing them through a recombination compartment (8). The clone-generator females could not evolve in response to evolutionary advance of the synthetic Y chromosomes because these females were taken anew each generation from an independent stock. The two control lines contained males and females with normal karyotypes (hence male-limited genomic haplotypes were absent), and had effective population size, densities and other environmental conditions matched with the experimental lines.
The earlier study (8) demonstrated that by generation 32, experimental males (lines EA and EB, which expressed a synthetic Y) had evolved a 24% increase in net fitness, compared with their controls (lines CA and CB), when mated to the stock of females (clone-generator) with which they had evolved. This increased male fitness may represent, (i) a response to release from counterselection in females, and/or (ii) adaptation to the specific phenotype of the clone-generator females. Here I specifically tests for the response to release from counterselection in females by measuring male fitness in response to tester-females, i.e., a stock unrelated to the clone-generator females (see below).
Because only part of the gain in fitness of the EA and EB experimental lines was expected to be due to release from counter selection in females, an increased signal to noise ratio was achieved by reciprocally crossing the two experimental lines (using males carrying a dominant-marker translocation and females from an expanded recombination compartment; ref. 8) to form a hybrid EAB line. This line contained a double dose of male-limited genes. In the same way the two control lines were crossed to make a CAB line.
Male Fitness Assays. Tester-females.
All male fitness assays were carried out with tester-females, carrying a singe genetic marker; pink-peach (pP). This line was constructed at the beginning of the experiment by backcrossing the pP marker three times through the wild-type stock used to begin the control and experimental lines. The pP stock was then cultured in parallel (same density, same culturing times, etc.) with the controls. The following four male fitness components were assayed.
Mating speed.
Mating speed measures how strongly a male stimulates virgin females to mate. This was measured in generation 38 by combining, in each of 11 vials and without anesthesia, 7 males with 5 virgin tester-females and then measuring the number of mating pairs 15 min later.
Remating rate.
The remating rate measures how strongly a male stimulates nonvirgin females to mate. Remating rate was measured three times: (i) Early measure: In generation 38, 30 males (EAB or CAB) were combined, without anesthesia, with 20 virgin tester-females (2–3 days old) in each of seven vials. After a 3-hr delay (during which time virtually every female mated), the number of remating pairs was observed four times, with each sequential observation separated by ≈25 min intervals (mating duration rarely exceeds 20 min). (ii) Late measure: The vials from the early measure group were again observed for rematings the next day in the same way. (iii) Remating by EAB or CAB males was also measured during the offense assay (see below). Remating was tallied as in the early measure group above, but there were 11 sequential observations beginning 4 hr after the pP competitor males had first mated the tester-females.
Defense assay.
Defense assay (EAB and CAB males mated first) measures the extent to which a male’s mate continues to utilize his sperm. In this assay tester-females, that had just previously mated to EAB or CAB males, were challenged with pP competitor males. The smaller the fraction of offspring sired by the pP competitor, the higher the defense ability of the EAB or CAB males. In generation 40, 30 EAB males were combined with 20 3-day old, virgin tester-females (eight vials of each) and then the males were removed after 1 hr via 50 sec of CO2 anesthesia. The same procedure was carried out with CAB males. Virtually every female mated during this time. Two hours after the males were removed, 30 pP competitor males were added to the females, without anesthesia. After an additional 24 hr, the pP competitor males were removed, and then each female was placed in an individual food vial and allowed to produce progeny. Ten days later, the progeny were scored as red eyed (from the CAB or EAB males) or pink eyed (from the pP competitor males).
Offense assay.
Offense assay (pP competitor males mated first) measures the extent to which a male remates a competitor male’s mate and then replaces the competitor’s sperm with his own. In this assay tester-females, that had just previously mated to pP competitor males, were challenged with EAB and CAB males. The smaller the fraction of the tester-female’s offspring sired by the pP competitors, the higher the offense ability of the EAB or CAB males. Tests followed the same protocol described above for defense but the order of males was reversed.
Female Fitness Assays. Development speed.
Retarded development is commonly associated with harmful mutations in D. melanogaster (14). This motivated an assessment of the development speed in females expressing the male-limited genomic haplotypes. In generations 29 and 30, development time was measured on females that expressed a synthetic Y chromosome or the corresponding X, II, and III chromosomes from controls. Control males (CA and CB) were crossed to clone-generator females to produce sons (CA′ and CB′) with the same translocation genetic background as the experimental males (8). Six yeasted vials of 7 EA, EB, CA′, or CB′ males were mated to 5 virgin pp tester-females (3-days old) and then the females were permitted to lay eggs for 24 hr. Daughters, expressing a synthetic Y chromosome (or an X-II-III haplotype from the controls), were distinguished by their red eye color and were counted as they eclosed.
Net productivity.
To resolve further the impact of the synthetic Y chromosomes on female fitness, CAB and EAB females were assayed for net fitness, independent of development time. Two EAB (or CAB) virgin females, 3 pp virgin tester-females, and 7 pp males were placed in each of 11 yeasted food vials. The females rapidly mated and were permitted to lay eggs for 24 hr. Offspring from the EAB (or CAB) females had red eyes and those from the pp tester-females had pink eyes. The ratio of red to pink adult daughters measures net female fitness of the synthetic Y chromosomes across one generation. To eliminate the impact of development time, the daughters were counted at the end of the 11th day when almost all offspring had eclosed.
Mortality and Fertility of Females Mated to Experimental Males.
Females may be harmed by the behavior and seminal fluid of their mates (15). To assess such harm, EAB and CAB males were mated to tester-females and the females’ survival and fertility was measured.
Survival. EAB and CAB males were mixed with 3-day old virgin pP tester-females (seven vials of 20 pP tester-females and 30 EAB or CAB males) and transferred, without anesthesia, daily (days 1–5) and then again on day 12. The number of dead females was measured at each transfer.
Mortality (dead or alive after 7 days) and fertility (some vs. no larvae produced) were also recorded on the individually housed females during the offense and defense assays described above.
RESULTS
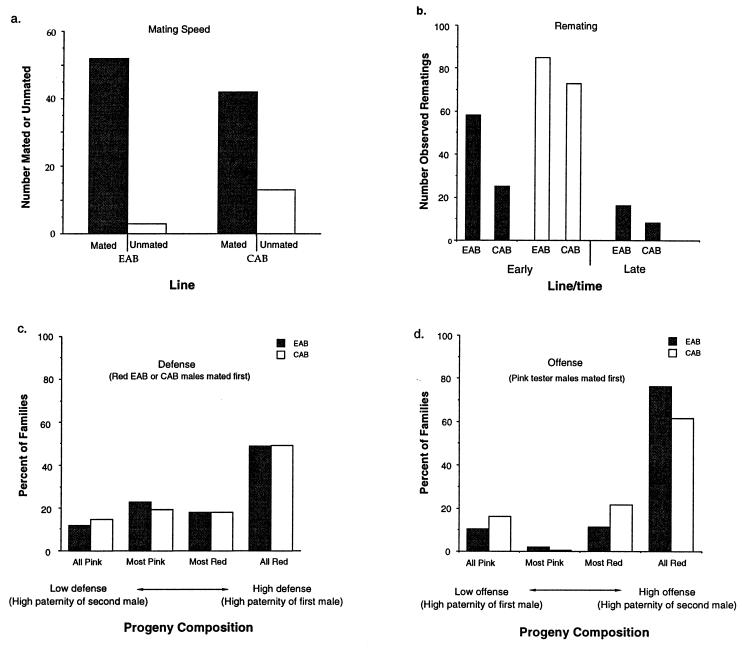
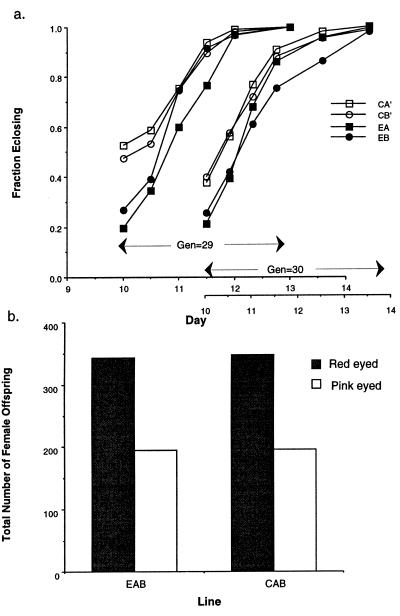
Experimental EAB males surpassed their controls in all fitness measures except defense (i.e., when EAB and CAB males mated tester females first), where there was no difference (Fig. 1). Clearly an important part of the gain in fitness of the synthetic Y chromosomes was due to release from counterselection in females. Yet additional fitness gain must be due to adaptation to the fixed phenotype of the clone-generator females, because in a previous assay males expressing a synthetic Y did demonstrate higher defense with these females (8). Females expressing a synthetic Y chromosome had substantially retarded development time (Fig. 2a). When female net fitness, independent of development time was measured (Fig. 2b), no difference between experimental and control lines was observed.
Figure 1.
Males expressing synthetic Y chromosomes (line EAB) surpass controls (CAB) in all fitness measures except defense. (a) Mating speed is faster for the experimental males (P = 0.008, directed (24) conditional binomial exact test (25, CBET). (b) Remating rate is higher for the experimental males [▪, histograms from generation 38; □, histograms from the net offense assay in generation 40; P = 0.0002, directed, consensus P value (26) for the three χ2 contingency tests]. (c) Defense is the same for experimental and control males (P = 0.40, directed χ2 contingency test). The histogram tallies families from individually cultured females that had been first mated to EAB (▪, n = 147) or CAB (□, n = 152) males for 1 hr and then housed with pP competitor males for 24 hr. Families were divided into four discrete categories grading from low defense (all pink, females remated to the pP competitor males and no progeny produced from the first male’s sperm) to high defense (all red, no progeny produced from the pP competitor males). (d) Offense is higher for experimental (EAB) than control (CAB) males (P = 0.016, directed χ2 contingency test). The histogram tallies percent of families from individually cultured females, that had been first mated to pP competitor males for 1 hr and then housed with EAB (▪, n = 145) or CAB (□, n = 149) males for 24 hr. Families were divided into four discrete categories grading from low offense (all pink, progeny exclusively derived from the pP competitor males’ sperm and none from remating with the experimental or control males) to high defense (all red, females remated to experimental or control males and no progeny produced from the pP competitor male’s sperm).
Figure 2.
Females expressing the experimental, synthetic Y chromosomes have retarded development, but no other reductions in fitness. (a) The fraction of females emerging vs. time since eggs were laid, for females expressing a synthetic Y from experimental EA (NG = 29 = 107, NG = 30 = 143) and EB (NG = 29 = 130, NG = 30 = 153), or control CA′ (NG = 29 = 114, NG = 30 = 141) and CB′ (NG = 29 = 126, NG = 30 = 148) males. The fraction of females emerging by day 10 is significantly lower in the experimental lines (PG = 29 = 0.0163, PG = 30 = 0.0161, directed Student’s t tests, df = 2). (b) The total number of adult offspring (measures net female fitness independent of development time) produced by EAB or CAB females (red-eyed offspring) in competition with tester-females (pink-eyed offspring). There is no significant difference between lines (P = 0.625, directed χ2 contingency test).
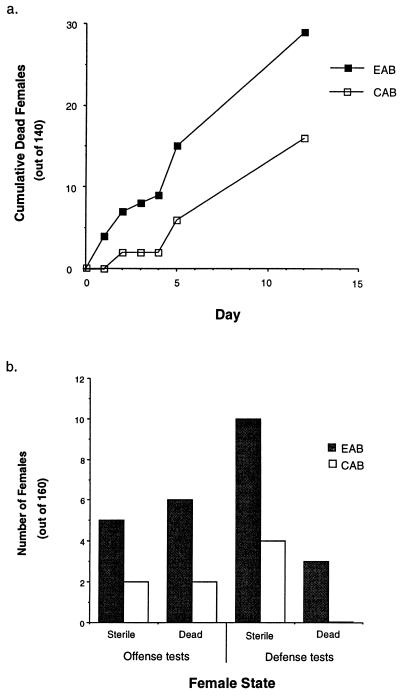
The mortality rate of the tester-females increased when their mates expressed a synthetic Y chromosomes (Fig. 3a). Data from the offense and defense assays corroborate this finding (Fig. 3b). When the defense data (i.e., when EAB and CAB males mated tester-females first) are considered alone, harm (mortality + sterility) to females increased due to a single mating with an experimental EAB male, indicating an increased toxicity of his seminal fluid.
Figure 3.
Mortality and sterility are higher for females mated to experimental EAB compared with control CAB males. (a) Cumulative numbers of dead tester-females when mated to, and continuously housed with, experimental or control males. Mortality is significantly different at day 4 and beyond (P ≤ 0.035 in all cases, directed CBET). (b) Number of sterile and dead females, from offense or defense assays (Fig. 2), that had previously been mated to EAB or CAB males. Statistical significance was determined via a multiple modulus test (27). This is a hierarchical-staged test that proceeds, in a priori order and conditioned on the previous test being significant, from global to more specific tests. Net harm (mortality + sterility) to females, pooling across offense and defense tests, was greater from experimental (EAB) than control (CAB) males (P < 10−6, directed CBET). This same pattern was manifest in both the defense test (P = 0.024, directed CBET) and the offense test (P = 0.05, directed CBET). Focusing next on specific types of harm to females, and first pooling data from both offense and defense tests, both sterility and mortality were higher for females exposed to EAB experimental males (P < 0.037 for both tests, directed CBET). Neither mortality nor sterility were individually significant when testing the offense and defense data individually.
DISCUSSION
SA Alleles.
In this study there was multifarious evidence that the male-limited genomic haplotypes evolved to increase male fitness, while only retarded development indicated reduced fitness in females. So only the benefit to the included sex is supported by multiple assays. But the earlier study (4) of female-limited chromosomal regions provides the converse; multifarious evidence for maladaptation to the excluded sex. Taken together these studies support the conclusion that the genome contains a nontrivial frequency of SA alleles (or tightly linked genes). The two studies also support the conclusion that SA selection and the constraint of sharing a common gene pool may substantially restrict the adaptive evolution of each sex.
Why were females that expressed the male-limited genomic haplotypes not harmed more? One hypothesis is that recessive X-linked alleles played an important role in the adaptation of the experimental males. Theory predicts that the X will be especially polymorphic for such recessive male-benefit SA alleles (16). If different recessive X-linked genes accumulated in the two experimental lines, then the hybrid EAB males would, and hybrid EAB females would not, express such male-benefit SA alleles.
Why should retarded development be the primary cost to females that expressed the male-limited genomic haplotypes? It may be that ontogeny is a major arena for conflict among SA alleles, because sexual differences in developmental pathways ultimately cause sexually dimorphic phenotypes. In this case, developmental canalization might ameliorate much of the cost of male-benefit SA alleles in adult females, but at a cost of retarded development. Alternatively, development speed itself may be a trait that is divergently selected between the sexes. In wild-type stocks of D. melanogaster, males develop slower than females (17). Lastly, development speed may be an easily measured indicator of other factors with smaller (hence statistically less easily resolved) effects on fitness.
Male Adaptation to Females.
Males expressing the male-limited genomic haplotypes reduced the survival and fecundity of their tester-female mates. This reinforces previous evidence (8) for the operation of antagonistic coevolution between the sexes. Each adaptation favoring one sex can be countered by an adaptation favoring the other, and an open-ended intraspecific Red Queen process can ensue (18).
The observation that experimental males expressed a strongly elevated defense advantage when mated to the clone-generator females to which they had specifically adapted (8), but none when paired with tester-females, indicates that adaptation of the defense phenotype may be highly idiosyncratic to specific characteristics of the female reproductive tract, physiology, and neurobiology.
Degeneration of the Y Chromosome.
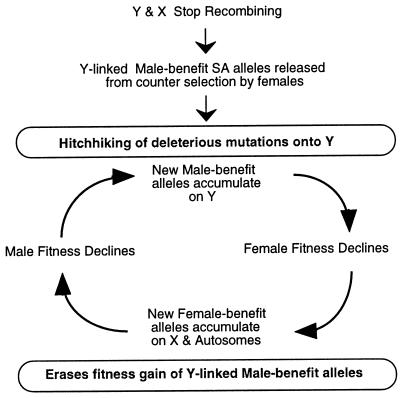
Theory and experiments indicate that a gender-limited, nonrecombining Y initially evolved in response to SA selection (20, 22). Prior experiments (4, 8), and data presented here, suggest that once a primitive Y (or part of it) stopped recombining: (i) a pulse of progressive evolution, favoring the heterogametic sex, would ensue, and (ii) this would reduce the fitness of the homogametic sex.
Loci residing outside a gender-limited Y are expected to evolve to ameliorate the fitness reduction of the homogametic sex. If such counteradaptations reduce the effectiveness of extant Y-linked SA alleles, this then will select for new alleles on the Y, leading to a new round of counteradaptation favoring the homogametic sex, etc. Hence perpetual “chase-away” antagonistic coevolution ensues between Y-linked vs. X-linked and autosomal loci (Fig. 4).
Figure 4.
Model for the degeneration of the Y via chase-away antagonistic coevolution between the sexes. Males are assumed to be the heterogametic sex. A parallel chase-away process can ensue that is driven by antagonistic coevolution between male offense and defense phenotypes or other forms of interlocus contest evolution (18).
This antagonistic coevolution will degenerate the Y in a way that augments Muller’s ratchet (9, 19) and background selection/background trapping (20–22). SA coevolution can perpetually recruit new mutations onto the Y that favor the heterogametic sex. During each such recruitment, mildly deleterious mutations may be dragged along to fixation via genetic hitchhiking (21, 22).
Other forms of interlocus antagonistic coevolution (e.g., genes coding for male offense vs. defense; see refs. 18 and 23 for additional examples) can also lead to such hitchhiking decay. Thus, counter intuitively, the evolution of a sex-limited Y chromosome is an ’evolutionary trap;’ it is initially selected by benefiting the heterogametic sex, but then perpetually eroded by this same selection process due to interlocus antagonistic coevolution and genetic hitchhiking of deleterious mutations.
Acknowledgments
I thank D. Gessler and B. Holland for technical assistance, and three anonymous referees who made many helpful suggestions. This work was funded by National Science Foundation Grant DEB-9509191.
ABBREVIATION
- SA
sexually antagonistic
References
- 1.Endler J A. Evolution (Lawrence, Kans) 1980;34:76–91. doi: 10.1111/j.1558-5646.1980.tb04790.x. [DOI] [PubMed] [Google Scholar]
- 2.Forsman A. J Evol Biol. 1995;8:53–70. [Google Scholar]
- 3.Sherman P W. Science. 1977;197:1246–1253. doi: 10.1126/science.197.4310.1246. [DOI] [PubMed] [Google Scholar]
- 4.Rice W R. Science. 1992;256:1436–1438. doi: 10.1126/science.1604317. -1439. [DOI] [PubMed] [Google Scholar]
- 5.Fisher R A. Biol Rev. 1930;6:345–368. [Google Scholar]
- 6.Bull J J. Evolution of Sex Determining Mechanisms. Menlo Park, CA: Benjamin/Cummings; 1983. [Google Scholar]
- 7.Rice W R. Evolution (Lawrence, Kans) 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 8.Rice W R. Nature (London) 1996;361:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 9.Rice W R. Science. 1994;263:230–232. doi: 10.1126/science.8284674. [DOI] [PubMed] [Google Scholar]
- 10.Manning J T, Thomas D J. Acta Biotheoretics. 1984;33:219–225. [Google Scholar]
- 11.Peck J R. Genetics. 1994;137:597–606. doi: 10.1093/genetics/137.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlesworth B. Genet Res. 1994;63:213–227. doi: 10.1017/s0016672300032365. [DOI] [PubMed] [Google Scholar]
- 13.Barton N. Genetics. 1995;140:821–841. doi: 10.1093/genetics/140.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsley D L, Grell E H. Genetic Variations of Drosophila melanogaster. Washington D.C.: Carnegie Institution of Washington; 1968. [Google Scholar]
- 15.Chapman T, Liddle L F, Kalb M J, Wolfner M F, Partridge L. Nature (London) 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 16.Rice W R. Evolution (Lawrence, Kans) 1984;38:735–743. [Google Scholar]
- 17.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Rice W R, Holland B. Behav Ecol Sociobiol. 1998;41:1–10. [Google Scholar]
- 19.Charlesworth B. Proc Natl Acad Sci USA. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charlesworth B. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 21.Rice W R. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice W R. Bioscience. 1996;46:331–343. [Google Scholar]
- 23.Hurst L D. Heredity. 1994;73:233–242. doi: 10.1038/hdy.1994.128. [DOI] [PubMed] [Google Scholar]
- 24.Rice W R, Gains S D. Trends Ecol Evol. 1994;9:235–237. doi: 10.1016/0169-5347(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 25.Rice W R. Biometrics. 1988;44:1–14. [Google Scholar]
- 26.Rice W R. Biometrics. 1990;46:303–308. [Google Scholar]
- 27.Miller R G., Jr . Simultaneous Statistical Inference. 2nd Ed. New York: McGraw–Hill; 1980. [Google Scholar]